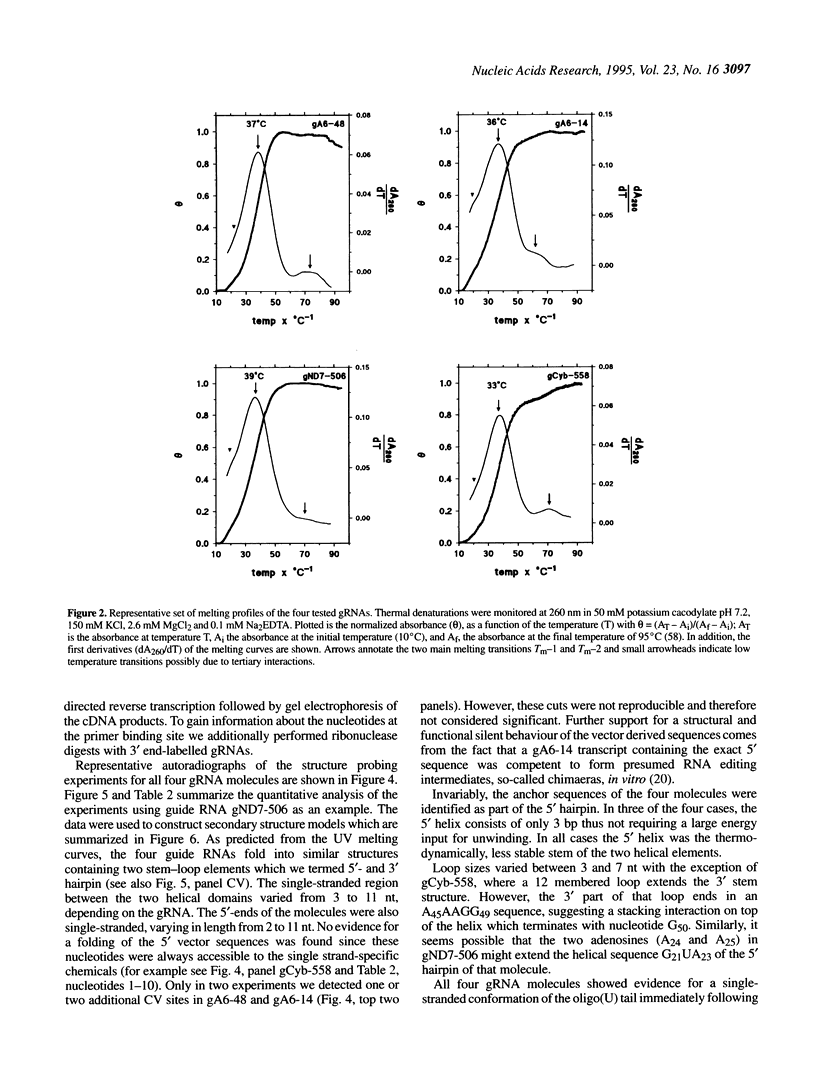
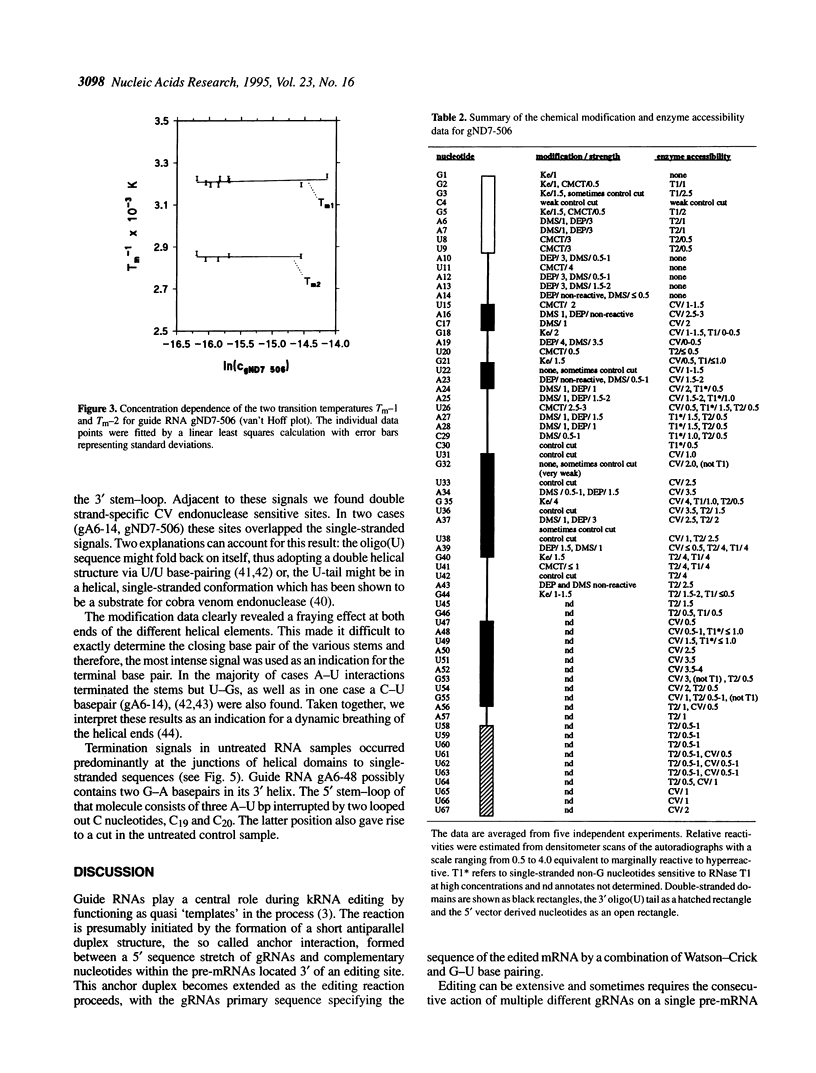
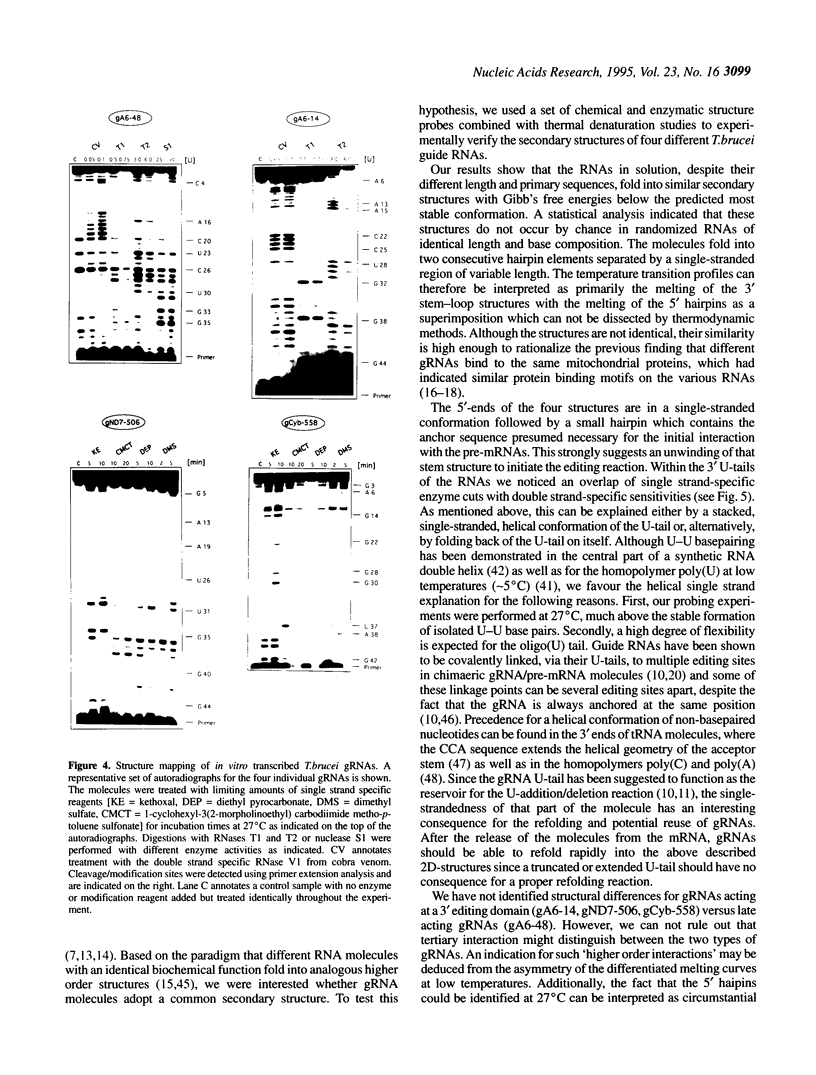
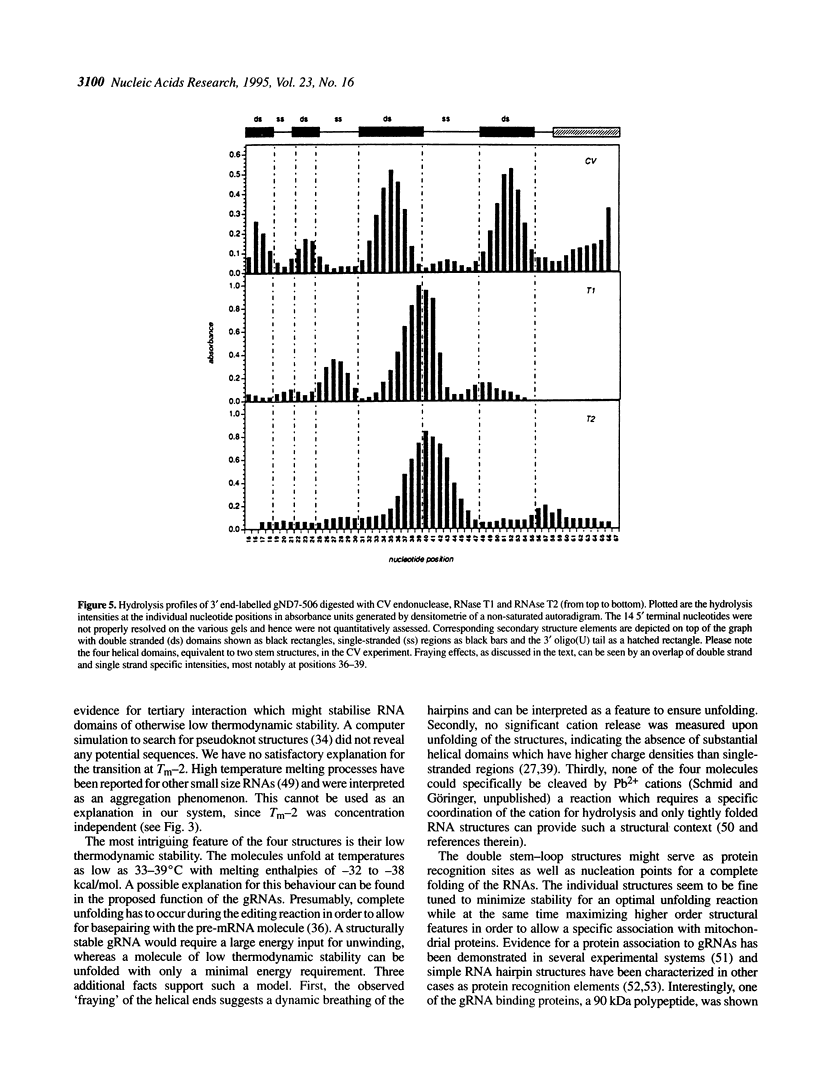
Abstract
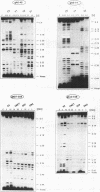
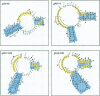
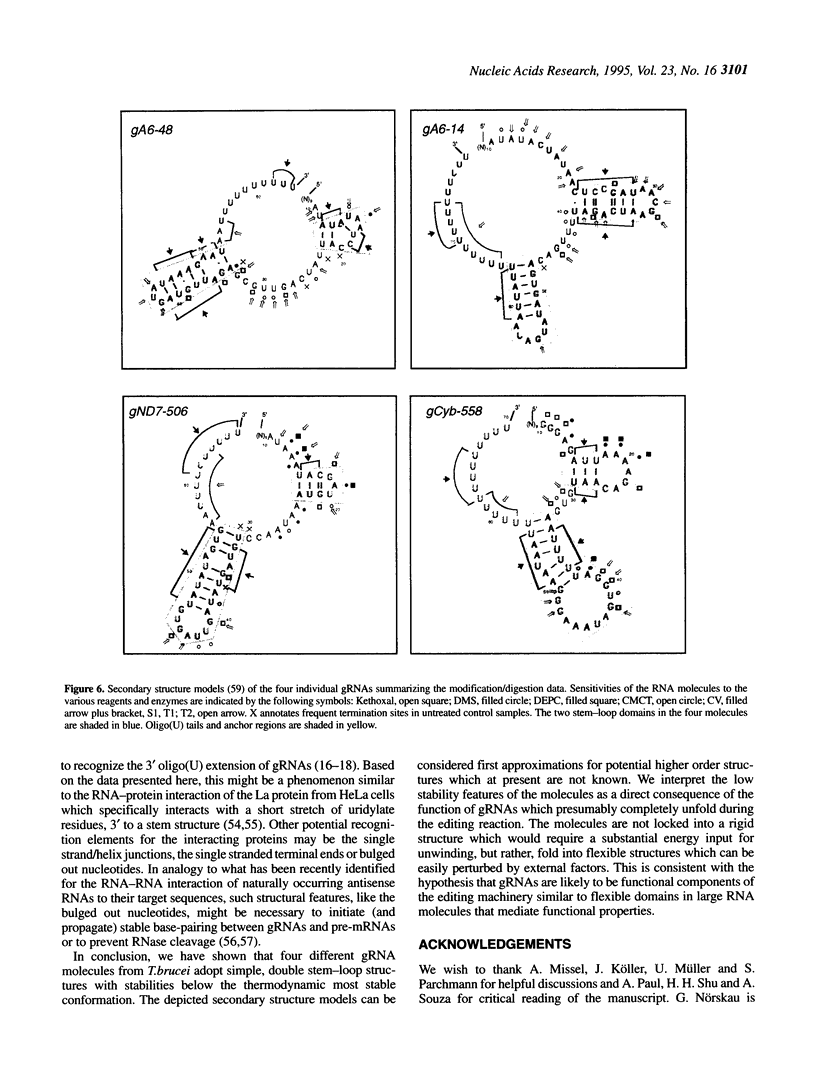
RNA editing in kinetoplastid organisms is a mitochondrial RNA processing phenomenon that is characterized by the insertion and deletion of uridine nucleotides into incomplete mRNAs. Key molecules in the process are guide RNAs which direct the editing reaction by virtue of their primary sequences in an RNA-RNA interaction with the pre-edited mRNAs. To understand the molecular details of this reaction, especially potential RNA folding and unfolding processes as well as assembly phenomena with mitochondrial proteins, we analyzed the secondary structure of four different guide RNAs from Trypanosoma brucei at physiological conditions. By using structure-sensitive chemical and enzymatic probes in combination with spectroscopic techniques we found that the four molecules despite their different primary sequences, fold into similar structures consisting of two imperfect hairpin loops of low thermodynamic stability. The molecules melt in two-state monomolecular transitions with Tms between 33 and 39 degrees C and transition enthalpies of -32 to -38 kcal/mol. Both terminal ends of the RNAs are single-stranded with the 3' ends possibly adopting a single-stranded, helical conformation. Thus, it appears that the gRNA structures are fine tuned to minimize stability for an optimal annealing reaction to the pre-mRNAs while at the same time maximizing higher order structural features to permit the assembly with other mitochondrial components into the editing machinery.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens K. J., De Bondt H. L., Holbrook S. R. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nat Struct Biol. 1995 Jan;2(1):56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990 Jan 26;60(2):189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Blum B., Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3' oligo(U) tail involved in recognition of the preedited region. Cell. 1990 Jul 27;62(2):391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- Blum B., Sturm N. R., Simpson A. M., Simpson L. Chimeric gRNA-mRNA molecules with oligo(U) tails covalently linked at sites of RNA editing suggest that U addition occurs by transesterification. Cell. 1991 May 17;65(4):543–550. doi: 10.1016/0092-8674(91)90087-f. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Warshaw M. M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9(9):1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Cech T. R. RNA editing: world's smallest introns? Cell. 1991 Feb 22;64(4):667–669. doi: 10.1016/0092-8674(91)90494-j. [DOI] [PubMed] [Google Scholar]
- Corell R. A., Feagin J. E., Riley G. R., Strickland T., Guderian J. A., Myler P. J., Stuart K. Trypanosoma brucei minicircles encode multiple guide RNAs which can direct editing of extensively overlapping sequences. Nucleic Acids Res. 1993 Sep 11;21(18):4313–4320. doi: 10.1093/nar/21.18.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse W. B., Saludjian P., Biala E., Strazewski P., Prangé T., Kennard O. Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4160–4164. doi: 10.1073/pnas.91.10.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast F. U. A Macintosh program for the versatile generation of random nucleic acid sequences and their structural analysis. Comput Appl Biosci. 1994 Feb;10(1):49–51. doi: 10.1093/bioinformatics/10.1.49. [DOI] [PubMed] [Google Scholar]
- Göringer H. U., Koslowsky D. J., Morales T. H., Stuart K. The formation of mitochondrial ribonucleoprotein complexes involving guide RNA molecules in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1776–1780. doi: 10.1073/pnas.91.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalt T. A., Wagner E. G. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 1995 Feb 25;23(4):580–587. doi: 10.1093/nar/23.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalt T. A., Wagner E. G. Bulged-out nucleotides protect an antisense RNA from RNase III cleavage. Nucleic Acids Res. 1995 Feb 25;23(4):571–579. doi: 10.1093/nar/23.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung S. H., Yu Q., Gray D. M., Ratliff R. L. Evidence from CD spectra that d(purine).r(pyrimidine) and r(purine).d(pyrimidine) hybrids are in different structural classes. Nucleic Acids Res. 1994 Oct 11;22(20):4326–4334. doi: 10.1093/nar/22.20.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmer D. P., Stuart K. Sequence organization in African trypanosome minicircles is defined by 18 base pair inverted repeats. Mol Biochem Parasitol. 1986 Mar;18(3):321–331. doi: 10.1016/0166-6851(86)90089-7. [DOI] [PubMed] [Google Scholar]
- Jin R., Chapman W. H., Jr, Srinivasan A. R., Olson W. K., Breslow R., Breslauer K. J. Comparative spectroscopic, calorimetric, and computational studies of nucleic acid complexes with 2',5"-versus 3',5"-phosphodiester linkages. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10568–10572. doi: 10.1073/pnas.90.22.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991 Jun;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Kim K. S., Teixeira S. M., Kirchhoff L. V., Donelson J. E. Transcription and editing of cytochrome oxidase II RNAs in Trypanosoma cruzi. J Biol Chem. 1994 Jan 14;269(2):1206–1211. [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Koslowsky D. J., Göringer H. U., Morales T. H., Stuart K. In vitro guide RNA/mRNA chimaera formation in Trypanosoma brucei RNA editing. Nature. 1992 Apr 30;356(6372):807–809. doi: 10.1038/356807a0. [DOI] [PubMed] [Google Scholar]
- Koslowsky D. J., Riley G. R., Feagin J. E., Stuart K. Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol Cell Biol. 1992 May;12(5):2043–2049. doi: 10.1128/mcb.12.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köller J., Nörskau G., Paul A. S., Stuart K., Göringer H. U. Different Trypanosoma brucei guide RNA molecules associate with an identical complement of mitochondrial proteins in vitro. Nucleic Acids Res. 1994 Jun 11;22(11):1988–1995. doi: 10.1093/nar/22.11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCuyer K. A., Crothers D. M. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993 May 25;32(20):5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- Le S. Y., Maizel J. V., Jr A method for assessing the statistical significance of RNA folding. J Theor Biol. 1989 Jun 22;138(4):495–510. doi: 10.1016/s0022-5193(89)80047-5. [DOI] [PubMed] [Google Scholar]
- Leegwater P., Speijer D., Benne R. Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata. Eur J Biochem. 1995 Feb 1;227(3):780–786. doi: 10.1111/j.1432-1033.1995.tb20201.x. [DOI] [PubMed] [Google Scholar]
- Lowary P. T., Uhlenbeck O. C. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res. 1987 Dec 23;15(24):10483–10493. doi: 10.1093/nar/15.24.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Maslov D. A., Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992 Aug 7;70(3):459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- Perochon-Dorisse J., Chetouani F., Aurel S., Iscolo N., Michot B. RNA-d2: a computer program for editing and display of RNA secondary structures. Comput Appl Biosci. 1995 Feb;11(1):101–109. doi: 10.1093/bioinformatics/11.1.101. [DOI] [PubMed] [Google Scholar]
- Pollard V. W., Hajduk S. L. Trypanosoma equiperdum minicircles encode three distinct primary transcripts which exhibit guide RNA characteristics. Mol Cell Biol. 1991 Mar;11(3):1668–1675. doi: 10.1128/mcb.11.3.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V. W., Rohrer S. P., Michelotti E. F., Hancock K., Hajduk S. L. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell. 1990 Nov 16;63(4):783–790. doi: 10.1016/0092-8674(90)90144-4. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Read L. K., Corell R. A., Stuart K. Chimeric and truncated RNAs in Trypanosoma brucei suggest transesterifications at non-consecutive sites during RNA editing. Nucleic Acids Res. 1992 May 11;20(9):2341–2347. doi: 10.1093/nar/20.9.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read L. K., Göringer H. U., Stuart K. Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins and gRNA domains but does not require preedited mRNA. Mol Cell Biol. 1994 Apr;14(4):2629–2639. doi: 10.1128/mcb.14.4.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzeperis D., Alessi K., Marky L. A. Thermodynamics of DNA hairpins: contribution of loop size to hairpin stability and ethidium binding. Nucleic Acids Res. 1993 Jun 11;21(11):2683–2689. doi: 10.1093/nar/21.11.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. N. In vitro synthesis of end-mature, intron-containing transfer RNAs. Methods Enzymol. 1989;180:63–69. doi: 10.1016/0076-6879(89)80092-8. [DOI] [PubMed] [Google Scholar]
- Riesner D., Steger G., Zimmat R., Owens R. A., Wagenhöfer M., Hillen W., Vollbach S., Henco K. Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis. 1989 May-Jun;10(5-6):377–389. doi: 10.1002/elps.1150100516. [DOI] [PubMed] [Google Scholar]
- Riley G. R., Corell R. A., Stuart K. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J Biol Chem. 1994 Feb 25;269(8):6101–6108. [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Seiwert S. D., Stuart K. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994 Oct 7;266(5182):114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- Sheardy R. D., Levine N., Marotta S., Suh D., Chaires J. B. A thermodynamic investigation of the melting of B-Z junction forming DNA oligomers. Biochemistry. 1994 Feb 15;33(6):1385–1391. doi: 10.1021/bi00172a014. [DOI] [PubMed] [Google Scholar]
- Stefano J. E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3' termini of RNA polymerase III transcripts. Cell. 1984 Jan;36(1):145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- Thrierr J. C., Dourlent M., Leng M. A study of polyuridylic acid. J Mol Biol. 1971 Jun 28;58(3):815–830. doi: 10.1016/0022-2836(71)90042-8. [DOI] [PubMed] [Google Scholar]
- Wang Y. X., Lu M., Draper D. E. Specific ammonium ion requirement for functional ribosomal RNA tertiary structure. Biochemistry. 1993 Nov 23;32(46):12279–12282. doi: 10.1021/bi00097a002. [DOI] [PubMed] [Google Scholar]