Abstract
Pacidamycins are a family of uridyl peptide antibiotics that inhibit the translocase MraY, an essential enzyme in bacterial cell wall biosynthesis not yet clinically targeted. The pacidamycin structural skeleton contains a doubly inverted peptidyl chain with a β-peptide and a ureido linkage, and a 3’-deoxyuridine nucleoside attached to DABA3 of the peptidyl chain via an enamide linkage. Although the biosynthetic gene cluster for pacidamycins was identified recently, the assembly line of this group of peptidyl nucleoside antibiotics remained poorly understood due to the highly dissociated nature of the encoded nonribosomal peptide synthetase (NRPS) domains and modules. In this work, a minimum set of enzymes were identified in generating the pacidamycin scaffold from amino acid and nucleoside monomers, highlighting a freestanding thiolation (T) domain (PacH) as a key carrier component in the peptidyl chain assembly, as well as a freestanding condensation (C) domain (PacI) catalyzing the release of the assembled peptide by a nucleoside moiety. Based on the substrate promiscuity of this enzymatic assembly line, several pacidamycin analogs were produced using in vitro total biosynthesis.
Pacidamycins are members of a large class of uridyl peptide antibiotics, which also includes mureidomycins, napsamycins and sansanmycins.1,2 They are sufficient structural mimics of UDP-N-acetylmuramoyl-pentapeptide intermediate in bacterial cell wall assembly to inhibit the translocase MraY, a clinically unexploited target in the development of new antibacterial drugs.3 Elucidation of the enzymatic mechanism for pacidamycin scaffold assembly will therefore facilitate the generation of new MraY-targeted peptidyl nucleoside antibiotics through combinatorial biosynthesis.
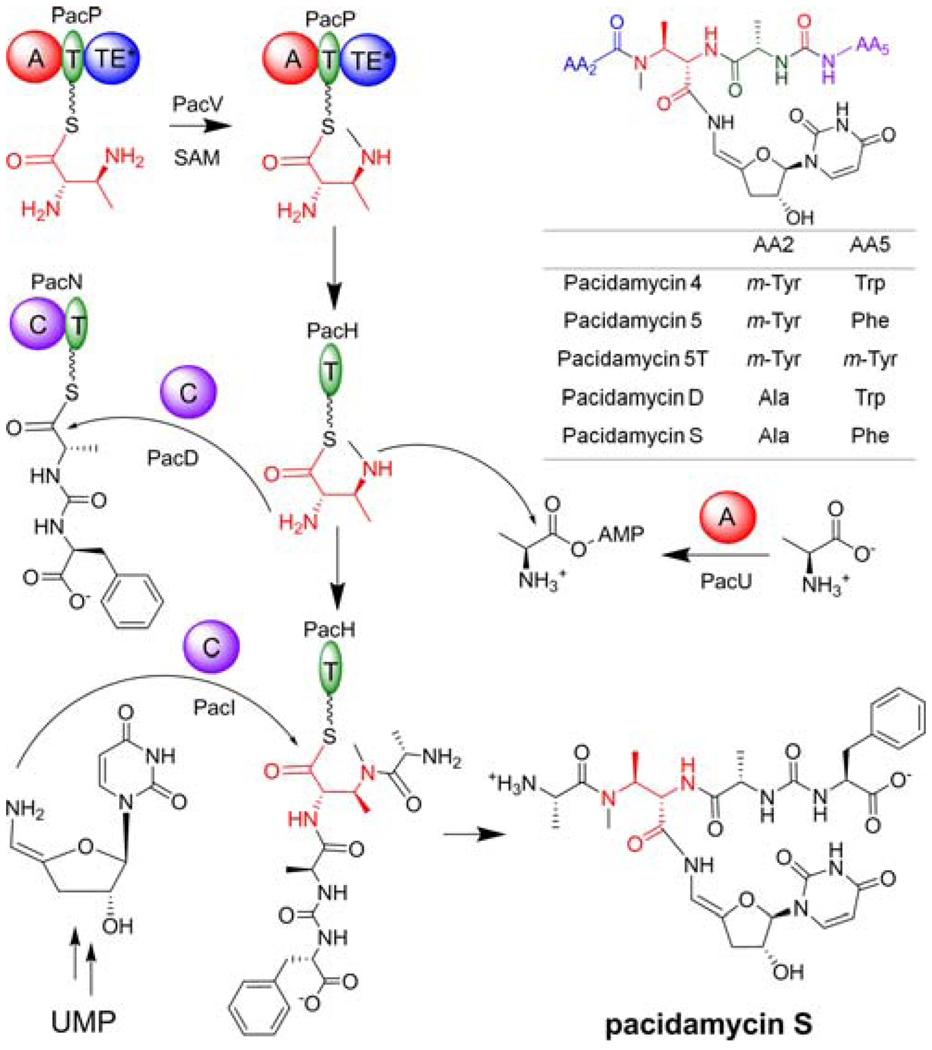
More than 10 related pacidamycin family compounds have been isolated from Streptomyces coeruleorubidus,1 all of which share a common structural skeleton with a 2S,3S-diaminobutyric acid (DABA) residue serving as a connection point for the 3’-deoxyuridine moiety via a 4’,5’-enamide linkage. A ureido dipeptide (Ala4-Phe/Trp/m-Tyr5) is attached to the α-amino of DABA at the C-terminus; a single amino acid (Ala/m-Tyr2), a bicyclic heterocycle or a dipeptide (Ala/Gly1-m-Tyr2) is linked to the β-amino of DABA at the N-terminus. Thus in the pacidamycin framework, the tetra/pentapeptidyl chain reverses direction twice at Ala/m-Tyr2-DABA3 and Ala4-Phe/Trp/m-Tyr5 via a β-peptide and a ureido linkage respectively (Figure 1). All of these nonstandard connectivities suggest novel chemical logic and enzymatic machinery for assembly of the pacidamycin scaffold.
Figure 1.
Structures of selected pacidamycins and proposed biosynthetic scheme for pacidamycin S. Domain notation: T, thiolation; A, adenylation; C, condensation; TE, thioesterase.
The biosynthetic gene cluster for pacidamycins was reported recently,4,5 allowing for the heterologous production of pacidamycin D/S, the uridyl tetrapeptides containing a single N-terminal Ala tethered to the β-amino of DABA (Figure 1). Highly dissociated NRPS modules were encoded, bringing a challenge in dissecting the peptidyl core assembly (Figure S1). We characterized the substrate specificities of several encoded adenylation (A) domains (PacLOP), and reconstituted the formation of C-terminal ureido dipeptide (Ala4-Phe/Trp/m-Tyr5) using PacJLNO.5 In this work, the functions of six additional proteins (PacDHIUVW) are defined by in vitro characterization, and a picture of the complete assembly line is provided in generating the unusual uridyl tetrapeptide scaffold.
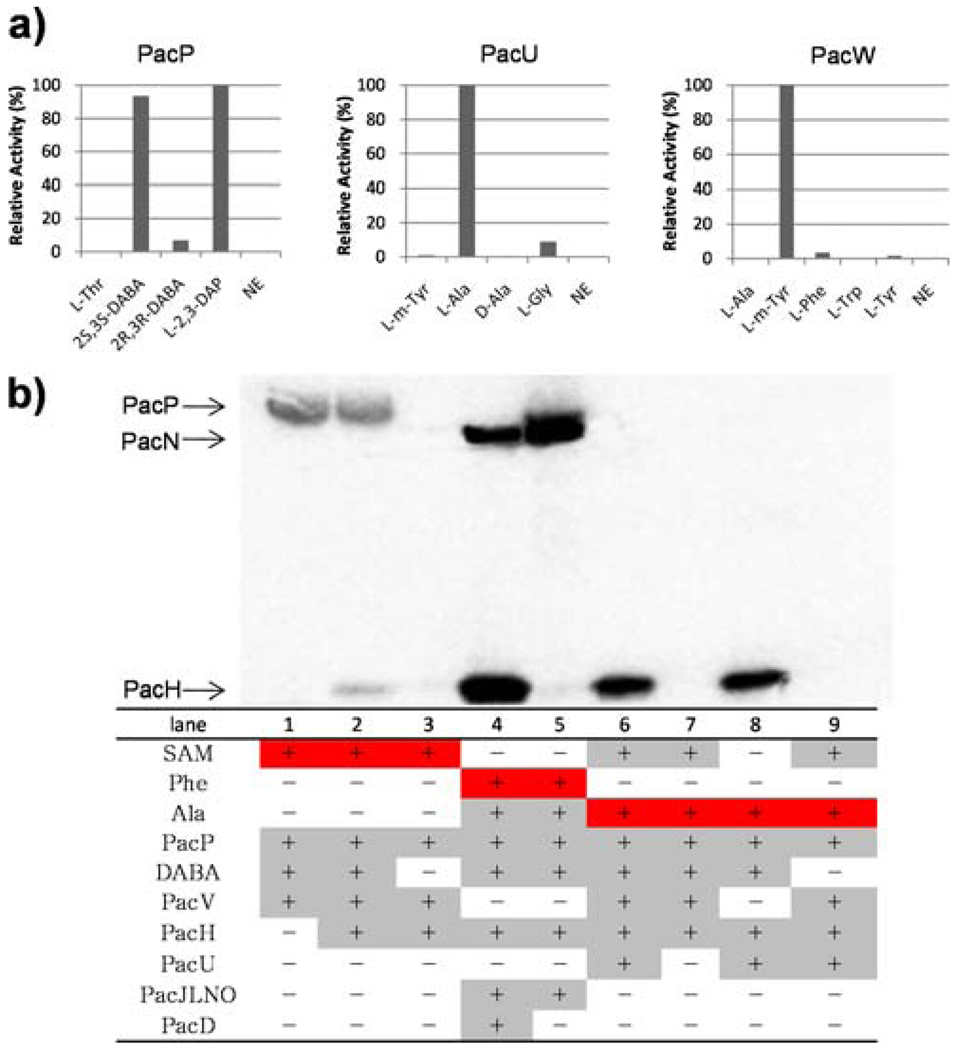
PacP has previously demonstrated a strong preference for the activation of l-2,3-diaminopropionate (DAP), indicating that it would be the DABA activation enzyme.5 Therefore 2S,3S- and 2R,3R-DABA were chemically synthesized based on the published methods6 (Figure S2) and tested in the ATP-[32P]PPi exchange assay. As expected, PacP exhibited a preference toward reversible formation of 2S,3S-DABA-AMP; no substantial activation of 2R,3R-DABA was detected, validating the stereoselectivity of PacP (Figure 2a). PacV, a stand-alone methyltransferase encoded by the gene cluster, was predicted to catalyze the N-methylation of DABA. The purified PacV indeed transferred the methyl group from S-adenosylmethionine (SAM) to holo-PacP (~90kD) as demonstrated by SDS-PAGE autoradiography, only after DABA had been activated by the A domain of PacP and presumably loaded onto the in cis thiolation (T) domain (Figure 2b). No methylation of free DABA/DAP catalyzed by PacV could be detected by LC-MS, and the preincubation of DABA/DAP with PacP significantly increased the initial rate of methylation (Figure S3), indicating that PacP-tethered DABA could be the natural substrate for the SAM-dependent methyltransferase PacV to install a methyl moiety on the β-amino of DABA.
Figure 2.
Characterization of NRPSs. a) A domain activities of PacP, PacU and PacW. NE, no enzyme; DABA, diaminobutyric acid; DAP, diaminopropionate; m-Tyr, meta-Tyrosine (DL). One hundred percent relative activity for l-2,3-DAP, l-Ala and m-Tyr-dependent exchange corresponds to 282k, 70k and 141k cpm respectively. b) Autoradiograph of one SDS-PAGE gel illustrating the covalent loading of 14C-labeled substrate (highlighted in red). Enzymes and substrates used in each lane are indicated in the table (highlighted in grey).
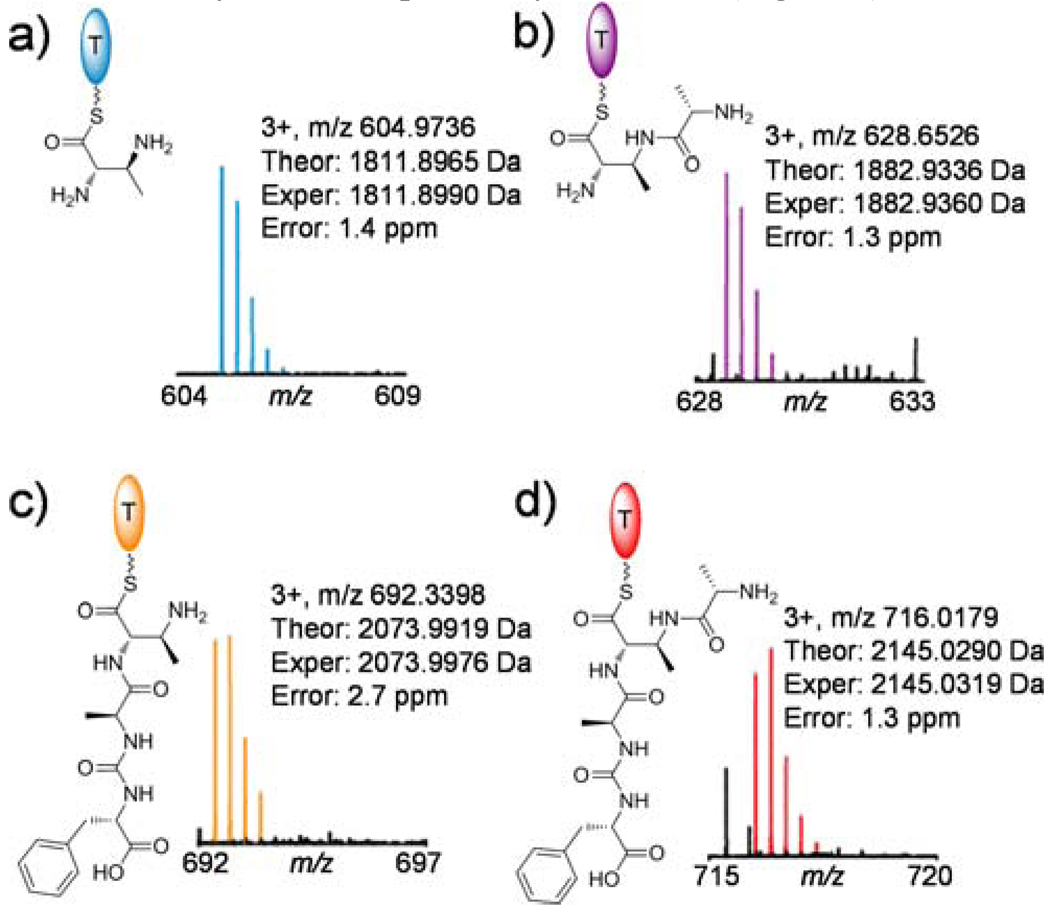
Upon the addition of a freestanding holo-T protein (PacH, ~10kD), the radioactive [14C]-CH3 derived from SAM was transferred from PacP to PacH (Figure 2b). The PacH homolog from S. roseosporus (88% similarity) was also purified in holoform, exhibited higher loading efficiency, and was used throughout subsequent studies. Transfer of aminoacyl groups between T domains to facilitate subsequent tailoring has been observed before, promoted by an acyltransferase of the α,β-hydrolase family, such as CmaE in coronamic acid biosynthesis.7 However, the thioesterase (TE) domain of PacP lacks the typical catalytic triad of this family, and no obvious in trans acyltransferase candidate could be found encoded in the gene cluster; the aminoacyl moiety may be shuttled between the HS-pantetheinyl prosthetic groups of PacP and PacH by spontaneous transthiolation. In addition, transfer of the DABA moiety was not dependent on the N-methylation, as DABA-S-PacH could be formed and detected by FTMS (Figure 3a).
Figure 3.
Detection of PacH-bound biosynthetic intermediates by FTMS after trypsin digestion. ATP, l-Phe, l-Ala, DABA, PacPH were present in all assays. PacU was present in assays b) and d), while PacDJLNO were present in assays c) and d). Theoretical mss calculations are shown in Figure S4.
We then probed the mechanism for attachment of the N-terminal Ala to the β-amino of thioester-tethered DABA in pacidamycin D/S biosynthesis. PacU, a stand-alone A domain protein, was indicated to be specifically related to the activation of N-terminal Ala by targeted gene disruption, albeit it failed to activate l-Ala in vitro in our initial trials.5 PacU was later repurified from E. coli, and exhibited a strong preference toward reversible formation of l-Ala-AMP (Figure 2a), correlating well with the in vivo PacU disruption result. [14C]l-Ala was further used in the loading assays to trace the covalent aminoacyl-S-thiolation intermediate. Radioactivity migrated with PacH but not PacP, and was completely dependent on the presence of PacU and DABA (Figure 2b). These results suggested that Ala was condensed to DABA-S-PacH but not DABA-S-PacP, after activation as Ala-AMP by PacU. The radioactivity signal remained the same in the absence of methyltransferase PacV and SAM, indicating that N-methylation of DABA is not a prerequisite for the aminoacylation of DABA-S-PacH by Ala. Indeed, the formation of Ala2-DABA3-S-PacH was confirmed by FTMS (Figure 3b). PacV and SAM were then excluded in the following assays to simplify the in vitro reconstitution system. It is notable that we also identified a di-gene cassette (designated pacWX) on the genome of S. coeruleorubidus (GenBank accession no. HQ874646) encoding a stand-alone A domain protein and a phenylalanine hydroxylase respectively. PacW shows high sequence similarity to PacU (87% similarity), and when purified demonstrated a strong preference for the activation of m-Tyr over other aromatic amino acids (Figure 2a). PacW is therefore hypothesized to be involved in the activation of N-terminal m-Tyr found in pacidamycin 4/5/5T (Figure 1).
The enzymatic mechanism and timing of the incorporation of ureido dipeptide onto the α-amino of DABA was also investigated using SDS-PAGE autoradiography and FTMS. Our prior work has shown that ureido dipeptide (Ala4-Phe/Trp/m-Tyr5) could be formed and tethered on the pantetheinyl arm of PacN upon the activity of PacJLNO.5 When PacN-S-Ala4-CO-[14C]Phe5 and DABA-S-PacH were combined, 14C-labeled l-Phe was transferred from PacN (~63kD) to PacH, only in the presence of a stand-alone condensation (C) domain protein PacD (Figure 2b). The PacH-S-DABA3-Ala4-CO-Phe5 species was further detected by FTMS (Figure 3c), confirming that PacD catalyzes the nucleophilic attack of the α-amino of DABA on the PacN tethered thioester of the C-terminal ureido dipeptide. No radioactive l-Phe was transferred to DABA-S-PacP, suggesting that PacH, not PacP, was one of the cognate loading domains for PacD recognition. It could also be deduced that the aminoacylation of α- and β-amino of DABA could take place independently.
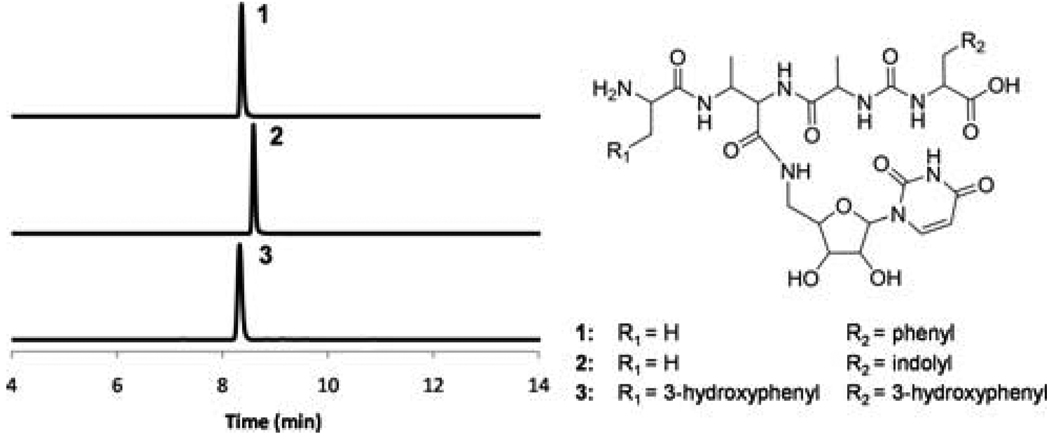
The tetrapeptidyl thioester of PacH (Figure 3d) was stable under incubation conditions, presumably awaiting offloading to the 5’-amino of a deoxyuridine species to generate pacidamycins. PacI, the remaining stand-alone C domain protein (~47kD), was anticipated to release the tetrapeptidyl chain from thioester linkage on PacH by a uridine derivative. Because 3’-deoxy-4’,5’-enamino uridine was not available, 5’-aminouridine was chemically synthesized8 and tested as a surrogate substrate in the in vitro assays to probe the function of PacI. In fact, PacI catalyzed the offloading of tetrapeptidyl chain from PacH by 5’-aminouridine via an amide linkage (Figure 4, S6–8, S19). PacI shows low sequence similarity to any other known proteins, and is the first identified C domain protein shown to condense peptide and nucleoside substrates. In addition, PacI could also utilize uridine and 3’-deoxy-uridine as substrate alternatives, forming an oxoester rather than the natural amide linkage (Figure 5, S9–18). The relaxed substrate specificities of PacI together with other catalytic domains in the assembly line could facilitate the enzymatic synthesis of pacidamycin analogs. For example, nine analogs (1–2, 5–8, 10–12) varied at the C-terminal aromatic amino acid, central diamino acid and uridine moieties were generated in vitro using nine proteins PacDHIJLNOPU (Figure 4 and 5). In addition, when PacU was replaced by PacW, only products with N-terminal m-Tyr (3–4, 9) were formed (Figure 4 and 5), confirming the orthogonal functions of PacU and PacW in the activation of N-terminal Ala and m-Tyr respectively.
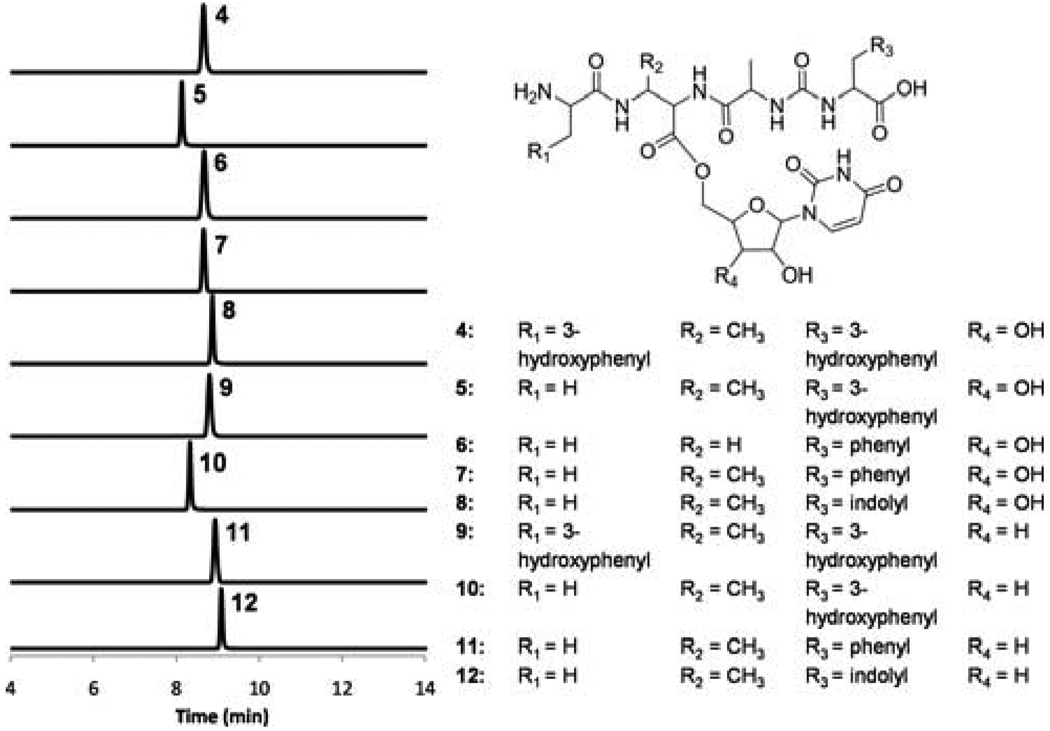
Figure 4.
Extracted ion chromatograms showing production of pacidamycin analogs using 5’-aminouridine as an offloading substrate. The calculated mass with 10-ppm mass error tolerance was used. The assay components are shown in Table S2; the HR-MS and HR-MS/MS analyses of each compound are shown in Figure S6–8. The MS/MS fragments confirmed the attachment of uridine moiety to DABA3 carbonyl through an amide linkage, as well as the regiospecific modifications of DABA3 amino (Nα vs. Nβ).
Figure 5.
Extracted ion chromatograms showing production of pacidamycin ester analogs. The calculated mass with 10-ppm mass error tolerance was used. The assay components are shown in Table S2; the HR-MS and HR-MS/MS analyses of each compound are shown in Figure S9–18.
In summary, we have dissected the distributed ten protein assembly line that builds the scaffold of tetrapeptidyl nucleoside pacidamycin antibiotics. The trifunctional 2S,3S-DABA is the central building block, activated by and loaded onto PacP, methylated by PacV, and transferred to PacH. The tetrapeptide framework was then assembled on PacH, perhaps reflecting the participation of that 10kD T domain protein scaffold in recognition by the downstream enzymes. The assembly included attachment of the N-terminal Ala (activated by PacU) or m-Tyr (activated by PacW) to the β-amino of DABA, and attachment of the C-terminal ureido dipeptide (formed by PacJLNO) to the α-amino of DABA promoted by PacD. Finally, PacI catalyzed the release of tetrapeptidyl intermediate from PacH by uridines (Figure 1 and S20). Our work provides the basis for rational reprogramming of various uridyl peptide antibiotics assembly lines, including pacidamycins, mureidomycins and napsamycins, as the recent identified biosynthetic gene cluster for napsamycins encodes the highly homologous discrete NRPSs.9 We will next scale up the in vitro total biosynthesis of various pacidamycin analogs to obtain material to test for antibiotic activities and evaluate structure-activity relationships.
Supplementary Material
Acknowledgement
This work was supported by NIH Grant GM49338 (C.T.W.), GM 067725-08 (N. L. K.) and GM066174 (D. K.).
Footnotes
Supporting Information Available: Experimental procedures, SDS-PAGE analysis of purified proteins, synthesis of DABA and aminouridine, methylation time course, MS calculation of PacH-bound biosynthetic intermediates, compound characterizations and biosynthetic pathway schemes. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Chen RH, Buko AM, Whittern DN, McAlpine JB. J. Antibiot. (Tokyo) 1989;42:512–520. doi: 10.7164/antibiotics.42.512. [DOI] [PubMed] [Google Scholar]; (b) Fronko RM, Lee JC, Galazzo JG, Chamberland S, Malouin F, Lee MD. J. Antibiot. (Tokyo) 2000;53:1405–1410. doi: 10.7164/antibiotics.53.1405. [DOI] [PubMed] [Google Scholar]
- 2.(a) Isono F, Inukai M, Takahashi S, Haneishi T, Kinoshita T, Kuwano H. J. Antibiot. (Tokyo) 1989;42:667–673. doi: 10.7164/antibiotics.42.667. [DOI] [PubMed] [Google Scholar]; (b) Chatterjee S, Nadkarni SR, Vijayakumar EK, Patel MV, Ganguli BN, Fehlhaber HW, Vertesy L. J. Antibiot. (Tokyo) 1994;47:595–598. doi: 10.7164/antibiotics.47.595. [DOI] [PubMed] [Google Scholar]; (c) Xie Y, Chen R, Si S, Sun C, Xu H. J. Antibiot. (Tokyo) 2007;60:158–161. doi: 10.1038/ja.2007.16. [DOI] [PubMed] [Google Scholar]
- 3.(a) Winn M, Goss RJ, Kimura K, Bugg TD. Nat. Prod. Rep. 2010;27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]; (b) Boojamra CG, Lemoine RC, Lee JC, Leger R, Stein KA, Vernier NG, Magon A, Lomovskaya O, Martin PK, Chamberland S, Lee MD, Hecker SJ, Lee VJ. J Am Chem Soc. 2001;123:870–874. doi: 10.1021/ja003292c. [DOI] [PubMed] [Google Scholar]; (c) Boojamra CG, Lemoine RC, Blais J, Vernier NG, Stein KA, Magon A, Chamberland S, Hecker SJ, Lee VJ. Bioorg. Med. Chem. Lett. 2003;13:3305–3309. doi: 10.1016/s0960-894x(03)00682-6. [DOI] [PubMed] [Google Scholar]; (d) Sun D, Jones V, Carson EI, Lee RE, Scherman MS, McNeil MR. Bioorg. Med. Chem. Lett. 2007;17:6899–6904. doi: 10.1016/j.bmcl.2007.09.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rackham EJ, Gruschow S, Ragab AE, Dickens S, Goss RJ. Chembiochem. 2010;11:1700–1709. doi: 10.1002/cbic.201000200. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Ostash B, Walsh CT. Proc. Natl. Acad. Sci. U S A. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Bunnage ME, Burke AJ, Davies SG, Millican NL, Nicholson RL, Roberts PM, Smith AD. Org. Biomol. Chem. 2003;1:3708–3715. doi: 10.1039/b306936m. [DOI] [PubMed] [Google Scholar]; (b) Davies SG, Garrido NM, Kruchinin D, Ichihara O, Kotchie LJ, Price PD, Price Mortimer AJ, Russell AJ, Smith AD. Tet. Asymm. 2006;17:1793–1811. [Google Scholar]
- 7.Strieter ER, Vaillancourt FH, Walsh CT. Biochemistry. 2007;46:7549–7557. doi: 10.1021/bi700243h. [DOI] [PubMed] [Google Scholar]
- 8.Winans KA, Bertozzi CR. Chem. Biol. 2002;9:113–129. doi: 10.1016/s1074-5521(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 9.Kaysser L, Tang X, Wemakor E, Sedding K, Hennig S, Siebenberg S, Gust B. Chembiochem. 2011;12:477–487. doi: 10.1002/cbic.201000460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.