Summary
The MR (Mre11 nuclease and Rad50 ABC ATPase) complex is an evolutionarily conserved sensor for DNA double-strand breaks, highly genotoxic lesions linked to cancer development. MR can recognize and process DNA ends even if they are blocked and misfolded. To reveal its mechanism, we determined the crystal structure of the catalytic head of Thermotoga maritima MR and analyzed ATP dependent conformational changes. MR adopts an open form with a central Mre11 nuclease dimer and two peripheral Rad50 molecules, a form suited for sensing obstructed breaks. The Mre11 C-terminal helix-loop-helix domain binds Rad50 and attaches flexibly to the nuclease domain, enabling large conformational changes. ATP binding to the two Rad50 subunits induces a rotation of the Mre11 helix-loop-helix and Rad50 coiled-coil domains, creating a clamp conformation with increased DNA binding activity. The results suggest that MR is an ATP controlled transient molecular clamp at DNA double-strand breaks
Keywords: Rad50, Mre11, DNA double-strand break repair, X-ray crystallography, protein complex, homologous recombination, ABC ATPases
Introduction
DNA double-strand breaks (DSBs) are a major threat to genome stability in all kingdoms of life and can lead to gross chromosomal aberrations and cancer (Lee et al., 2008; Putnam et al., 2009; Wang et al., 2009). DSBs arise during chromosomal replication but can also be products of ionizing radiation and genotoxic chemicals (Costanzo et al., 2001; Ward, 1988). In addition, enzymatically induced DSBs are intermediates in V(D)J recombination, immune globulin class switching and meiosis. Both sporadic and programmed DSBs need careful and prompt repair to maintain genome stability and allow cell survival.
DSBs elicit a complex cellular response and induce different repair pathways (Harper and Elledge, 2007). Repair proceeds predominantly by homologous recombination (HR), non-homologous end joining (NHEJ) or microhomology mediated end joining (MMEJ). In HR, DSBs are repaired error-free using the sister-chromatid as template. In NHEJ, DNA ends are directly religated, often at microhomologies after limited, potentially mutagenic, processing of the DNA ends (MMEJ).
The central response factor for DSBs is the Mre11:Rad50:Nbs1 (MRN) complex (Mre11:Rad50:Xrs2 (MRX) in Saccharomyces cerevisiae). It is involved in most, if not all DNA end associated processes including damage checkpoint signaling, HR, NHEJ/MMEJ, telomere maintenance and meiotic recombination (Borde, 2007; Lavin, 2007; Lee and Paull, 2007; Stracker et al., 2004; Williams et al., 2007). Hypomorphic mutations in human Mre11, Rad50 and Nbs1 are linked to the cancer predisposition diseases Nijmegen breakage syndrome (NBS), Ataxia-telangiectasia like disorder (A-TLD) and NBS-like disorder (Carney et al., 1998; Petrini, 2000; Stewart et al., 1999; Varon et al., 1998).
The multifunctional MRN possesses both enzymatic and architectural activities. MRN activates the ATM kinase, resulting in activation and coordination of subsequent checkpoint and repair processes. The ATP stimulated nuclease activity of MRN together with Sae2/CtIP then removes short oligonucleotides from the 5′ end of the DSBs (Budd and Campbell, 2009; Hopkins and Paull, 2008; Mimitou and Symington, 2008; Zhu et al., 2008). MRN additionally possesses a 3′ to 5′ exonuclease, ssDNA endonuclease and hairpin-opening activities, presumably to clean misfolded and blocked DNA ends (Paull and Gellert, 1998, 1999). Finally, MRN is implicated in tethering of DNA ends and chromatids via coiled-coil tails of Rad50 (Hopfner et al., 2002).
Homologs for Mre11 and Rad50 - but not Nbs1 - are found in all three phylogenetic domains and are also known as SbcC (Rad50 homolog) and SbcD (Mre11 homolog) in bacteria (Sharples and Leach, 1995). The prokaryotic MR complex shares enzymatic activities and morphological features with eukaryotic MRN and is implicated in processing and repair of DNA double-strand breaks, interstrand-crosslinks and replication fork associated hairpins (Bentchikou et al., 2007; Cromie et al., 2001; Eykelenboom et al., 2008; Leach et al., 1997; Mascarenhas et al., 2006; Zahradka et al., 2009). MR is a large, bipolar complex containing two Mre11 and two Rad50 polypeptides (Hopfner et al., 2001). The Mre11 dimer and the two Rad50 nucleotide binding domains (NBDs) form a “catalytic head” that harbors ATP stimulated nuclease and DNA binding activities. Two long Rad50 coiled-coil domains protrude from this head and can bind other MR complexes via their apical zinc-hook dimerization motifs (de Jager et al., 2001; Hopfner et al., 2002), thereby forming large molecular bridges to transiently tether broken chromosomes (Lobachev et al., 2004).
The functional interplay of Rad50’s ATPase and the Mre11 nuclease in DSB repair remained unclear, mostly because there is not a structural framework for the catalytic head complex. As a consequence, the role of Rad50’s ATPase function has been unclear, although it is essential for MR(N/X) function (Bhaskara et al., 2007). Likewise, it is unknown why DSB recognition by MRN/MR(N/X) has no clear biochemical preference for DNA ends or hairpins and binds DNA also at internal sites. This is important because MRN can sense and clear protein bound DNA ends in vitro (Connelly et al., 2003) and in vivo (Lobachev et al., 2002; Neale et al., 2005). The remarkable ability to clean up blocked and misfolded ends using an ATP-driven nuclease activity distinguishes MR(N/X) from e.g. the NHEJ protein Ku (Walker et al., 2001).
To provide a structural framework for the complex between Mre11 and Rad50, we determined the X-ray crystal structure of the catalytic head of the Thermotoga maritima MR (SbcCD) complex as well as the crystal structure of the AMPPNP bound Rad50 NBD dimer in complex with the Rad50 binding helix-loop-helix motif (HLH) of Mre11. We also analyzed ATP dependent conformational changes of the MR head module by small angle X-ray scattering (SAXS) and chemical crosslinking assays. The structure revealed that Mre11 and Rad50 form a large ATP controlled molecular clamp, suited to recognize even blocked DSBs. The observed interfaces were tested by mutating homologous regions in eukaryotic MRX in Saccharomyces cerevisiae. Finally, we demonstrate that ATP binding to Rad50 induces conformational changes in the Mre11 dimer, providing a mechanistic role for Rad50’s ATP binding activity in controlling DNA processing by Mre11. This unifies the functional architecture of MR with other ABC type molecular machines such as ABC transporters.
Results and Discussion
Structure determination
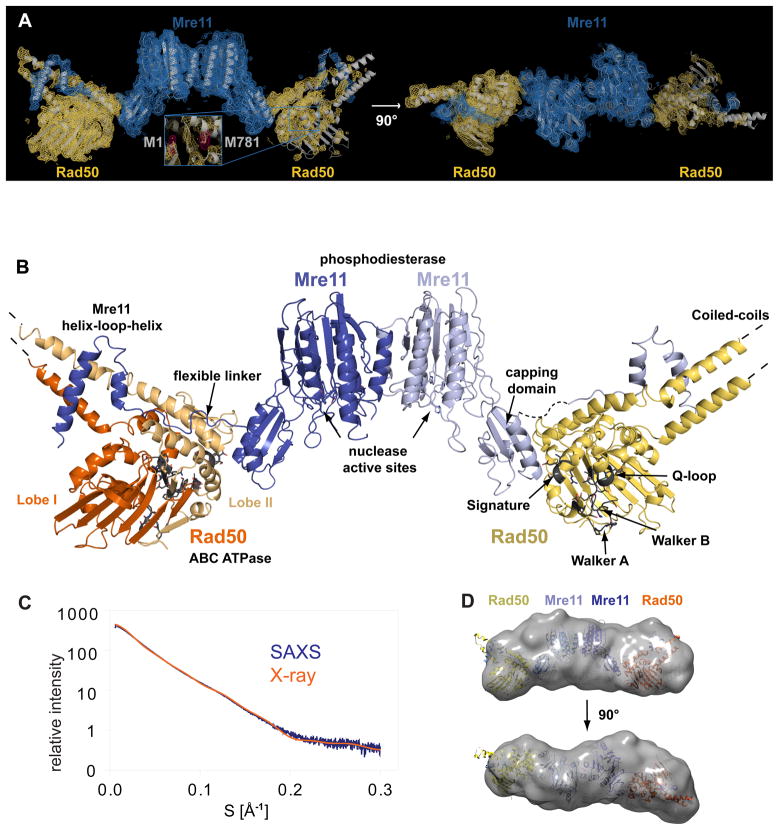
We co-purified and crystallized Mre11 from Thermotoga maritima (Tm) and a shortened Rad50 (see Methods) that comprises the nucleotide binding domain and approx. 50 amino acids of the Mre11 binding coiled-coil (Rad50NBD). The two ends of the shortened coiled-coil are fused by a short linker. Both proteins form a stoichiometric complex with ATPase as well as DNA binding activity (Fig. S1). Crystals of space group I222 contained one full catalytic head (M2:RNBD 2) per asymmetric unit and diffracted X-rays to 3.4 Å resolution. Single wavelength anomalous dispersion phasing and density modification produced an interpretable electron density for both Mre11’s and one Rad50NBD (Fig. 1A). Electron density for the second Rad50 was less defined, presumably because this domain is more mobile due to lack of additional crystal contacts. However, we could rigid body dock the model obtained from the well-defined Rad50NBD into the density of the second Rad50NBD using unambiguous anomalous difference density markers and some secondary electron density. Both Rad50NBDs interact with Mre11 in a similar manner. The resulting M2:RNBD 2 heterotetramer could be refined with acceptable geometry and R-values (suppl. Table S1).
Figure 1. Overall structure of the crescent-shaped Mre11:Rad50 catalytic head.
A) Experimental electron density map (contoured at 1.0 σ and colored blue for Mre11 and yellow for Rad50) superimposed with a ribbon representation of the Thermotoga maritima Mre11:Rad50NBD complex (gray). Two views are shown. Inset: anomalous difference electron density map for the selenium atoms (pink, contoured at 5.0 σ).
B) Ribbon representation of the bacterial Rad50 Mre11 catalytic head. Individual domains and important motifs are highlighted and annotated.
C) Experimental small angle X-ray scattering (SAXS) profile of Mre11:Rad50NBD (blue) compared to the theoretical scattering curve calculated from the crystal structure of the complex (orange).
D) Two orthogonal views of the average SAXS envelope of the Mre11:Rad50NBD assemblies (calculated with DAMMIN, superimposed with a ribbon representation of the atomic model of the complex) highlight the similarity of crystals structure and solution conformation.
Crystal structure of the bacterial Mre11:Rad50 catalytic head
The TmMR catalytic head (TmMRNBD) is an elongated crescent shaped complex with approx. 60Å × 70Å × 210Å dimensions (Fig. 1A and B). Its core is formed by a dimer of the two Mre11 nuclease domains, with the two nuclease active sites located near the center of the concave face. The Rad50 NBDs each attach to the outside of the nuclease dimer and form the tips of the crescent. The Rad50 coiled-coils protrude from the convex side of the catalytic head, opposite to the nuclease active sites, at an angle of approx. 120° from each other. This architecture fits well the bipolar shapes of full prokaryotic MR and eukaryotic MRN complexes previously visualized by electron and scanning force microscopy (Connelly et al., 1998; de Jager et al., 2001; Hopfner et al., 2001).
The NBDs of Rad50 possess the typical bi-lobed ABC fold, while Mre11 shows an extended structure, composed of two functional modules, the “nuclease module” and the Rad50 binding domain. The “nuclease module” comprises the phosphodiesterase and accessory DNA binding “capping” domains. The Rad50 binding domain contains a helix-loop-helix (HLH) domain that binds to the root of Rad50’s coiled-coil. Surprisingly, the “nuclease module” and the HLH domain are widely separated and connected by a long, poorly structured linker, that wraps around Rad50’s NBD and places the capping and HLH domains of Mre11 on opposite sites of it. A second interaction is formed between Mre11’s capping domain and Lobe II of Rad50’s NBDs, stabilizing the observed extended domain arrangement between Mre11 and Rad50 in the catalytic head.
To verify the domain arrangements found in the crystal structure, we performed small angle X-ray scattering (SAXS). The experimentally derived SAXS intensities of MRNBD closely match the scattering intensities calculated from its crystal structure and individual as well as averaged ab initio SAXS models exhibit the elongated shape of the atomic model (Fig. 1C, D). This supports the conclusion that the MR crystal structure we present here closely resembles the conformation of the complex in solution.
The overall structure explains two poorly understood functional characteristics of MR. The widely separated, outward placement of the Rad50 NBDs allows unobstructed access of DNA to the Mre11 active sites, even if DNA ends are blocked by large proteins. In addition, our structure shows that the complex has the potential to undergo major conformational changes, consistent with the observation of large conformational changes identified by scanning force microscopy of human MRN (Moreno-Herrero et al., 2005).
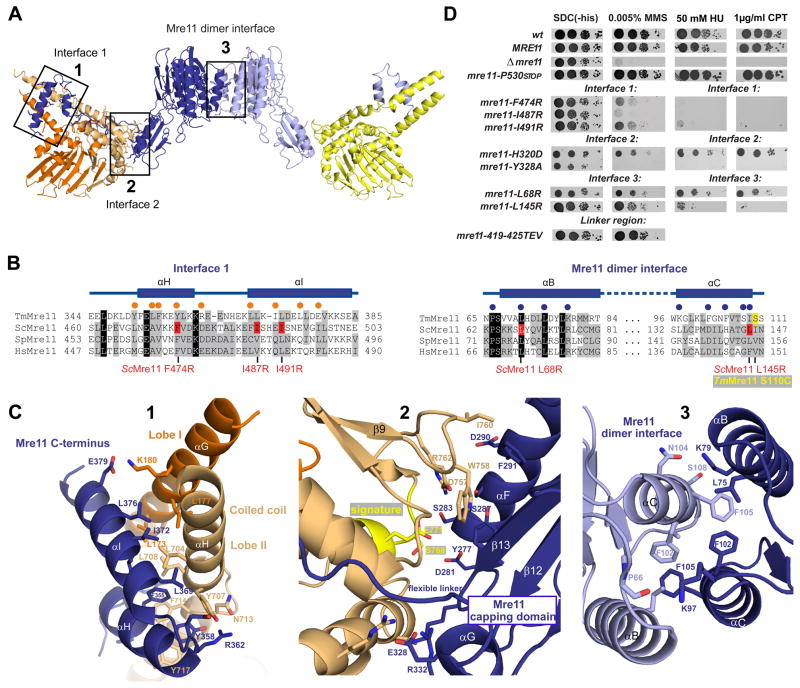
Details of Mre11:Rad50 interfaces
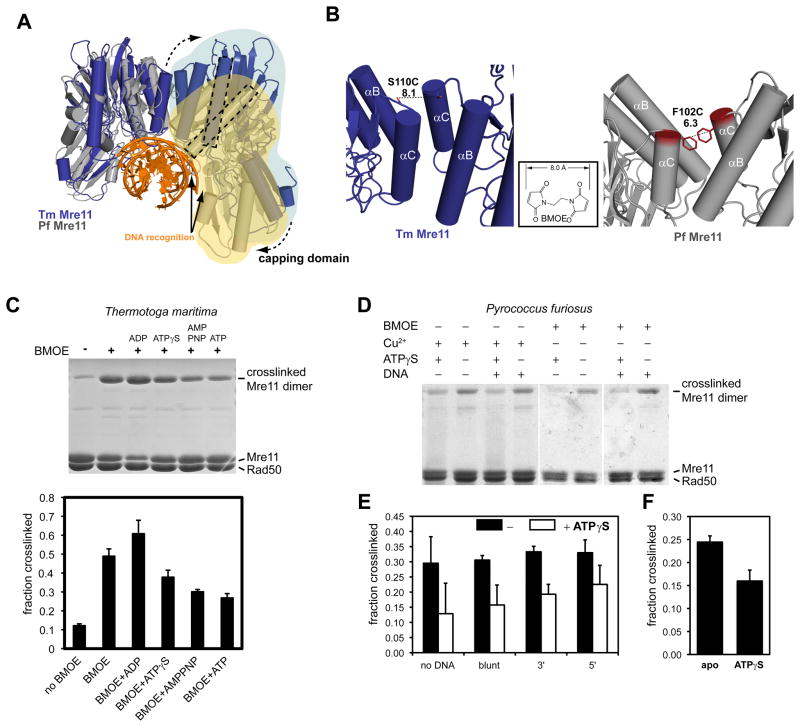
There are three types of macromolecular interfaces in MRNBD, one between the two Mre11 subunits and two between Mre11 and Rad50 (Fig. 2A–C). A fourth interface between the two Rad50 NBDs in the presence of ATP will be described below. The two Mre11 phosphodiesterase domains interact by forming a four-helix bundle (αB and αC from both protomers) around central hydrophobic/aromatic residues (F102, F105 and L75 from both protomers) (Fig. 2C3). This interface is related to that seen in the crystal structure of the catalytic domain of P. furiosus Mre11, showing that the architectural principles of MR are conserved.
Figure 2. Details of Mre11:Rad50NBD and the Mre11 dimer interfaces.
A) Ribbon representation of the Mre11:Rad50NBD complex colored according to figure 1B. The boxed interfaces (1,2,3) are shown in C).
B) Sequence alignment of the Mre11 HLH domain (αH and αI) and the Mre11 dimer interface (αB and αC). Yeast mutations analyzed in D) are highlighted in red. Spheres represent residues implicated in Mre11HLH:Rad50NBD (orange) and Mre11:Mre11 interaction (blue).
C) Details of macromolecular interfaces. Selected side chains are shown as color-coded sticks and are annotated. 1) The Mre11 helix-loop-helix (HLH) motif (blue) and its interaction with the base of the Rad50 coiled coil (orange and yellow). 2) The interface between the capping domain of Mre11 (blue) and the opposite Rad50 (orange) region in close proximity to the signature motif (yellow). 3) The Mre11 dimerization interface.
D) Yeast survival assays by serial dilutions show that mutations in Mre11 predicted to affect interface 3 (L145R, L68R), mutations within the helix-loop-helix motif predicted to affect interface 1 (L474R, I487R, I491R), and mutations in the capping domain predicted to affect interface 2 (H320D, Y328A) result in sensitivity to Methyl-methanesulfonate (MMS), hydroxyurea (HU) and camptothecin (CPT). Changing the flexible linker residues (419–425 TEV) has no strong effect on MMS sensitivity. Left: SDC medium without histidine alone. Right: media supplemented with indicated concentrations of MMS, HU or CPT.
See also Figure S2.
Rad50 binds Mre11 with two interfaces (1 and 2) comprising areas of 1334 Å2 and 686 Å2, respectively. Interface 1 (Fig. 2A–C) is formed by binding of the Mre11 C-terminal HLH domain to the coiled-coil root of Rad50, with αH and αI of the HLH domain binding perpendicularly across Rad50’s coiled-coil. αHMre11 binds only one coiled-coil helix (αHRad50), while αIMre11 reaches across both coiled-coil helices (αGRad50 and αHRad50). Interestingly, αHMre11 binds to αHRad50 at a pronounced kink and extended loop, located directly opposite the start of αGRad50 at the other side of the coiled-coil. Thus, the coiled-coil is attached to the NBD not with a continuous helix, but with a structure that may allow for movements between the coiled-coil and Mre11 binding site and the globular part of the ATPase.
The HLH motif and coiled-coil domains bind to each other via an array of aromatic and hydrophobic residues. αHMre11 and αHRad50 interact through eight aromatic residues (Mre11: Y351, F352, F355, Y358; Rad50: Y707, F714, Y717, F718). The interface is further stabilized mainly by hydrophobic interactions between αIMre11 and αGRad50 but also some specific hydrogen bonds and salt bridges, e.g. between E379Mre11 and K180Rad50 (Fig. 2C).
The linker preceding αHMre11 and connecting the HLH domain with the nuclease module runs along the outside of the NBD at Lobe II until it reaches the capping domain at the second interface between Rad50 and Mre11. Although parts of the linker adjacent to the HLH domain are ordered and we observe a few, dispersed hydrophobic and ionic contacts to the NBD, some parts appear to be disordered, suggesting that the linker acts as a flexible leash.
Interface 2 (Fig. 2C) is formed between one face of Mre11’s capping domain and the “bottom” side of Rad50’s Lobe II (the side opposite the coiled-coil). This interface is predominantly polar, although W758Rad50 and I760Rad50 bind a small hydrophobic patch at the core of this interface (F291Mre11), located at the edge of the β-sheet of the capping domain, with contributions from the flanking αFMre11. Interestingly, a part of the interface is mediated by the signature motif, which becomes bound by ATP in the engaged conformation of the NBDs. Likewise, the Mre11 capping domain also participates in DNA binding (Williams et al., 2008). Consequently, interface 2 could transiently stabilize the “open” conformation with separated NBDs until DNA and/or ATP is bound at Mre11 nuclease active sites, whilst also allowing the NBDs mobility for ATP induced conformational changes (see below).
Mutational analysis of Mre11:Rad50 interfaces in S. cerevisiae in vivo
To verify the functional significance of the observed interfaces in vivo, we tested mutants in all three interfaces of Saccharomyces cerevisiae MRX for their ability to complement the methyl methanesulfonate (MMS), hydroxyurea (HU) and camptothecin (CPT) sensitivity of the Δmre11 strain (Fig. 2D). Mutations were chosen on the basis of sequence conservation (Fig. 2B) or using a structure of S. pombe Mre1 as guide (to be published). We also tested a replacement of seven residues of the linker between catalytic and Rad50 binding domains of Mre11 with a TEV protease target sequence (419-425TEV) and a Mre11 truncation variant (P530STOP). All proteins were expressed at normal levels according to Western blot analysis (Fig. S2). Consistent with previous data, wild-type MRE11 and the mre11-P530STOP (resulting in a protein truncated after the HLH region) confers resistance to MMS, HU and CPT in the Δmre11 strain (Chamankhah and Xiao, 1999). The Mre11 dimer interface mutants (L145R and L68R) can only partially rescue MMS, HU and CPT sensitivity, similar to what has been observed in the case of S. pombe Rad32 (Williams et al., 2008). In addition, we found that F474R, I487R, I491R in Mre11:Rad50 interface 1 (HLHMre11:coiled-coilRad50) do not rescue MMS, HU and CPT sensitivity. Finally, the Mre11:Rad50 interface 2 (CapMre11:NBDRad50) mutations H320D and Y328A partially or completely fail to rescue MMS, HU and CPT sensitivity. Previously it was found that an 11 amino acid deletion mutant of the S. cerevisiae Mre11 at the predicted interface 2 (Mre11-6) also shows MMS sensitivity and DSB processing defects (Usui et al., 1998).
In contrast, mre11 419-425TEV complements the Δmre11 strain. This indicates that the sequence of the “linker” region is not important for the activity of MRX on homologous recombination. Co-expression of TEV protease did not result in significantly increased MMS sensitivity, although most Mre11 was cleaved, according to Western blot analysis (data not shown). It is possible that low levels of residual or newly synthesized Mre11 are sufficient to complement MMS sensitivity. In support of this, co-expression of the catalytic and Rad50 binding domains of S. cerevisiae Mre11 could not complement Δmre11 (data not shown). In summary, all observed interfaces of TmMRNBD are functionally significant for the yeast MRX complex in vivo.
ATP and DNA induced engagement of Rad50 NBDs
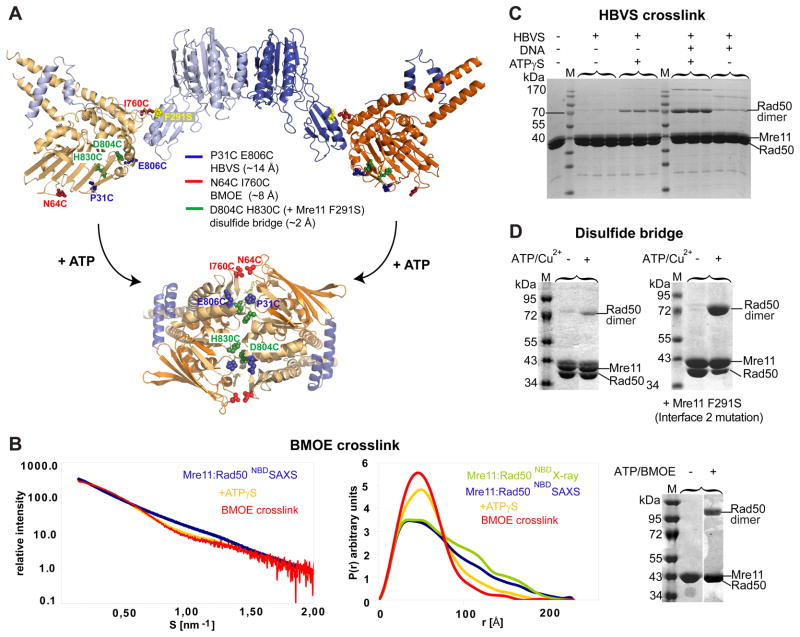
Since ATP hydrolysis by Rad50NBD requires formation of a tightly engaged NBD dimer with sandwiched ATP molecules (Hopfner et al., 2000) (Fig. 3A) we tested whether NBDs from a single-head engage by a large conformational change, or alternatively NBDs from different heads assemble in the presence of ATP by recording SAXS intensities of TmMRNBD in the absence and presence of ATPγS (Fig. 3B). Adding ATPγS indeed resulted in a substantial decrease of the radius of gyration (Rg) from 230 Å to 193 Å as well as a more pronounced peak at shorter vectors and a significant decrease of the long vectors in the pair distribution function P(R) (Fig. 3B). Thus, ATPγS renders the complex more compact, consistent with an expected engagement of the two NBDs within a single catalytic head, but not with ATP mediated clustering of multiple MR catalytic heads.
Figure 3. ATP engages Rad50NBDs in the catalytic head.
A) Cysteine mutations introduced in TmRad50NBD to test formation of the ATP bound Rad50 dimer. Sites are widely separated in the open form (upper crystal structure), but closely spaced for crosslinking or disulfide bonding in the ATP bound form (below, see Fig. 4).
B) Left panel: Superposition of experimental SAXS curves of Mre11:Rad50NBD with and without ATPγS indicate a more compact, globular shape in the presence of ATPγS. SAXS of the BMOE crosslinked Mre11:Rad50NBD,N64C,I760C results in a shape similar to the ATPγS bound form. Middle panel: the electron pair distance distribution function P(r) in the absence of nucleotides corresponds well to the crystal structure derived P(r). ATPγS increases the short distances and decreases the long distances. Residual long distances suggest a heterogeneous mixture between the open form and closed ATPγS complex. The BMOE crosslinked complex has a similar shape to the ATPγS complex, but appears to be more homogenous for the compact form. Right panel: Non-reducing Coomassie stained SDS PAGE of the MRNBD,N64C,I760C crosslinking experiment using BMOE. The crosslinking forms a covalently connected Rad50 dimer in an ATP dependent manner.
C) Chemical crosslinking by HBVS of MRNBD,P31C,E806C creates a covalently connected Rad50 dimer in an ATPγS and DNA dependent manner. The identity of the corresponding gel band was confirmed by mass spectrometry.
D) The formation of ATP bound engaged Rad50NBD,D804C,H830C is tested by using ATP/Cu2+ dependent disulfide bond formation. Modulating the Mre11:Rad50 interface 2 by Mre11F291S results in dramatically increased disulfide bond formation efficiency, consistent with the idea that interface 2 stabilizes the open form and is disrupted in the closed form.
See also Figure S3.
Although ATPγS induces a compact conformation, the residual long vectors in P(R) indicate that the population still contains a substantial amount of the open form. Attempts to obtain a more homogeneous population by addition of various amounts of ATPγS, or AMPPNP, ADP plus BeF3 or AlF3− or using a Walker B E798Q mutation in combination with ATP, have been unsuccessful.
We therefore investigated the ATP dependent engagement of the two Rad50 NBDs by site-specific crosslinking. We engineered 3 pairs of suitable cysteine residues at positions that are predicted to be far apart in the nucleotide free form but close together in the ATP bound dimer of Rad50NBD (Fig. 3A). P31C plus E806C were introduced to allow crosslinking analysis by the thiol reactive agent 1,6-Hexane-bis-vinylsulfone (HBVS). While 110 Å apart in the open form, their estimated distance of 10 Å in the closed form matches the range of HBVS. In the absence of ATPγS and DNA, we do not see substantial crosslinking, in agreement with the large separation of the two NBDs (Fig. 3C). However, ATPγS increases the crosslinking efficiency. The composition of the crosslinked species was verified by mass spectrometry. We also observe some increased crosslinking efficiency in the presence of plasmid DNA, suggesting that DNA has an influence on the NBD arrangement. Some additional bands arise presumably from nonspecific crosslinking. Significantly, the crosslinking efficiency substantially increases further in the presence of both ATPγS and DNA, suggesting that ATP and DNA cooperate in forming the engaged NBD form. Gel filtration or SAXS on the crosslinked species we never revealed evidence of higher molecular weight complexes, ruling out crosslinking of adjacent MR complexes on DNA (Fig. S3A).
While the low to moderate crosslinking efficiency (10–20%) of the HBVS crosslinker enables to detect allosteric effects of DNA addition, it is not suited to analyze the structural and biochemical properties of the crosslinked protein. Using I760C plus N64C (approx. 110 Å apart in the nucleotide free form but approx. 8 Å apart in the ATP bound form) we could very efficiently (up to 70%) crosslink the two NBDs in the presence of ATP, but not in the absence of ATP (Fig. 3B, right panel and 3D). This independently verifies the clamping movement of the NBDs. The proposed clamp movement can also be seen by ATP induced formation of a disulfide bond between D804C and H830C, which are ideally positioned at opposite interface loops in the ATP bound form of NBDs (Fig. 3D). The high efficiency of I760C/N64C in the presence of BMOE (Bis-Maleimidoethane) compared to the disulfide bond could possibly arise from the fact that I760Rad50 is located in interface 2. If interface 2 stabilizes the open form, as seen in the crystal structure, the prediction is that destabilizing interface 2 will increase the efficiency of formation of the closed form. To test this idea, we introduced F291S (F291Mre11 interacts with I760Rad50 in interface 2) into Mre11 and analyzed disulfide bond formation (Fig. 3D). Consistently, while still no efficient disulfide bond formation is observed in the absence of ATP, we can now efficiently crosslink the NBDs by disulfide bond formation. This is corroborated by a slight, but statistical significant increase in ATP hydrolysis activity of the mutants that disrupt interface 2 (Fig. S3G).
To see whether the crosslinked protein has a structure similar to the ATP bound protein, we prepared BMOE crosslinked protein in the presence of ATP (about 65% crosslinked), purified the crosslinked species by gel filtration to remove ATP and further enrich the crosslinked species and performed SAXS. Indeed, the solution structure of the crosslinked protein is very similar to the ATPγS bound protein (Fig. 3B), validating the idea that the crosslink stabilizes the closed conformation. We conclude that ATPγS induces a conformation in MR’s catalytic head with engaged NBDs, thereby the transient interface 2 is disrupted. Besides forming interface 2, the Mre11 capping domains are also involved in DNA binding (Williams et al., 2008). This favors a model in which DNA allosterically helps to modulate interface 2, thus explaining why it promotes formation of the closed state.
To analyze, whether the closed MRNBD structure is formed around DNA we performed the HBVS crosslinking and disulfide bridging in the presence of single-stranded and/or double-stranded plasmid DNA (pBSII KS+, ΦX174 Virion and ΦX174 RF II) under conditions where in EMSA most of the DNA is shifted by bound protein (Fig. S3A–F). In all cases subsequent gel filtration failed to detect comigrating DNA and protein, i.e. the protein is not crosslinked around internal DNA.
ATP bound Rad50 NBDs in complex with Mre11
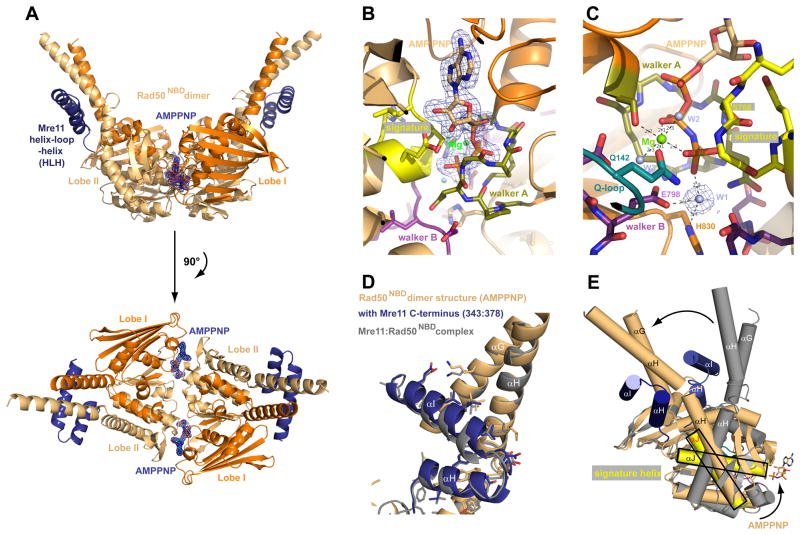
We crystallized TmRad50NBD in complex with the TmMre11 HLH domain (residues 343–385) in the presence of the non-hydrolysable ATP analogue adenylyl-imidodiphosphate (AMPPNP) (Fig. 4A, Fig. S4, Table S2). Although the archaeal Rad50 ATPase domain has been crystallized as ATP bound dimer before (Hopfner et al., 2000), this structure lacked the coiled-coil domain and could not give insights into how ATP might impact on the orientation of the coiled-coil domain and Mre11 interaction.
Figure 4. Structure of the AMPPNP bound Rad50NBD dimer in complex with the HLH domain of Mre11.
A) Two perpendicular views of a ribbon representation of the AMPPNP bound Rad50NBD dimer in complex with the C-terminal helix-loop-helix region of Mre11 color coded according to Fig. 1B. AMPPNP is highlighted and shown with 2FoFc electron density (1σ contour).
B), C) Two views of the ATP binding site with 1.9 Å 2Fo−Fc electron density around AMPPNP (B) or the nucleophilic water (“W1” in C) and highlighted and color coded ATP binding motifs signature motif (yellow), Walker A (olive), Walker B (purple), Q-loop (turquoise) and Mg2+ ion (green sphere).
D) Superposition of Rad50NBD from apo (from MR catalytic head, grey) and AMPPNP (blue/orange) crystal structures at Lobes II shows that for T. maritima MR, coiled-coil structure and interaction interface 1 is not directly modulated by AMPPNP binding.
E) Superposition of apo and AMPPNP bound forms of Rad50NBDs via Lobe I shows that AMPPNP binding induces a large, approx. 50° rotation between Lobe I and Lobe II, leading to a rigid body movement (arrow) of the HLH and coiled coil with respect to the ATP binding interface of Rad50.
The AMPPNP bound structure was determined to a resolution of 1.9 Å and offers a much more detailed view of nucleotide coordination and hydrolysis (Fig. 4B,C). The Walker A motifs bind the three phosphates of the AMPPNP moieties, while the Signature motifs of the opposing NBDs coordinates the γ-phosphates. Mg2+ is coordinated by oxygens from β and γ phosphate, two water molecules and side chain oxygens of S37 (Walker A) and Q142 (Q-loop). A bound water molecule (W1) is suitably located for nucleophilic attack on the γphosphate, positioned and activated by hydrogen bonds to E798 (Walker B), H830 (His-switch) and the main chain oxygen of S802 (opposing D-loop) (Fig. 4C). In addition, the helix following the Q-loop and the subsequent coiled-coiled domains are nicely visualized and well ordered, allowing comparison of nucleotide-bound and -free states of the interaction site between Mre11 and Rad50 (Fig. 4D,E).
ATP binding not only tightly engages the two NBDs, but induces a second conformational change within the NBDs, resulting in an approx. 50° rotation of the Signature motif helix (Fig. 4E) with respect to the Walker motifs. This rotation is the result of Q142 (Q-loop) binding to Mg2+, inducing a conformation within the NBDs that enables tight NBD-NBD engagement. As a consequence, the coiled-coil and the interacting HLH domain of Mre11 undergo a “rigid body” rotation by approx. 50° with respect to ATP binding Lobe I.
Formation of the engaged NBDs strongly affects the angle between the two coiled-coils protruding from the DNA binding catalytic head, consistent with scanning force microscopy of human MRN (Moreno-Herrero et al., 2005), where DNA binding was shown to alter the angle between two coiled-coils of about 60°. Comparing the angle between coiled-coils in the “open” conformation (~120°) and the ATP-bound “closed” state (~60°), we also see a difference of about 60°. Our data therefore suggest that the identified clamp movement in the catalytic head is the molecular basis for the observed mesoscale movements of the MRN coiled-coils upon DNA binding. We envision that DNA binding to the capping domains of Mre11 help to displace the NBDs from interface 2, allowing them to adopt a closed conformation in the presence of ATP.
Rad50 induces a conformational change in the Mre11 dimer
The NBDs within a single MR catalytic head undergo an engagement-disengagement cycle similar to that seen in ABC transporter (Hollenstein et al., 2007; Oldham et al., 2008). In ABC transporter, ATP dependent NBD engagement drives conformational changes in transmembrane domains (TMD), a “powerstroke” that transports solutes across the membrane. To see whether Rad50 ATP binding can alter the Mre11 dimer, in analogy to the function of NBDs in ABC transporter, we tested the conformation of the Mre11 dimer using a crosslinking approach. In support of such a function, the “angles” between Mre11 protomers in T. maritima and P. furiosus Mre11 dimers are quite different and the TmMre11 dimer needs to undergo a pivoting rotation to be able to bind DNA similar to the PfMre11 nuclease dimer (Fig. 5A) (Williams et al., 2008).
Figure 5. ATP binding to Rad50 changes the Mre11 dimer interface.
A) Comparison of the nuclease dimer of DNA bound PfMre11 (grey, yellow, orange) with that of TmMre11 (blue, lightblue). Both complexes are superimposed via the left protomer. In T. maritima, the right protomer has to undergo a substantial rigid body motion to adopt the same DNA binding contacts (solid arrows) and conformation as seen for PfMre11 (dashed arrow). This can be seen by the different orientations of the minor groove binding helix (dashed rectangles).
B) To probe for conformational changes, we mutated S110C in TmMre11 (left panel) and F102C in PfMre11 at a position that is expected to undergo movements when switching between different pivot angles. The distances are suitable for crosslinking with a short bifunctional sulfhydryl directed crosslinker (inset: BMOE), or by forming disulfide bonds.
C) Crosslinking analysis of TmMre11 dimer structure. Upper panel, non-reducing Coomassie stained SDS PAGE of TmMRNBD crosslinked under different conditions (lane labels see lower panel); Lower panel, quantification of the crosslinking efficiency (mean +/− standard deviation of three independent experiments) for different conditions and adenosine nucleotides. A crosslinked band corresponding to the Mre11 dimer in the absence of BMOE crosslinker indicates disulfide bond formation.
D) Coomassie stained non-reducing SDS PAGE of crosslinked PfMRNBD, showing different conditions and replicates. Cu2+ was used to increase disulfide bond formation.
E) and F) Quantification of crosslinking efficiency by BMOE (E) or disulfide bonds (F). Error bars depict standard deviations. In F) the effect of ATPγS is tested. In E) the effect of dsDNA 50mer with either blunt ends or 3′ or 5′ 10 dT ssDNA overhangs is shown. Black bars are without ATPγS, white bars with ATPγS.
We mutated Mre11 dimer interface residues S110 in TmMre11 and F102 in the PfMre11 to cysteines. The introduced cysteines in both protomers are separated by approx. 6–8 Å, close enough to be able to form a disulfide bond or be substrates for BMOE (8 Å) in a manner that might be sensitive to changes in the Mre11 dimer. Both mutants form M2R2NBD heterotetramers and otherwise behave like wild-type protein during purification and the Mre11 interface is not physically disrupted. Indeed, the cysteine engineered Mre11s could be efficiently crosslinked with either BMOE or by forming disulfide bonds (Fig. 5C–F), while the longer HBVS crosslinker did not efficiently crosslink Mre11 (data not shown).
ATP, AMPPNP and ATPγS reduced the ability of BMOE to crosslink Mre11 S110C dimers in MRNBD up to approx. 50% and the most plausible explanation is that the structure of the Mre11 dimer is altered such that the position of the opposing engineered cysteines is changed. As control, ADP did not reduce but seems to slightly increase crosslink efficiency as if stabilizing a different conformation than ATP. DNA did not largely impact on the crosslinking efficiency but perhaps DNA induced changes that are not sensitive to our method (Fig. 5E). To rule out crosslinker specific effects, we also tested the formation of a disulfide bond. While TmMRS110C formed disulfide bonds already during purification (Fig. 5C, lane 1), PfMre11F102C did not form spontaneous disulfide bonds but could be linked by the disulfide bond promoter Cu2+(Fig. 5D). Using Cu2+ we again tested the effect of ATPγS and found that disulfide bond formation is significantly reduced in the presence of ATPγS (Fig. 5F).
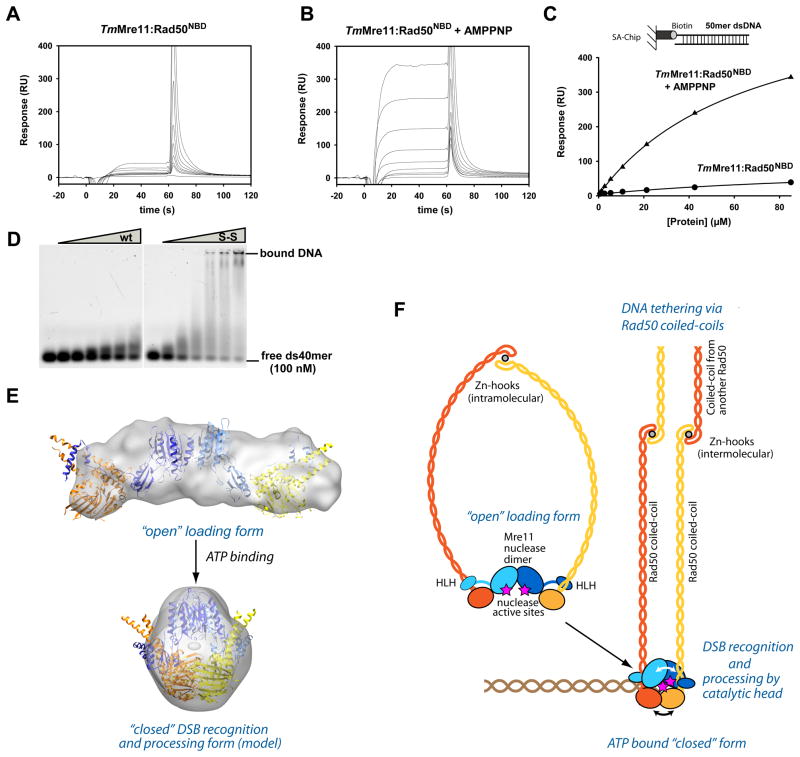
To test whether ATP leads to a structure with increased DNA binding affinity we used Surface Plasmon Resonance (SPR) and Electrophoretic Mobility Shift Assays (EMSAs). Indeed, SPR showed a strong effect of AMPPNP on dsDNA and hairpin DNA binding by TmMre11:Rad50NBD (Fig. 6A–C, Fig. S6A). Interestingly, the disulfide bonded TmMre11:Rad50NBD,H830,D804C,F291S complex showed even higher DNA affinity in the presence of AMPPNP but we also observe some nonspecific interaction with the matrix, making the interpretation difficult (data not shown). For that reason we compared TmMre11:Rad50NBD and TmMre11:Rad50NBD,H830,D804C,F291S also in EMSA and find a strongly increased binding of dsDNA oligonucleotides even in the absence of AMPPNP (Fig. 6D). An EMSA using double-stranded plasmid φX174 RF II DNA verified the increased DNA binding of the MRNBD complex in the presence of AMPPNP and of the disulfide bridged closed protein complex (S-S) (Fig. S6B). These data suggests that the engaged, clamp like form is the DNA binding conformation.
Figure 6. ATP stimulated dsDNA binding and clamp model for DSB sensing.
A) and B) Surface Plasmon sensograms for binding of TmMre11:Rad50NBD to 50mer dsDNA in absence (A) and presence of AMPPNP (B).
C) Corresponding binding curves reveals AMPPNP stimulated binding to dsDNA. AMPPNP also increases hairpin recognition (Fig. S6A) and we also observe increased binding affinity of the protein with crosslinked Rad50 NBDs (ATP mimic state) (data not shown).
D) Electrophoretic mobility shift assays show that the MRNBD,H830C,D804C complex with a disulfide bond between the NBDs (S-S) has strongly increased dsDNA oligonucleotide binding affinity compared to the wildtype MRNBD complex (wt). Following concentrations of the protein (0, 1.0, 2.5, 5.0, 7.5, 10.0 and 15.0 μM respectively) were used.
E) Model for the closed, clamp like complex with engaged NBDs by combining X-ray structures and SAXS analysis. “Open” (experimental) and “closed” (rigid body docked model) forms are displayed with corresponding SAXS envelopes.
F) Proposed model for ATP dependent DSB sensing and processing by MR by formation of a transient clamp at DNA end structures.
See also Figure S5.
Implications for DSB detection and ABC enzyme architecture
We provide here a structural framework for the architecture and ATP dependent conformational changes of the bacterial MR complex. In the absence of ATP, the NBDs are widely separated, while ATP promotes a DNA binding conformation by stabilizing an engaged form of the NBDs. Using SAXS data of the open and the 65% crosslinked form we can generate ab initio reconstructions as well as rigid body docked models of the ATP bound, clamp like structure (Fig. 6E, Fig. S6C–G and Supplemental Data for the SAXS analysis and modeling). The model is consistent with all crosslinking data as well as the high- and low-resolution structural data presented here. The structure draws interesting parallels to the Ku DNA end sensor, which forms a constitutive ring. This ring form makes Ku specific for DNA ends and prevents internal DNA binding (Walker et al., 2001). Other DNA associated rings like PCNA or helicases are associated with ATP dependent loading factors to allow encircling of DNA (Bowman et al., 2004). Our data suggest that MR forms a transient clamp, controlled by ATP binding. A proposed mechanism for DSB sensing is shown in figure 6F. DNA binding and nuclease active sites are likely located in the central hole of the clamp, which might provide specificity for DNA end or hairpin structures, as in the case of Ku, but also allow controlled and limited DNA end processing by Mre11. In striking contrast to the constitutive ring shape of Ku, the open conformation of MR could explain how the complex can still bind to meiotic breaks with covalently attached Spo11.
The precise interaction of MR with DNA ends remains to be seen, but we were unable to detect crosslinked MR around plasmid DNA (Fig. S3A–F). Additionally, we find that the crosslinked form can still bind to DNA ends that are blocked on both 5′ ends by a fluoresceine specific single chain Fab antibody fragment (data not shown). It appears that the closed ATP bound complex does not entrap dsDNA like e.g. the PCNA ring and that ends are not simply recognized by a topological ring. This does not exclude the possibility that the open form of the MR complex can still bind to internal stretches of DNA. Indeed a partially open form of the complex could bind to the DNA, diffusing along it to rapidly identify DSBs, where the NBD heads could fully engage in the presence of ATP to more strongly bind to DNA, and perhaps melt and process ends (Fig. 6).
Our finding, that ATP binding to Rad50 alters the crosslink sensitivity at the Mre11 dimer interface (Fig. 5C–F) and that the ATP bound and/or crosslinked form of MR binds DNA much more tightly than the “open” form, draws an interesting parallel to the mechanism of the related ABC transporter ATPases. The possible explanation is that Rad50 promotes a structural change between the Mre11 protomers to induce a conformation with increased DNA binding activity, but Rad50 contributes to DNA binding also directly in the presence of ATP (Hopfner et al., 2000; Raymond and Kleckner, 1993). Structural changes in Mre11 are appealing because it may provide not only a model how ATP helps to trigger nuclease or DNA melting activities of MR, but also unify the functional architecture of MR with ABC transporters, where the conformational changes between the NBDs trigger conformational changes in TMDs to transport solutes (Hollenstein et al., 2007).
Biochemically it has been found that some nuclease functions such as hairpin opening and ssDNA endonuclease activity do not require ATP, while others such as endonucleolytic cleavage of blocked DNA, dsDNA exonuclease and 5′ dsDNA endonuclease require ATP (Paull and Gellert, 1998; Trujillo and Sung, 2001). It can be argued that latter activities might require partial melting of the DNA duplex to reach the active site of Mre11 and indeed MRN also possesses an ATP stimulated dsDNA melting activity (Paull and Gellert, 1999). Since Mre11 dimers cooperate to bind DNA and DNA is also bound across both Mre11 protomers (Williams et al., 2008), a structural modulation in the Mre11 dimer would be an ideal mechanism to provide the dsDNA melting activity and position DNA into the active site for endo/exonucleolytic cleavage.
In summary, we provide a first structural framework for the architecture of the Mre11-Rad50 catalytic head module and reveal the ATP dependent clamp formation responsible for DSB recognition and processing.
Experimental Procedures
Proteins
Rad50NBD was engineered by fusing N-terminal (residues 1–190) and C-terminal (residues 686–852) segments by an 8 amino acid linker (GGAGGAGG) in a single open reading frame, and coexpressed with Mre11 (residues L7M-385). For crystallization of the AMPPNP Rad50 NBD dimer, Rad50NBD was co-purified with TmMre11 HLH (residues 343–385). Protein synthesis and purification procedures are provided in Supplemental Data.
Crystallization and structure determination of the Mre11:Rad50NBD complex
Crystals of TmMRNBD were grown after mixing 2 μl of protein solution at 9.6 mg/ml protein concentration with 2 μl of the reservoir solution containing 9% (w/v) PEG-6000, 5% (v/v) MPD, 1 mM TCEP and 0.1 M HEPES pH 7.9. Prior to flash freezing in liquid nitrogen, crystals were transferred to reservoir solution supplemented with 10% (v/v) 2-methyl-2,4-pentanediole. Anomalous data to 3.4 Å were collected at the X06SA beamline (Swiss Light Source) on selenium-containing crystals. Details and statistics of data analysis and model building are provided in Supplemental Data.
Crystallization and structure determination of the Mre11HLH:Rad50NBD:AMPPNP complex
Crystals of TmMre11HLH(aa 343–385):Rad50NBD were grown by mixing 200 nl of protein solution at 10.5 mg/ml protein concentration with 200 nl of the reservoir solution containing 20% (w/v) PEG-2000 MME, 0.2 M Trimethylamine N-oxide and 0.1 M Tris pH 8.5. Prior to flash freezing in liquid nitrogen, crystals were transferred to reservoir solution supplemented with 10% (v/v) 2,3-butandiol. Data to 1.9 Å were collected at the ID14-1 (European Synchrotron Radiation Facility). Details and statistics of data analysis and model building are provided in Supplemental Data.
Small angle X-ray scattering
SAXS data were acquired at the EMBL X33 beamline (Deutsches Elektronensynchrotron) using a MAR345 two-dimensional image plate detector and at beamline BL12.3.1 (Advanced Light Source at Lawrence Berkeley National Laboratories). Scattering patterns were collected from solutions of TmMRNBD in 50 mM Tris pH 7.7, 100 mM NaCl, 5 mM MnCl2 and 10 mM MgCl2 at concentrations between 2 and 30 mg/ml. For details, see Supplemental Data.
Crosslinking analysis
Rad50 crosslinking reactions were performed either with MRNBD,P31C,E806C and HBVS (Pierce), MRNBD,N64C,I760C and BMOE (Pierce) or by formation of disulfide bonds (MRNBD,H830,D804C,(F291S)). Crosslinking of the Mre11:Mre11 dimer was performed either by BMOE (Pierce) (PfMre11F102C, TmMre11S110C) or by formation of disulfide bonds of Cu2+ (PfMre11F102C). Details are provided in Supplemental Data.
Cloning, yeast manipulation and MMS sensitivity assays
The MRE11 shuffle strain (Mat a; his3Δ1; leu2Δ0; ura3Δ0; YMR224c::kanMX4, pRS316-MRE11) was generated by transformation of the MRE11/Δmre11 heterozygous knockout strain (Euroscarf) with pRS316-MRE11. Point mutations of MRE11 were generated by quickchange site directed mutagenesis of pRS313-MRE11. Plate survival assays of the mre11 mutant strains were assessed by spotting 10-fold serial dilutions on SDC(–his) plates and SDC(–his) plates containing 0.005% MMS, 50 mM hydroxyurea or 1 μg/ml camptothecin. For detailed information see Supplemental Data.
DNA binding analysis
DNA binding reactions were carried out by Surface Plasmon Resonance and Electrophoretic Mobility Shift Assay. For detailed information see Supplemental Data.
Supplementary Material
Acknowledgments
We thank John Petrini for his gift of an α-Mre11 antibody. We thank Britta Coordes for help with the yeast work and members of the Hopfner lab, especially Matthew Bennett, Gregor Witte and Alfred Lammens for discussions. We thank the staffs of the Swiss Light Source (Villingen), European Synchrotron Radiation Facility (Grenoble), German Electron Synchrotron (Hamburg) and Advanced Light Source (Berkeley) for technical support. We thank the Max-Planck-Crystallization Facility (Martinsried) for crystallization trials. This work was funded by grants from the German Research Council (SFBs 684, 646 and TR5), the German Excellence Initiative (CIPSM), European Commission (IP DNA repair), and NIH U19AI83025.
Footnotes
Accession numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3QG5 for the Mre11:Rad50NBD complex and 3QF7 for the Mre11HLH:Rad50NBD:AMPPNP complex.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bentchikou E, Servant P, Coste G, Sommer S. Additive effects of SbcCD and PolX deficiencies in the in vivo repair of DNA double-strand breaks in Deinococcus radiodurans. J Bacteriol. 2007;189:4784–4790. doi: 10.1128/JB.00452-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Chamankhah M, Xiao W. Formation of the yeast Mre11-Rad50-Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res. 1999;27:2072–2079. doi: 10.1093/nar/27.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst) 2003;2:795–807. doi: 10.1016/s1568-7864(03)00063-6. [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci U S A. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- Eykelenboom JK, Blackwood JK, Okely E, Leach DR. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- Leach DR, Okely EA, Pinder DJ. Repair by recombination of DNA containing a palindromic sequence. Mol Microbiol. 1997;26:597–606. doi: 10.1046/j.1365-2958.1997.6071957.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J, Sanchez H, Tadesse S, Kidane D, Krisnamurthy M, Alonso JC, Graumann PL. Bacillus subtilis SbcC protein plays an important role in DNA inter-strand cross-link repair. BMC Mol Biol. 2006;7:20. doi: 10.1186/1471-2199-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr Opin Struct Biol. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond WE, Kleckner N. RAD50 protein of S.cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Zahradka K, Buljubasic M, Petranovic M, Zahradka D. Roles of ExoI and SbcCD nucleases in “reckless” DNA degradation in recA mutants of Escherichia coli. J Bacteriol. 2009;191:1677–1687. doi: 10.1128/JB.01877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.