Abstract
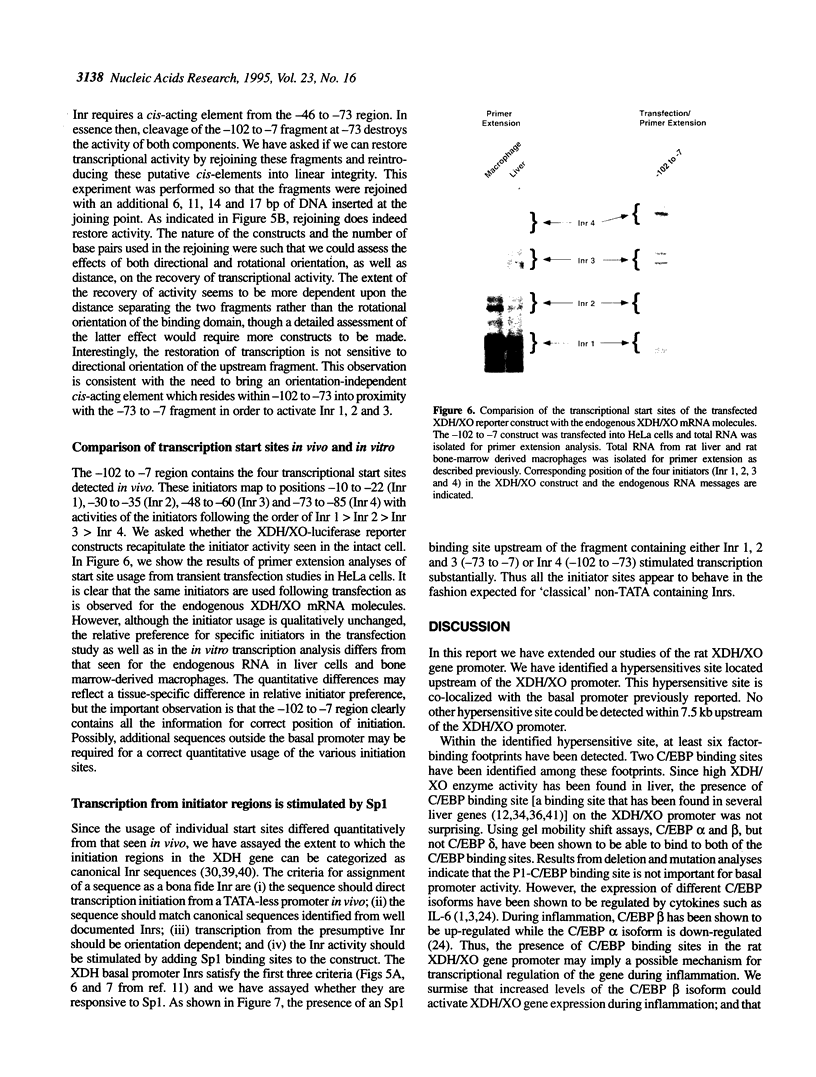
In the present study, we have explored further the organization of the TATA-less rat xanthine dehydrogenase/oxidase gene (XDH/XO). A DNase I hypersensitive site has been identified which it colocalizes with the basal promoter reported previously [Chow et al. (1994) Nucleic Acids Res., 22, 1846-1854]. Gel mobility shift assays indicate the presence of multiple binding factors located in the promoter. At least six footprints were detected of which two have been shown to be C/EBP binding sites. Members of the C/EBP-alpha and C/EBP-beta, but not C/EBP-delta, family are able to bind to these two sites. Deletional and mutational studies revealed that C/EBP binding is not essential for the basal level of transcription initiation of this promoter. Much of the transcriptional activity resides in the -102 to -7 DNA fragment, which contains all initiator activity which acts unidirectionally. Within this fragment, four putative initiator elements could be identified; interestingly, the linear integrity of these initiators is important for efficient transcription of the XDH/XO gene. Separation of the initiators leads to a complete loss of transcription activity; however, this loss could be partially restored by the introduction of an Sp1 binding site upstream of the separated initiators. Despite a difference in usage/frequency of initiation at the various initiators, primer extension analyses reveal similar positions for transcription initiations in both XDH/XO reporter constructs and in the endogenous XDH/XO gene. The differential usage of initiators may imply a possible post-transcriptional regulation for the XDH/XO gene.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alam T., An M. R., Papaconstantinou J. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J Biol Chem. 1992 Mar 15;267(8):5021–5024. [PubMed] [Google Scholar]
- Beckman J. S., Parks D. A., Pearson J. D., Marshall P. A., Freeman B. A. A sensitive fluorometric assay for measuring xanthine dehydrogenase and oxidase in tissues. Free Radic Biol Med. 1989;6(6):607–615. doi: 10.1016/0891-5849(89)90068-3. [DOI] [PubMed] [Google Scholar]
- Birkenmeier E. H., Gwynn B., Howard S., Jerry J., Gordon J. I., Landschulz W. H., McKnight S. L. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989 Aug;3(8):1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- Bonifer C., Hecht A., Saueressig H., Winter D. M., Sippel A. E. Dynamic chromatin: the regulatory domain organization of eukaryotic gene loci. J Cell Biochem. 1991 Oct;47(2):99–108. doi: 10.1002/jcb.240470203. [DOI] [PubMed] [Google Scholar]
- Brass C. A., Narciso J., Gollan J. L. Enhanced activity of the free radical producing enzyme xanthine oxidase in hypoxic rat liver. Regulation and pathophysiologic significance. J Clin Invest. 1991 Feb;87(2):424–431. doi: 10.1172/JCI115013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Umek R. M., McKnight S. L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991 Sep;5(9):1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Cheyette T. E., Ip T., Faber S., Matsui Y., Chalkley R. Characterization of the factors binding to a PEPCK gene upstream hypersensitive site with LCR activity. Nucleic Acids Res. 1992 Jul 11;20(13):3427–3433. doi: 10.1093/nar/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Chow C. W., Clark M., Rinaldo J., Chalkley R. Identification of the rat xanthine dehydrogenase/oxidase promoter. Nucleic Acids Res. 1994 May 25;22(10):1846–1854. doi: 10.1093/nar/22.10.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descombes P., Chojkier M., Lichtsteiner S., Falvey E., Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990 Sep;4(9):1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- Dupont G. P., Huecksteadt T. P., Marshall B. C., Ryan U. S., Michael J. R., Hoidal J. R. Regulation of xanthine dehydrogenase and xanthine oxidase activity and gene expression in cultured rat pulmonary endothelial cells. J Clin Invest. 1992 Jan;89(1):197–202. doi: 10.1172/JCI115563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciani F., Ghezzi P., Terao M., Cazzaniga G., Garattini E. Interferons induce xanthine dehydrogenase gene expression in L929 cells. Biochem J. 1992 Aug 1;285(Pt 3):1001–1008. doi: 10.1042/bj2851001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl H. P., Till G. O., Ryan U. S., Ward P. A. Mediator-induced activation of xanthine oxidase in endothelial cells. FASEB J. 1989 Nov;3(13):2512–2518. doi: 10.1096/fasebj.3.13.2806779. [DOI] [PubMed] [Google Scholar]
- Gorski K., Carneiro M., Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986 Dec 5;47(5):767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hassoun P. M., Yu F. S., Shedd A. L., Zulueta J. J., Thannickal V. J., Lanzillo J. J., Fanburg B. L. Regulation of endothelial cell xanthine dehydrogenase xanthine oxidase gene expression by oxygen tension. Am J Physiol. 1994 Feb;266(2 Pt 1):L163–L171. doi: 10.1152/ajplung.1994.266.2.L163. [DOI] [PubMed] [Google Scholar]
- Hille R., Massey V. Studies on the oxidative half-reaction of xanthine oxidase. J Biol Chem. 1981 Sep 10;256(17):9090–9095. [PubMed] [Google Scholar]
- Houde M., Tiveron M. C., Brégégère F. Divergence of the nucleotide sequences encoding xanthine dehydrogenase in Calliphora vicina and Drosophila melanogaster. Gene. 1989 Dec 28;85(2):391–402. doi: 10.1016/0378-1119(89)90432-0. [DOI] [PubMed] [Google Scholar]
- Ichida K., Amaya Y., Noda K., Minoshima S., Hosoya T., Sakai O., Shimizu N., Nishino T. Cloning of the cDNA encoding human xanthine dehydrogenase (oxidase): structural analysis of the protein and chromosomal location of the gene. Gene. 1993 Nov 15;133(2):279–284. doi: 10.1016/0378-1119(93)90652-j. [DOI] [PubMed] [Google Scholar]
- Isshiki H., Akira S., Sugita T., Nishio Y., Hashimoto S., Pawlowski T., Suematsu S., Kishimoto T. Reciprocal expression of NF-IL6 and C/EBP in hepatocytes: possible involvement of NF-IL6 in acute phase protein gene expression. New Biol. 1991 Jan;3(1):63–70. [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- Juan T. S., Wilson D. R., Wilde M. D., Darlington G. J. Participation of the transcription factor C/EBP delta in the acute-phase regulation of the human gene for complement component C3. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2584–2588. doi: 10.1073/pnas.90.7.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnitz L., Poon D., Weil P. A., Chalkley R. Purification and properties of the Rous sarcoma virus internal enhancer binding factor. Mol Cell Biol. 1989 May;9(5):1929–1939. doi: 10.1128/mcb.9.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith T. P., Riley M. A., Kreitman M., Lewontin R. C., Curtis D., Chambers G. Sequence of the structural gene for xanthine dehydrogenase (rosy locus) in Drosophila melanogaster. Genetics. 1987 May;116(1):67–73. doi: 10.1093/genetics/116.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R. D., Harford J. B. cis-trans models for post-transcriptional gene regulation. Science. 1989 Nov 17;246(4932):870–872. doi: 10.1126/science.2683086. [DOI] [PubMed] [Google Scholar]
- Kollmar R., Farnham P. J. Site-specific initiation of transcription by RNA polymerase II. Proc Soc Exp Biol Med. 1993 Jun;203(2):127–139. doi: 10.3181/00379727-203-43583. [DOI] [PubMed] [Google Scholar]
- Kooij A., Bosch K. S., Frederiks W. M., Van Noorden C. J. High levels of xanthine oxidoreductase in rat endothelial, epithelial and connective tissue cells. A relation between localization and function? Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;62(3):143–150. doi: 10.1007/BF02899676. [DOI] [PubMed] [Google Scholar]
- Kooij A., Schijns M., Frederiks W. M., Van Noorden C. J., James J. Distribution of xanthine oxidoreductase activity in human tissues--a histochemical and biochemical study. Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;63(1):17–23. doi: 10.1007/BF02899240. [DOI] [PubMed] [Google Scholar]
- Kuppusamy P., Zweier J. L. Characterization of free radical generation by xanthine oxidase. Evidence for hydroxyl radical generation. J Biol Chem. 1989 Jun 15;264(17):9880–9884. [PubMed] [Google Scholar]
- Li X. X., Huang J. H., Rienhoff H. Y., Jr, Liao W. S. Two adjacent C/EBP-binding sequences that participate in the cell-specific expression of the mouse serum amyloid A3 gene. Mol Cell Biol. 1990 Dec;10(12):6624–6631. doi: 10.1128/mcb.10.12.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord J. M. Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med. 1985 Jan 17;312(3):159–163. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- Parks D. A., Granger D. N. Xanthine oxidase: biochemistry, distribution and physiology. Acta Physiol Scand Suppl. 1986;548:87–99. [PubMed] [Google Scholar]
- Rinaldo J. E., Gorry M. Protection by deferoxamine from endothelial injury: a possible link with inhibition of intracellular xanthine oxidase. Am J Respir Cell Mol Biol. 1990 Dec;3(6):525–533. doi: 10.1165/ajrcmb/3.6.525. [DOI] [PubMed] [Google Scholar]
- Sears R. C., Sealy L. Characterization of nuclear proteins that bind the EFII enhancer sequence in the Rous sarcoma virus long terminal repeat. J Virol. 1992 Nov;66(11):6338–6352. doi: 10.1128/jvi.66.11.6338-6352.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S. T., Schmidt M. C., Berk A. J., Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson E. C., Conley P. B., Paulson J. C. Regulated expression of alpha 2,6-sialyltransferase by the liver-enriched transcription factors HNF-1, DBP, and LAP. J Biol Chem. 1992 Feb 15;267(5):3466–3472. [PubMed] [Google Scholar]
- Terao M., Cazzaniga G., Ghezzi P., Bianchi M., Falciani F., Perani P., Garattini E. Molecular cloning of a cDNA coding for mouse liver xanthine dehydrogenase. Regulation of its transcript by interferons in vivo. Biochem J. 1992 May 1;283(Pt 3):863–870. doi: 10.1042/bj2830863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil E. C. Regulation of ferritin and transferrin receptor mRNAs. J Biol Chem. 1990 Mar 25;265(9):4771–4774. [PubMed] [Google Scholar]
- Waud W. R., Rajagopalan K. V. The mechanism of conversion of rat liver xanthine dehydrogenase from an NAD+-dependent form (type D) to an O2-dependent form (type O). Arch Biochem Biophys. 1976 Feb;172(2):365–379. doi: 10.1016/0003-9861(76)90088-6. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- van Ooij C., Snyder R. C., Paeper B. W., Duester G. Temporal expression of the human alcohol dehydrogenase gene family during liver development correlates with differential promoter activation by hepatocyte nuclear factor 1, CCAAT/enhancer-binding protein alpha, liver activator protein, and D-element-binding protein. Mol Cell Biol. 1992 Jul;12(7):3023–3031. doi: 10.1128/mcb.12.7.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]