Abstract
microRNAs function as genetic rheostats to control gene output. Based on their role as modulators, it has been postulated that microRNAs canalize development and provide genetic robustness. Here, we uncover a novel regulatory layer of chemokine signaling by microRNAs that confers genetic robustness on primordial-germ-cell (PGC) migration. In zebrafish, PGCs are guided to the gonad by the ligand Sdf1a, which is regulated by sequestration receptor Cxcr7b. We find that miR-430 regulates sdf1a- and cxcr7-mRNAs. Using Target Protectors, we demonstrate that miR-430-mediated regulation of endogenous sdf1a and cxcr7b (i) facilitates dynamic expression of sdf1a by clearing its mRNA from previous expression domains, (ii) modulates the levels of the decoy receptor Cxcr7b to avoid excessive depletion of Sdf1a and (iii) buffers against variation in gene dosage of chemokine signaling components to ensure accurate PGC migration. Our results indicate that losing microRNA-mediated regulation can expose otherwise buffered genetic lesions leading to developmental defects.
Introduction
Biological systems can compensate for genetic and environmental perturbations. Genetic buffering allows invariance of the phenotype in the face of perturbations. This endows the organism with reduced susceptibility to mutations and results in robustness1,2. External perturbations may arise from changes in the environment while internal factors derive from inexact processes within a cell, such as transcriptional bursts, changes in gene dosage, and leaky transcription3–5. This variation in gene expression may lead to fluctuation in activity, limiting the accuracy of specific cellular processes. In order to prevent these fluctuations from having a severe impact on the phenotype, several mechanisms for providing robustness have evolved. First, creating networks of feed-back and feed-forward loops can reduce the effects of variation by stabilizing different phenotypic states, such that minor variation will not be sufficient to alter the current state6. Second, functional redundancy within regulatory networks provides stability because multiple changes need to occur to perturb the system1. Third, genetic buffering prevents small changes at the genetic level from influencing the phenotype. For example, the chaperone protein HSP90 facilitates proper folding of mutant proteins, masking the effects of these mutations by preventing variation in expression and activity7,8. Finally, additional mechanisms might be in place at the RNA level to provide robustness and guard against transcriptional fluctuations. It has been proposed that high levels of transcription coupled with inefficient translation can lower intrinsic noise in protein output4,5. This is supported both by theoretical models and experimental evidence in unicellular organisms4. Because of their ability to regulate a wide variety of genes9, their function as rheostats that modulate the mRNA and protein output of their target genes10,11, and their position within network motifs (reviewed in3, 6,12) it has been postulated that microRNAs (miRNAs) may play an important role in buffering biological systems against genetic variation. A prominent example in Drosophila is miR-7, which functions within a feed-forward loop to guard against the consequences of temperature fluctuation to stabilize a phenotypic state13. However, few studies have experimentally addressed the role of miRNAs in genetic robustness in vertebrates.
Results
Loss of miRNAs leads to mislocalized PGCs
In this study we investigate the role of miRNAs in buffering against genetic variation in vertebrates using long-distance cell migration as a model. In this context, slight perturbations in protein levels or distribution of guidance cues might lead to erroneous cell migration because the chance that such small fluctuations will affect cell migration accumulates with the distance the cells need to migrate. Primordial germ cell (PGC) migration in zebrafish is a prominent example of such a long-distance migratory process that is likely evolutionarily reinforced (reviewed in14). PGCs express the chemokine receptor Cxcr4b and follow the shifting expression domain of its ligand Sdf1a (Cxcl12a) as they migrate through the gastrulating embryo to reach the site of the future gonad. The dynamic expression of the ligand in the somatic tissues plays a crucial role in determining the migratory path and must be tightly regulated at the protein and the mRNA levels (reviewed in15). It has been proposed that the Sdf1a protein gradient is refined by Cxcr7, a “decoy” receptor that is thought to act as an Sdf1a sink16,17. At the mRNA level, new transcriptional domains must be coupled to rapid removal of the transcripts that remains in previous domains of expression, yet the mechanisms underlying this regulation are elusive. One possibility involves rapid removal of sdf1a transcripts from tissues where they are no longer needed through miRNA-mediated degradation. To test this idea, we examined PGC localization in the absence of miRNAs. We have generated mutant embryos lacking both the maternal and the zygotic contribution of the miRNA-processing enzyme dicer (MZdicer)18,19. Intriguingly, in such embryos we find that PGCs are frequently mislocalized. This mismigration is not due to disruption of the migratory path, as these somatic tissues are properly specified in MZdicer. Importantly, re-introducing miR-430 or one of its mammalian homologues, miR-302, into MZdicer mutants rescued the localization of PGCs (Supplementary Fig. 1). These results suggest that miR-430-mediated repression of specific targets mRNAs may play a role in PGC migration.
miR-430 can regulate expression of chemokine signaling genes
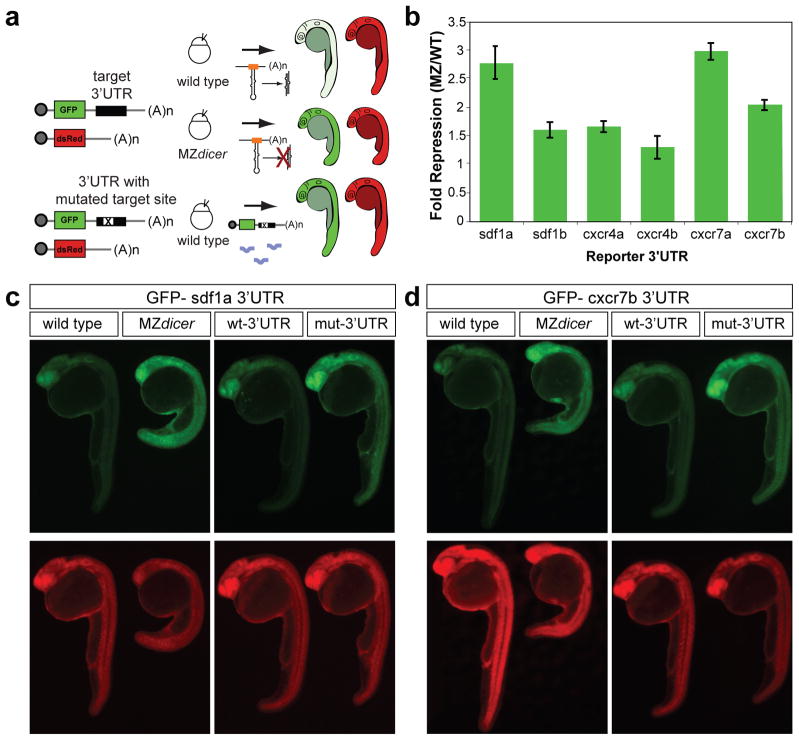
Because miRNAs accelerate the decay of their target mRNAs, we have previously used microarrays to identify miRNA targets by comparing gene expression in wild type, MZdicer and MZdicer injected with miR-43019. Analysis of these data revealed that sdf1a and cxcr4b might be repressed by miR-430 (Supplementary Fig. 1). To determine whether additional genes in the chemokine signaling pathway are directly regulated by miR-430, we searched their 3′UTRs for sequences complementary to the miR-430 “seed” sequence (reviewed in9). This analysis revealed that 3′UTRs of both ligands (sdf1a and sdf1b) and receptors (cxcr4a, cxcr4b, cxcr7a, and cxcr7b) contain putative miR-430 target sites (Supplementary Fig. 2 and 3). If these transcripts are regulated by miR-430, then the target site in each 3′UTR should influence their translation. To test this hypothesis, we co-injected reporter mRNAs encoding the GFP ORF and each full length 3′UTR with a dsRed control mRNA (Fig. 1a). To analyze the level of regulation of each target, we quantified the fluorescence of the GFP reporter relative to the dsRed control in the presence or absence of miRNAs (Fig. 1b). These 3′UTRs conferred repression of GFP in wild type but not in MZdicer mutants that lack mature miR-430 (Fig. 1c, d and Supplementary Fig. 2). Next, we tested whether miR-430 sites play a direct role in the regulation. Mutating the miR-430 sites in each 3′UTR (GCACUU to GCUGAU) prevented miRNA-mediated repression of reporter mRNAs in wild type embryos (Fig. 1c, d and Supplementary Fig. 2). To further characterize the regulation of the sdf1a 3′UTR, we mutated individual target sites in this reporter. We found that mutation of the first target site was necessary and sufficient to relieve repression of the GFP reporter. However, mutating the second or third target site did not affect regulation of the reporter (Supplementary Fig. 3). These results indicate that the ligand (sdf1a) and the decoy receptors cxcr7a/b are more strongly regulated by miR-430 (>2-fold) than cxcr4a/b and sdf1b (<2-fold). On the basis of the regulation provided by the sdf1a and cxcr7 3′UTRs and their essential role in PGC migration, we focused our analysis on these genes.
Figure 1.
miR-430 target validation of chemokine signaling genes. (a) Schematic representation of the experimental set up. Expression of GFP reporters with the 3′UTRs of putative targets is compared in wild type and MZdicer embryos. dsRed mRNA lacking a target site was co-injected as a control. To test whether the miR-430 target site plays a role in the regulation, wild type embryos are also injected with wild type reporters (wt-3′UTR) or mutant reporters where three bases in the miR-430 target site were mutated (mut-3′UTR). All three miR-430 target sites in the sdf1a 3′UTR were mutated to generate the mutant reporter. (b) Quantification of the GFP fluorescence in MZdicer compared to wild type embryos injected with each GFP reporter mRNA and dsRed control. GFP fluorescence is normalized to the dsRed control. Data are shown as mean ± s.d. (c, d) Fluorescence microscopy shows GFP (green) and dsRed (red) expression in 24–28 hpf embryos injected with GFP reporters with sdf1a or cxcr7b 3′UTR. Endogenous miR-430 represses the expression of each reporter in wild type but not MZdicer embryos. Similarly, wild type embryos injected with the mut-3′UTR reporter fail to repress GFP. The sequence of the wild type or the mutant target site and miR-430 are shown in Supplementary Fig. 2.
Target Protectors specifically relieve miR-430-mediated repression
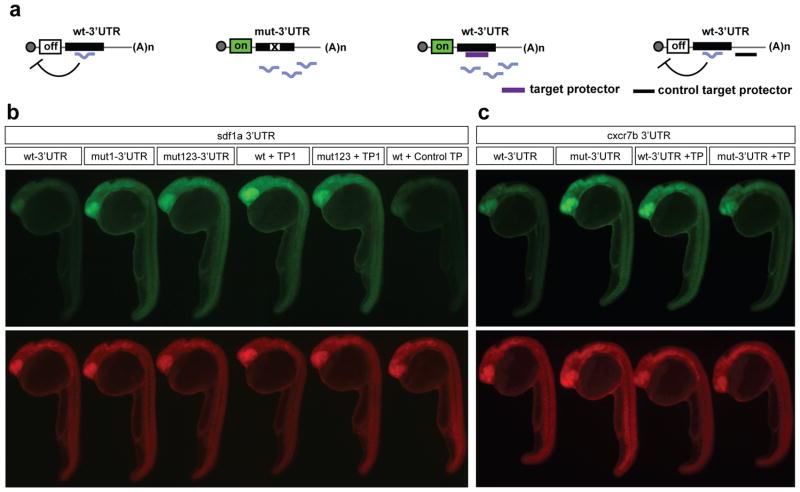
The analysis of individual miRNA-mRNA interactions in animals that are depleted of most miRNAs poses a challenge because many miRNA targets are misregulated19,20. To investigate the physiological function of miR-430-mediated regulation of sdf1a and cxcr7a/b, we employed Target Protectors (TPs). TPs are morpholino antisense oligonucleotides that are complementary to the miRNA recognition site in the transcript and interfere with miRNA-mRNA interactions, thus protecting a specific target from its miRNA (Fig. 2a)21. Three lines of evidence indicate that TPs interfere with the regulation of sdf1a and cxcr7 by miR-430. First, TPs for sdf1a, cxcr7a and cxcr7b restore expression of the GFP reporter to a level similar to that observed in the absence of miR-430-mediated repression (mutated reporter; Fig. 2b, c, and Supplementary Fig. 3 and 4). Individual TPs for each target site in the sdf1a 3′UTR were injected. Consistent with the results obtained during the mutagenesis of individual target sites, injection of a TP complementary to the first site (TP1) was sufficient to relieve repression to the level of the mutated reporter (Fig. 2b and Supplementary Fig. 3). Conversely TPs for the second and third sites did not have a noticeable effect, suggesting that the first miR-430 target site in sdf1a confers most of the regulation. Thus in all following experiments sdf1a-TP refers to the TP complementary to the first target site in sdf1a. Second, injection of the TPs into MZdicer mutants did not affect the expression level of the endogenous target mRNA compared to control MZdicer mutants (Supplementary Fig. 4), suggesting that each TP does not cause stabilization of the target mRNA independent of the miRNA. Third, a TP designed to bind to a region of the sdf1a 3′UTR approximately 200 bases downstream of the first miR-430 binding site (sdf1a-control-TP) did not affect repression of the reporter, indicating that target protection depends on the overlap with the miRNA target site (Fig. 2b). These results indicate that sdf1a-TP, cxcr7a-TP, and cxcr7b-TP provide specific tools to interfere with miRNA-mediated regulation of endogenous sdf1a and cxcr7a/b.
Figure 2.
Target Protectors prevent miRNA-mediated repression of target GFP reporters. (a) Schematic shows repression of targets by a miRNA and loss of repression of mutated reporters. GFP reporters are protected by TPs (purple) while binding of the control target protector (black) downstream of the miR-430 target site does not prevent miRNA-mediated repression. (b, c) GFP and dsRed fluorescence in 24-28 hpf embryos injected with GFP reporters. Expression of a wild type GFP-sdf1a 3′UTR reporter is repressed. Injection of sdf1a-TP, but not a control TP, blocks miR-430-mediated repression of the GFP reporter. Expression of the mut-3′UTR GFP reporter with the first (mut1-3′UTR) or all three target sites mutated (mut123-3′UTR) is shown for comparison. (c) Derepression of the GFP-cxcr7b wt-3′UTR reporter is also observed upon injection of the cxcr7b-TP. Predicted Watson-Crick pairing of the 3′UTR target sites with each TP (blue) and miR-430 are shown in Supplementary Fig. 3.
Regulation of sdf1a and cxcr7b by miR-430 promotes accurate PGC migration
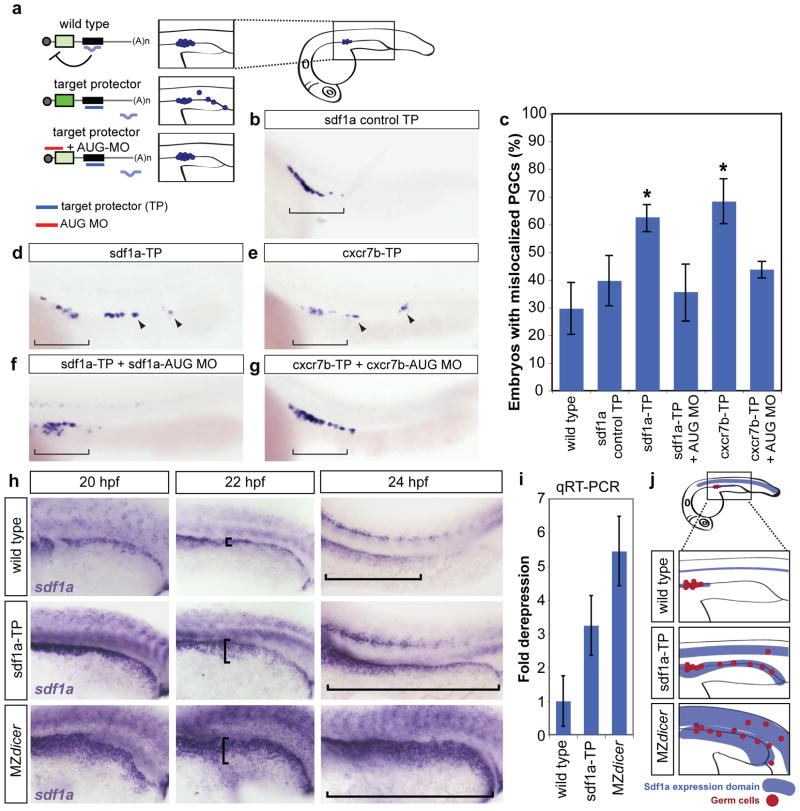
To investigate the physiological role of each miRNA-target interaction, we quantified PGC localization in TP-injected embryos. Blocking miR-430 regulation of sdf1a or cxcr7b by injection of sdf1a-TP or cxcr7b-TP but not cxcr7a-TP increased the percentage of embryos with mislocalized germ cells at 24 hpf (from 30% in wild type to more than 60%)(Fig. 3 and Supplementary Fig. 5). Injection of sdf1a-TP resulted in an increase in germ cells that were outside the location of the future gonad (bracket, Fig. 3d) and remained in the posterior pronephric regions (Supplementary Fig. 6). Blocking the regulation of sdf1a by miR-430 increased both the number of mislocalized cells and the fraction of embryos with mislocalized cells posterior to the future gonad. Conversely, protecting cxcr7b from miR-430 regulation caused mislocalization of a significantly higher number of cells to ectopic positions in the tail (Fig. 3e and Supplementary Fig. 6). These results are consistent with a model in which misregulation of Cxcr7b depletes the Sdf1a protein, causing the PGCs to lose the correct migratory path. Two controls support the specificity of these effects. First, injection of sdf1a-control-TP did not significantly increase the percentage of embryos with mislocalized PGCs (Fig. 3b, c). Second, we asked whether the TP phenotypes are caused by the binding of the TPs to their cognate target 3′UTR rather than off-targets. Since TPs block the repression of the target by the miRNA, increasing the protein expression from the mRNA, we reasoned that if the TP phenotype is due to an increase in expression of the target gene, it should be rescued by lowering the translation of this target (Fig. 3a). Indeed, a low level of a translation blocking morpholino complementary to sdf1a AUG start site22 rescued the mislocalization seen upon injection of sdf1a-TP alone (Fig. 3c, d, f). Similar results were observed for cxcr7b-TP and its AUG blocking morpholino17(Fig. 3c, e, g). These results suggest that miR-430 regulation of endogenous sdf1a and cxcr7b plays a role in PGC migration.
Figure 3.
Blocking miR-430-mediated repression of sdf1a and cxcr7b causes PGC mislocalization and expanded sdf1a expression. (a) Schematic representation of the experimental setup. Injecting the TP (purple) blocks miRNA-mediated repression, increasing mRNA expression and leading to mislocalization of cells. Co-injecting a morpholino to reduce translation of the target gene (red, AUG MO) rescues the mislocalization phenotype. The inset shows the region of the embryo depicted in the panels (b, d-h). (b, d-g) Whole mount in situ of nanos mRNA, labeling PGCs in 24 hpf embryos. Bracket shows correct localization of PGCs. Arrowheads identify mislocalized PGCs. (c) Quantification of the percentage of embryos with mislocalized PGCs in each experimental condition as indicated. A significantly increased number of TP-injected embryos have mislocalized PGCs (*, p=1.185 ×10−7, sdf1a-TP; p=2.52×10−7, cxcr7b-TP; two-sided Fisher’s exact test). Error bars show ± s.d. (d, e) Representative images of PGC mislocalization are shown. (f, g) Co-injection of a low level of the corresponding AUG MO rescues the TP phenotype (sdf1a AUG MO, 0.01 pmol; cxcr7b AUG MO, 0.045 pmol). (h) In situ hybridization to detect sdf1a mRNA. The trunk of embryos at 20 hpf, 22 hpf, and 24 hpf are wild type, injected with sdf1a-TP, or MZdicer. Brackets illustrate the extension of the sdf1a expression domain along the pronephric region. (i) qPCR for sdf1a in 24 hpf wild type, sdf1a-TP-injected embryos, and MZdicer. An increase in expression was observed in the absence of miR-430-mediated repression. (j) Schematic summary of Sdf1a tail expression and the resulting PGC mislocalization.
miR-430 promotes clearance of sdf1a from previous domains of expression
Germ cell migration depends on the dynamic expression of the ligand Sdf1a that elicits local attraction of the PGCs to a series of intermediate positions before reaching the future gonad14,23. Guidance is therefore achieved in part by modifying the position of the attractive domain. This process requires tightly controlled expression of sdf1a in new domains and rapid degradation of sdf1a transcripts lingering in old expression domains. Initially, sdf1a is expressed along the lateral mesoderm region and later becomes restricted to the anterior domains to attract the cells to the future gonad (Fig. 3h). However, the molecular mechanisms responsible for this dynamic expression are largely unknown. Based on the germ cell mismigration observed in sdf1a-TP injected embryos we hypothesized that miR-430 could facilitate clearance of previously expressed transcripts and, therefore, sharpen the domain of ligand expression. Using in situ hybridization, we found that sdf1a 3′UTR can enhance degradation of the GFP reporter mRNA, an effect that is dependent on the first miR-430 target site. (Supplementary Fig. 7). We then examined the impact of miR-430 on endogenous sdf1a transcripts in MZdicer and TP-injected embryos. Throughout PGC migration, sdf1a expression is stronger in sdf1a-TP and MZdicer embryos (Fig. 3h, and Supplementary Fig. 8). Consistent with our previously published microarray data (Supplementary Fig. 1)19, qPCR shows that sdf1a mRNA levels are increased in MZdicer embryos (Fig. 3i). Interfering with miR-430 regulation resulted in an expansion of the sdf1a expression domain, with a failure to clear it from more posterior regions of the trunk mesoderm (Fig. 3h). The regions that fail to clear sdf1a mRNA coincide with the position of mislocalized germ cells in sdf1a-TP injected embryos. Thus, miR-430 is required to modulate the amount of sdf1a RNA and to shape its expression pattern by clearing transcripts from old expression domains.
Cxcr7b and miR-430 negatively regulate sdf1a
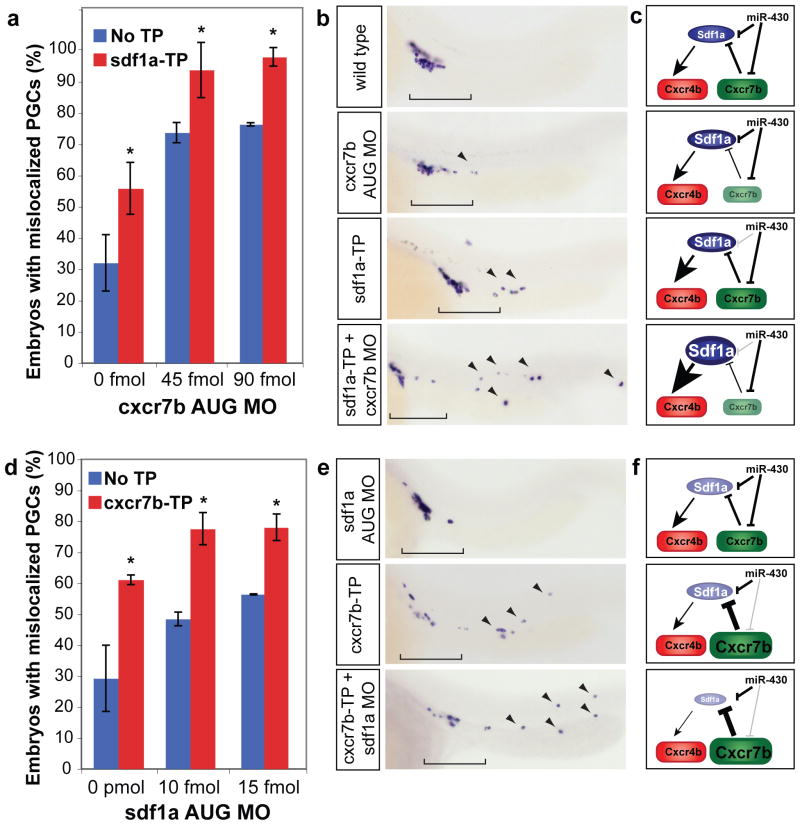
To test whether Cxcr7b and miR-430 function to regulate sdf1a in a partially redundant manner, we asked whether cxcr7b interacted genetically with miR-430-mediated regulation of sdf1a. Decreasing the level of sdf1a regulation by miR-430 enhanced the germ cell migration defect observed in a hypomorphic condition cxcr7b caused by injecting subthreshold levels of a cxcr7b translation blocking morpholino (cxcr7b AUG MO) (Fig. 4a–c). This phenotype was more severe than the sdf1a-TP alone in regards to the number of embryos affected as well as the mislocalization of the germ cells to the tail and head regions (Fig. 4b). A similar result was seen when injecting the cxcr7b-TP with an AUG MO for sdf1a (Fig. 4d–f). These results indicate that miR-430 and Cxcr7b act together to modulate sdf1a mRNA expression and protein levels, respectively.
Figure 4.
miR-430 and Cxcr7b act in a functionally redundant manner to refine Sdf1a expression. (a, d) Quantification of PGC mislocalization. Embryos were injected at the one-cell stage with a morpholino targeting the start site (AUG MO) of cxcr7b (a) or sdf1a (d). These AUG MOs were injected at low concentrations, which were insufficient to completely knockdown the transcript and caused a weak mislocalization phenotype. (a) Co-injecting sdf1a-TP and cxcr7b AUG MO causes significantly more mismigration that injection of sdf1a-TP or the same amount of the AUG MO alone (*, p=4.08×10−3, sdf1a-TP + 45 fmol to 45 fmol alone; p=4.87×10−3, sdf1a-TP + 90 fmol to 90 fmol alone; two-tailed Fisher’s exact test), suggesting that miR-430 regulation of sdf1a mRNA can partially compensate for a reduction of cxcr7b. Similarly, co-injecting cxcr7b-TP and sdf1a AUG MO significantly enhances the mislocalization phenotype (*, p=8.93×10−4, cxcr7b-TP + 10 fmol to 10 fmol alone; p=0.016, cxcr7b-TP + 10 fmol to 10 fmol alone; two-tailed Fisher’s exact test), suggesting that regulation of cxcr7b by miR-430 prevents excessive clearance of the sdf1a. Data are shown as mean ± S.D. (b, e) nanos in situ at 24 hpf to visualize the location of germ cells. Brackets indicate correctly localized PGCs, and arrowheads show mislocalized cells. (c, f) Scheme representing the predicted effect of the experimental conditions on Sdf1a and Cxcr7b shown in panels (b and e). The added effect of removing miR-430 targeting and modulation by Cxcr7b supports a functional redundancy of miR-430 and Cxcr7b.
miR-430 buffers against variation in gene dosage
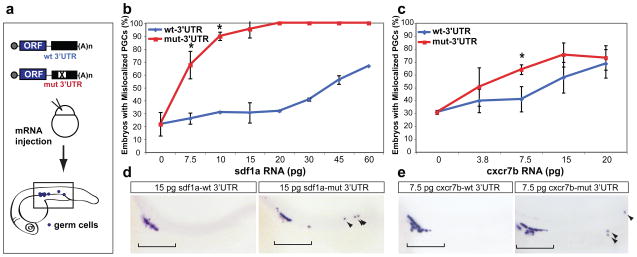
Because miRNAs modulate the protein output of their target mRNAs, it has been hypothesized that they may guard against genetic variation3. By dampening the expression of both the ligand and the decoy receptor, miR-430 may provide robustness to chemokine signaling. To test this hypothesis, we assessed the ability of miR-430 targeting to buffer against higher levels of sdf1a and cxcr7b (Fig. 5a). Embryos injected with increasing amounts of sdf1a or cxcr7b mRNAs with the mut-3′UTR, but not the wt-3′UTR, show an increase in germ cell mismigration (Fig. 5b–e). For instance, injecting 15 pg of sdf1a-mut-3′UTR significantly increases the percentage of embryos with mislocalized PGCs, but embryos receiving this amount of sdf1a-wt-3′UTR show a wild type phenotype (Fig. 5b, d). This suggests that repression by miR-430 is able to protect PGC migration from elevated levels of gene expression.
Figure 5.
miR-430 buffers against overexpression of the chemokine signaling components. (a) Schematic representation of the experiment. mRNA encoding the open reading frame for the target gene with either the wt-3′UTR or a mutant 3′UTR with the miR-430 target site mutated is injected at the one-cell stage. PGC localization is assayed at 24 hpf using an in situ for nanos. The rectangle illustrates the region of the embryos shown in panels d, e and g. (b, c) Quantification of the percentage of embryos with PGCs outside of the gonad region upon injection of sdf1a mRNA (b) or cxcr7b mRNA (c). A significant difference is seen between transcripts with the wt-3′UTR (blue) and those with the mut-3′UTR (red) for sdf1a (*, p=7×10−6) and cxcr7b (*, p=6.3×10−3; two-tailed Fisher’s exact test). (d, e) Representative images of injected embryos are shown. The bracket illustrates the PGCs that are correctly localized. Arrowheads indicate mislocalized PGCs. Error bars indicate ± s.d.
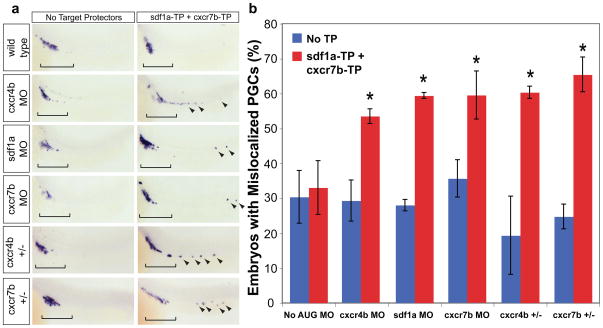
We then tested the role of miR-430 in compensating for decreases in expression. We began by eliminating miR-430-mediated regulation of chemokine signaling by co-injecting sdf1a-TP and cxcr7b-TP. This combination results in a wild-type phenotype, presumably because the increase in Cxcr7b can compensate for the increase in Sdf1a (Fig. 6, No AUG MO). To mimic genetic variation, we co-injected a low level of a morpholino against the chemokine signal or either of its receptors in the presence (No TP) or the absence of miR-430 mediated regulation (sdf1a-TP and cxcr7b-TP). Reducing the gene dosage of sdf1a, cxcr7b, or cxcr4b increased PGC mismigration in the absence of miR-430 regulation (with 60% of the embryos presenting mislocalized germ cells) but not in wild type embryos (Fig. 6, Supplementary Fig. 9). To support these experiments, we tested the effect of heterozygous mutations for cxcr7b and cxcr4b24,25. The incidence of germ cell mismigration for either of these heterozygous mutant embryos is similar to wild type. However, blocking miR-430 regulation of sdf1a and cxcr7b in either cxcr4b+/− or cxcr7b+/− mutant background increased the number of embryos with mislocalized PGCs (60%) (Fig. 6, Supplementary Fig. 9). Together, these experiments indicate that miR-430 lowers expression of sdf1a and cxcr7b such that minor perturbations in gene dosage do not cause mismigration (Fig. 7). Upon specific removal of the miRNA regulation, the system is more sensitive to small changes caused by a reduction in protein level or gene copy number. Together, these observations suggest that miR-430 provides genetic robustness and protects against alterations in gene dosage.
Figure 6.
Regulation by miR-430 guards against variation in gene dosage. (a) Quantification of the percentage of embryos with mislocalized PGCs in different experimental conditions as shown. Co-injection of sdf1a-TP, cxcr7b-TP, and a low level of translation blocking morpholino (MO) against cxcr7b (0.02 pmol), sdf1a (0.005 pmol), or cxcr4b (0.005 pmol) causes a significant increase in PGC mismigration (*, p=8.89×10−3, cxcr7b MO; p=2.87×10−3, sdf1a MO; p=0.011, cxcr4b MO; two-tailed Fisher’s exact test). A similar effect is seen upon injection of sdf1a-TP and cxcr7b-TP in cxcr4b heterozygous mutants (*, p=7.50×10−6) and cxcr7b heterozygous mutants (*, p=1.64×10−6) Error bars show ± s.d. (b) Representative examples of PGC mislocalization, shown by nanos in situ. Brackets show correctly localized PGCs, and arrowheads indicate mislocalized cells.
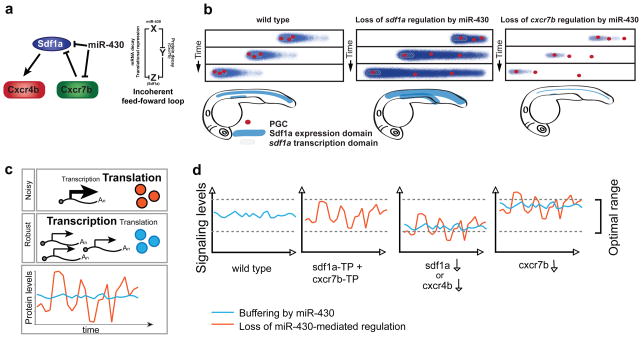
Figure 7.
Model of miR-430-mediated repression of chemokine signaling. (a) Our results are consistent with a model in which miR-430 regulates sdf1a at the RNA level while previous results indicate that Cxcr7b, a decoy receptor, restricts the spatial expression pattern of Sdf1a protein. (b) Model adapted from17 to represent how miR- 430 regulates the dynamic expression of Sdf1a (blue gradient). miR-430-mediated regulation of sdf1a facilitates the formation of a sharp Sdf1a gradient by accelerating the clearance of sdf1a mRNA, concentrating expression on the actively transcribing domains (gray box) (middle panel). miR-430 modulates the levels of cxcr7b to prevent excessive clearance of the Sdf1a protein (right panel). (c) Model for generating robustness by regulating translation of abundantly transcribed genes. High levels of transcription coupled with inefficient translation can lower intrinsic noise in protein output4,5. (e) By dampening the expression of chemokine signaling genes, miR-430 buffers against changes in gene dosage (blue). We postulate that injecting TPs to block miR-430-mediated repression of sdf1a and cxcr7b increases the variability of gene expression (red). This reduces the ability of the system to compensate for minor perturbations in expression (See also Fig. 4, 5, and Supplementary Fig. 9)
Discussion
Secreted signaling molecules are potent developmental regulators, and therefore their expression must be tightly controlled. This is achieved not only at the transcriptional level but also at the level of transcript degradation. This ensures that transcripts are made where they are needed and cleared from domains where they should no longer be present. Our results indicate that miR-430 functions in an incoherent feed-forward loop (FFL) to regulate chemokine signaling by targeting sdf1a and cxcr7b (Fig. 7a). In this network motif, a factor X (miR-430), represses Z (sdf1a) and Y (cxcr7b), which also represses Z. Indeed, mathematical models indicate that incoherent FFLs provide a mechanism for speeding transcriptional networks response times that are generally slow26,27 (reviewed in28). miR-430 is ubiquitously expressed19, and it sharpens the sdf1a expression domain by accelerating degradation of the transcript in all cells. In this manner, only those cells actively transcribing the gene will express it and once transcription is turned off perduring transcripts are rapidly cleared. This regulation is necessary for accurate PGC migration, since loss of miR-430 (MZdicer mutants) or blocking miRNA target sites (Target Protectors) leads to an expansion of the sdf1a expression domain and mislocalized PGCs (Fig. 3). While regulation of cxcr7b and sdf1a explains some of the phenotypes observed in MZdicer, the mismigration phenotype observed in these embryos is consistently stronger than that observed when the miRNA-mediated regulation of each individual target is blocked (Fig. 3 and Supplementary Fig. 1), suggesting that there might be additional targets and other miRNAs that regulate PGCs migration (Supplementary Note).
mRNA degradation not only clears previous expression domains of sdf1a transcripts, but also facilitates spatial separation of different Sdf1a expression domains. Sdf1 signaling is used to guide multiple migratory events during early development, including migration of the trigeminal sensory neurons29, the posterior lateral line primordium30, vasculature31 and endoderm cells32,33. Employing the same guidance cue for each of these migratory processes is possible because the embryo maintains spatially restricted expression domains. The separation of these domains confines the cells to their respective paths so that one type of migrating cell does not follow the path intended for another cell type. Preserving distinct Sdf1a regions is in part accomplished by sharpening of the protein expression by Cxcr7b17. Here we identify a novel regulatory mechanism, acting at the RNA level, in which miR-430 helps to maintain the separation between different migratory paths by (i) sharpening the expression domain of the ligand mRNA and by (ii) dampening the levels of the decoy receptor (Cxcr7b) to prevent excessive clearance of the guidance cue Sdf1a (Fig. 7b).
The interaction of Sdf1 with Cxcr4 and Cxcr7 is conserved among vertebrates including mice and humans16. In addition to its roles in development, Sdf1 signaling is involved in tissue homeostasis by guiding immune cell migration34, neo-vascularization35, stem cell proliferation36 and homing37. Because of their potent activities, misexpression of these molecules has been associated with tumorigenesis and metastasis38,39. miR-430 is a member of a large family of miRNAs, many of which are expressed in mammals (reviewed in 40). Interestingly, human Sdf1 also contains a putative miRNA target site for the homolog of miR-430, miR-30218, in its 3′UTR (Supplementary Fig. 10). Indeed, our results indicate that miR-302 can rescue the germ cell migration in MZdicer (Supplementary Fig. 1), suggesting that this regulatory mechanism could be conserved in other vertebrate systems where it may regulate cell migration and contribute to metastasis.
By dampening the expression of both the ligand and its decoy receptor, miR-430 fine-tunes the amount of signaling. This interaction provides robustness by guarding against genetic variation that arises from increases or reductions in gene dosage. We observe that the embryo can withstand alterations in the levels of sdf1a and cxcr7b within a certain range, but this capacity is reduced when miR-430 function is compromised. miR-430 has previously been shown to regulate nodal signaling by dampening and balancing the expression of both the signal (nodal)21 and its antagonist (lefty)21,41. As miR-430 plays a similar regulatory role in both of these contexts in the form of incoherent feed-forward loops, a more general function for miR-430 may be to balance and lower the expression of positive and negative signaling molecules, reducing the ability of perturbations to have a deleterious effect. The evidence presented here suggests that miRNAs can act as effective elements to buffer genetic variation and maintain homeostasis. Although this role of miRNAs has been proposed, supporting experimental evidence is limited, particularly in vertebrates. Several studies in invertebrates have shown a role of specific miRNAs in providing robustness based on (i) the integration of the miRNA within a network motif (feed-forward and feed-back loops)13,42,43; (ii) the stochastic nature of the loss-of-function phenotype43,44 reviewed in6. For example, in Drosophila, miR-263a/b regulates the proapoptotic gene hid in sensory organs to protect against the stochastic cell death that is triggered throughout the eye44; (iii) and the protection against environmental factors, as seen in the buffering effect of miR-7 protecting against the effect of temperature fluctuations during eye development13. Here we show that in addition to the stochastic nature of the phenotype in the TP-injected embryos, where few PGC fail to migrate to the gonad, miR-430-mediated regulation of sdf1a and cxcr7b buffers against genetic variation, providing robustness to the system.
Recent studies of the human genome have uncovered potential sources of genetic variation, most notably a large number of genetic lesions such as copy number variations, heterozygous, and homozygous mutations. Because many of these lesions have no immediately appreciable phenotype45, regulatory mechanism must have evolved to maintain homeostasis. miRNAs are predicted to regulate over 70% of the protein coding genes in mammals, making them prime candidates for this role9,46. Based on our results, we propose that miRNAs like miR-430 might play a widespread role in buffering the genetic variation borne by common genetic lesions, providing biological robustness to the organism.
Methods
Fish Strains
MZdicer fish were generated as previously described18. Embryos used were dicerhu896/hu896 or dicer(hu896/hu715)47. cxcr4b heterozygous fish were generated by crossing cxcr4b(t26035/t26035)25 with wild type fish. cxcr7b heterozygotes were a cross between cxcr7b (sa0016/sa0016) and wild type24.
Target prediction and microarray analysis
The 3′UTRs of genes with a known role in PGC migration were analyzed for sites complementary to the miR-430 seed sequence (GCACTT). Both paralogs of sdf1 as well as their receptors contained complementary sequences. The expression of putative targets was examined using previously reported microarray data for cDNA from 8.5 hpf embryos hybridized to Affymetrix GeneChip Zebrafish Genome Array19.
GFP reporter constructs
The 3′UTRs of sdf1a, sdf1b, cxcr4a, cxcr4b, cxcr7a, and cxcr7b were identified using EST data available from Ensembl. cxcr7a did not have a predicted 3′UTR when this project was initiated, so 1.5 kb downstream of the predicted coding sequence was used. Currently the estimated 3′UTR based on mRNA seq is 1.3kb (RNASEQDART 00000015292, ZV8 ensemble genome assembly). Analysis of zebrafish RNAseq data at 6 and 24h revealed that reads in the sdf1a 3′UTR extended beyond the published RefSeq mRNA NM_178307. This refseq lacks a polyA tail and instead has a genome encoded stretch of seven adenosines, suggesting that the shorter 3′UTRs for sdf1a might be due to mispriming in a genome-encoded A rich region. Inspection of the zebrafish EST database indicate that there are multiple ESTs downstream of NM_178307 supporting a longer 3′UTR. See in situ hybridization and Supp Fig. 3 for additional supporting information.
These 3′UTRs were amplified from cDNA of 24 hpf embryos (Supplementary Table 1). 3′UTR PCR products were digested and ligated into pCS2+GFP (xhoI-xbaI). The long sdf1a 3′UTR was constructed by cloning the product amplified by the sdf1a oligos, cut the PCR fragment with nhe-xba and ligated downstream of the sdf1a short utr vector cut with xba. To clone mutant reporter constructs, the seed sequence in the wild type reporter was changed from GCACTT to GCTGAT, creating three mismatches in the putative miRNA target site.
Constructs to express sdf1a (ENSDART00000053946, ZV8) and cxcr7b (ENSDART00000063665, ZV8) were generated by amplifying the open reading frame from cDNA of 24 hpf embryos. PCR products were digested and ligated into wild type or mutated GFP reporter vectors substituting GFP for the ORF of interest (BamHI-XhoI).
Oligonucleotide sequences used in this manuscript are shown in the supplementary file.
mRNA and morpholino injection
Target validation: mRNA was transcribed from reporter constructs using mMessage mMachine kit (Ambion). 1 nL of a solution of GFP reporter mRNA at 0.1 μg/μL and 0.08μg/μL dsRed mRNA was injected into wild type or MZdicer embryos at the one-cell stage.
Target repression was quantified by comparing the average pixel intensity of GFP in injected wild type and MZdicer embryos, as previously described 19. Briefly, pixel intensity in a rectangle of the trunk of a GFP-injected embryo (subtracting the background intensity from a rectangle next to the trunk) was normalized to dsRed intensity in the same area (subtracting the background). This intensity value in MZdicer was divided by the value in wild type to obtain fold repression in wild type compared to MZdicer mutant.
Target protector morpholinos were designed to bind with perfect complementarity to 25 nucleotides in the 3′UTR including the miRNA seed sequence. The control target protector binds to a sequence 200 nt downstream of the target site. Unless otherwise noted, 1 nL of 0.2 mM TP was injected in one-cell stage embryos.
All morpholinos were ordered from Gene Tools and dissolved in water. Morpholinos to bind the translation start site (AUG MOs) of sdf1a22, cxcr7b48, and cxcr4b25 were injected at a concentration insufficient to completely knock down expression. These low levels were 1 nL of 0.02 mM (cxcr7b AUG MO) and 0.005 mM (sdf1a and cxcr4b AUG MO). Sequences of these AUG MOs are shown in the supplementary file.
In situ hybridization
In situ hybridization was performed as previously described49. For sdf1a in situ, wild type, TP-injected and MZdicer mutant embryos were combined in the same tube to eliminate variability as described in 49. Before imaging, embryos were dehydrated in methanol after in situ and transferred to benzl benzoate/benzyl alcohol. sdf1a in situs were flat mounted in Permount. Images were taken on a Zeiss Axioimager M1.
To score germ cell mislocalization, embryos were fixed at 24 hpf in 4% paraformaldehyde and PGCs were labeled by in situ for nanos. Locations of germ cells in each embryo were recorded as gonad, pronephros, tail, or head. To quantify the mislocalization phenotype, embryos with at least one mislocalized PGC were counted. To determine the percentage of embryos with mislocalized PGCs, sets of 25–40 embryos were scored. Standard deviations were calculated for multiple repetitions of each experiment. Statistical significance was determined using a two-tailed Fisher’s Exact Test or Wilcoxon rank-sum test as indicated.
qPCR
RNA was extracted from 10 embryos each of wild type, sdf1a-TP-injected, and MZdicer at 24hpf using Trizol reagent according to manufacturer’s instructions (Invitrogen). cDNA was made using Invitrogen SuperScript III kit with oligo dT. FastStart SYBR Green Mastermix (Roche) was used to amplify sdf1a and eif1α. The fold change in sdf1a expression was calculated by ddCt method using eif1α as a control.
Rescue of MZdicer by injecting mature miRNA duplex
MZdicer embryos were rescued by injecting at the one-cell stage 1 nL of 50 μM miR-430c duplex or 1 nL of 10 μM miR-302b (purchased from IDT). Rescue was assessed by ventricle inflation, mid-hind brain boundary formation, and decreased tail curvature18.
Supplementary Material
Acknowledgments
We thank H. Patnode for fish husbandry. C. Takacs and C. Stahlhut for discussion and critical reading of the manuscript, H. Xue for analysis of the sdf1a 3′UTR and D. Stemple for cxcr7b mutant zebrafish. This work was supported by NRSA NIH/NIGMS T32 GM007223 Training Grant (AAS), NIH grants R01GM081602-03/03S1, the Yale Scholar program, and the Pew Scholars Program in Biomedical Sciences (AJG.) and a Whitehead Fellowship Award (HK).
Footnotes
Contribution: AAS and AJG designed the experiments, interpreted the results. AAS preformed all experiments but the genetic interactions in the cxcr7b and cxcr4b mutant backgrounds, which were performed by HK. AAS, wrote the manuscript with input from HK and AJG.
References
- 1.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 2.Maheshri N, O’Shea EK. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu Rev Biophys Biomol Struct. 2007;36:413–34. doi: 10.1146/annurev.biophys.36.040306.132705. [DOI] [PubMed] [Google Scholar]
- 3.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38 (Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 4.Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–3. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24:1339–44. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sangster TA, et al. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Nat Acad Sci. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 12.Wu CI, Shen Y, Tang T. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res. 2009;19:734–43. doi: 10.1101/gr.084640.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–82. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raz E. Primordial germ-cell development: the zebrafish perspective. Nature reviews Genetics. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- 15.Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: a fisheye view. Development. 2009;136:1223–1229. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- 16.Naumann U, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–73. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 18.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–8. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 19.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 20.Nakahara K, et al. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci U S A. 2005;102:12023–8. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi WY, Giraldez AJ, Schier AF. Target Protectors Reveal Dampening and Balancing of Nodal Agonist and Antagonist by miR-430. Science. 2007 doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 22.Doitsidou M, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–59. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 23.Weidinger G, et al. Regulation of zebrafish primordial germ cell migration by attraction towards an intermediate target. Development. 2002;129:25–36. doi: 10.1242/dev.129.1.25. [DOI] [PubMed] [Google Scholar]
- 24.Busch-Nentwich E, et al. SangerInstitute Zebrafish Mutation Resource targeted knock-out mutants phenotype and image data submission. ZFIN Direct Data Submission. 2010 [Google Scholar]
- 25.Knaut H, Werz C, Geisler R, Nusslein-Volhard C. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–82. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- 26.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–5. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wall ME, Dunlop MJ, Hlavacek WS. Multiple functions of a feed-forward-loop gene circuit. J Mol Biol. 2005;349:501–14. doi: 10.1016/j.jmb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 28.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 29.Knaut H, Blader P, Strähle U, Schier AF. Assembly of trigeminal sensory ganglia by chemokine signaling. Neuron. 2005;47:653–66. doi: 10.1016/j.neuron.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 30.David NB, Sapède D, Saint-Etienne L, Thisse C, Thisse B, Dambly-Chaudière C, Rosa FM, Ghysen A. Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proceedings of the National Academy of Sciences. 2002;99:16297–16302. doi: 10.1073/pnas.252339399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND. Chemokine signaling guides regional patterning of the first embryonic artery. Genes & Development. 2009;23:2272–2277. doi: 10.1101/gad.1813509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair S, Schilling TF. Chemokine signaling controls endodermal migration during zebrafish gastrulation. Developmental Biology. 2008 doi: 10.1126/science.1160038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizoguchi T, Verkade H, Heath JK, Kuroiwa A, Kikuchi Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development. 2008;135:2521–9. doi: 10.1242/dev.020107. [DOI] [PubMed] [Google Scholar]
- 34.Luster AD. Chemokines --Chemotactic Cytokines That Mediate Inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 35.Petit I, Jin D, Rafii S. The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends in Immunology. 2007;28:299–307. doi: 10.1016/j.it.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broxmeyer HE, et al. Transgenic Expression of Stromal Cell-Derived Factor-1/CXC Chemokine Ligand 12 Enhances Myeloid Progenitor Cell Survival/Antiapoptosis In Vitro in Response to Growth Factor Withdrawal and Enhances Myelopoiesis In Vivo. J Immunol. 2003;170:421–429. doi: 10.4049/jimmunol.170.1.421. [DOI] [PubMed] [Google Scholar]
- 37.Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. The Chemokine SDF-1 Is a Chemoattractant for Human CD34+ Hematopoietic Progenitor Cells and Provides a New Mechanism to Explain the Mobilization of CD34+ Progenitors to Peripheral Blood. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer letters. 2008;267:226–44. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 39.Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 40.Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010 doi: 10.1038/embor.2010.102. advance online publication 23 July 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–27. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hilgers V, Bushati N, Cohen SM. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 8:e1000396. doi: 10.1371/journal.pbio.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–8. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 48.Boldajipour B, Mahabaleshwar H, Kardash E. Control of Chemokine-Guided Cell Migration by Ligand Sequestration. Cell. 2008 doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 49.Mishima Y, et al. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–32. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.