Abstract
Glutamate receptors are important potential drug targets for cognitive enhancement and the treatment of schizophrenia in part because they are the most prevalent excitatory neurotransmitter receptors in the vertebrate central nervous system. One approach to the application of therapeutic agents to the AMPA subtype of glutamate receptors is the use of allosteric modulators, which promote dimerization by binding to a dimer interface thereby reducing desensitization and deactivation. AMPA receptors exist in two alternatively spliced variants (flip and flop) that differ in desensitization and receptor activation profiles. Most of the structural information on modulators of the AMPA receptor target the flip subtype. We report here the crystal structure of the flop-selective allosteric modulator, PEPA, bound to the binding domains of the GluA2 and GluA3 flop isoforms of AMPA receptors. Specific hydrogen bonding patterns can explain the preference for the flop isoform. This includes a bidentate hydrogen bonding pattern between PEPA and N754 of the flop isoforms of GluA2 and GluA3 (the corresponding position in the flip isoform is S754). Comparison with other allosteric modulators provides a framework for the development of new allosteric modulators with preferences for either the flip or flop isoforms. In addition to interactions with N/S754, specific interactions of the sulfonamide with conserved residues in the binding site are characteristics of a number of allosteric modulators. These, in combination, with variable interactions with five subsites on the binding surface lead to different stoichiometries, orientations within the binding pockets, and functional outcomes.
Membrane receptors are the cell's gatekeepers, allowing chemical signals access to the cell's pathways. Through the binding of endogenous ligands, receptors identify relevant environmental cues and facilitate cell-cell communication. The regulation of membrane receptors has become an important goal of drug discovery efforts (1, 2). By targeting the physiological (orthosteric) ligand-binding site, agonists and antagonists control the function of membrane receptors. Unfortunately, exogenously induced agonist-activation at the orthosteric site can cause toxic effects from overstimulation. Allosteric modulator binding sites use a distinct avenue for altering the natural response of a receptor. The ability of some allosteric modulators to enhance receptor stimulation, while not actually providing the trigger for stimulation, is a clear advantage that conserves the endogenous signaling pathway. Being important mediators of higher-order processes such as learning and memory, ionotropic glutamate receptors (iGluRs) have attracted a great deal of interest as allosteric modulator targets (3–6). Of clear therapeutic importance, various neurodegenerative disorders such as Parkinson's and Alzheimer's diseases, Huntington's chorea, and neurologic disorders including epilepsy and ischemic brain damage have been linked to iGluRs (7).
The crystal structure of GluA2 (8) clarifies years of speculation on the complex arrangement of the glutamate receptor's four subunits (9). The GluA2 can be dissected into 3 functionally distinct layers. Farthest from the membrane, the amino terminal domain (ATD) can act as a peripheral regulatory domain but is also involved in assembly and trafficking (10, 11). Sandwiched between the ATD and the membrane domain, the ligand-binding domain (LBD) recognizes the neurotransmitter signal and directly regulates receptor activation (12). Structures for both isolated extracellular domains (ATD and LBD) reveal a dimeric organization (13–15). At the membrane interface, two alternative linker conformations transition the 2-fold symmetry, which is adopted by both extracellular domains, into the 4-fold symmetry of a membrane-traversing cation-selective channel (8, 16). For iGluRs, the ion channel domain confers functional relevance with its ability to selectively conduct the flow of ions across the cell's membrane. The layers of extracellular domains, each with the potential for multiple control points, allosterically regulate the ion channel domain's function (8). Therefore it is not surprising that the ATD, the LBD, and the LBD-channel linker have all been shown to be effective targets of allosteric modulators (13, 17, 18).
Since the structures of the ATD and the full iGluR channel have only recently been solved, allosteric drug-binding sites external to the LBD have not been fully explored in molecular detail. However, the decade-old LBD structure has proved to be indispensable as a heavily exploited scaffold for understanding agonist, partial agonist, and antagonist binding interactions as well as their ability to regulate channel gating behavior (12, 19, 20). Although the dimeric organization is consistent across all iGluR subtypes, the molecular details of LBD-agonist specificity define the subtype families into N-methyl-D-aspartic acid (NMDA) receptors (21), α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptors (12), and kainate receptors (22). Because all subtypes are constrained by their conserved sensitivity to glutamate stimulation, diversity at the orthosteric site is evolutionarily limited and most agonists display cross-subtype activity. An allosteric modulator-binding site within the quaternary LBD structure is located along the dimer interface (18) and offers improved discrimination by modulators. Drugs that bind to the allosteric sites on the LBD dimer interface can enhance the activity of iGluRs (23) and increase performance on tests of memory (24). Except for the LBD structures with modulatory ions bound to the dimer interface (25–27), only LBD structures from the AMPA receptor subtype, GluA2, have been reported with bound allosteric modulators (18, 28–31). Within the structures, the bound modulatory drugs stabilize the LBD dimer interface, which is required for activation of the ion channel and is dissociated during desensitization (18).
Although the residues that line the allosteric modulator-binding pocket do not differ between AMPA receptors subtypes (GluA1–4), the ability of allosteric modulators to stabilize the activated state still varies (32, 33). Also, AMPA receptors can be alternatively spliced into what is referred to as flip and flop isoforms (34). Modulator selectivity (23), desensitization (35), and channel closing rates (36) differ between flip and flop. Although several of the amino acid differences between the two forms are located in or near the allosteric modulator-binding site, the difference at position 754 (serine in flip, asparagine in flop) seems to be entirely responsible for the functional differences between allosteric modulator regulation of the flip and flop variants (23, 28, 32). Cyclothiazide (CTZ) and some other thiazide derivatives have improved binding to the flip form due to a hydrogen bond between S754 and the NH of the fused thiazide ring (28). In the case of the flop form, the alternatively spliced sequence places an asparagine in the 754 position, which is not optimally positioned to form a hydrogen bond. Sekiguchi et al. (33) introduced an allosteric modulator of AMPA receptors (4-[2-(phenylsulphonylamino)ethylthio]-2,6,-difluorophenoxyacetamide, PEPA) with a preference for the flop form. In fact, the relative sensitivity of CTZ to PEPA has been used as a diagnostic for the prevalence of flip vs. flop versions of AMPA receptor in particular cell types (37). PEPA shows potential in treatment of post ischemic memory impairment (38) and contextual fear (39) but despite PEPA's unique flop sensitivity, the modulator has not yet been used as a lead compound in SAR studies.
For drug discovery to be guided by structures, understanding the possible molecular interactions between modulators and the dimer interface is essential. We have shown previously (31) that changes in the structures of CTZ derivatives can reorient the modulator within the binding site. Subsequently, we proposed that the allosteric modulator site is comprised of 5 subsites (Figure 1C). In the present study, we determine the three dimensional structures of PEPA bound to the GluA2o and GluA3o LBDs (flop forms), and use PEPA's binding interactions to further characterize the subsite specific binding properties displayed by allosteric modulators. The amide group of PEPA makes a direct hydrogen bond to N754, explaining the preferential action of PEPA on the flop form of AMPA receptors. Another key structural element, the sulfonamide group of PEPA, is conserved with the biarylsulfonamide class of allosteric modulators (6) and interacts with the same residues of the dimer interface (8, 30). Although previously classified as unrelated, PEPA and the large group of biarylsulfonamide have similarities, which suggest that specific PEPA groups (particularly the unique flop-interacting amide) can be strategically integrated into biarylsulfonamide SAR studies.
Figure 1.
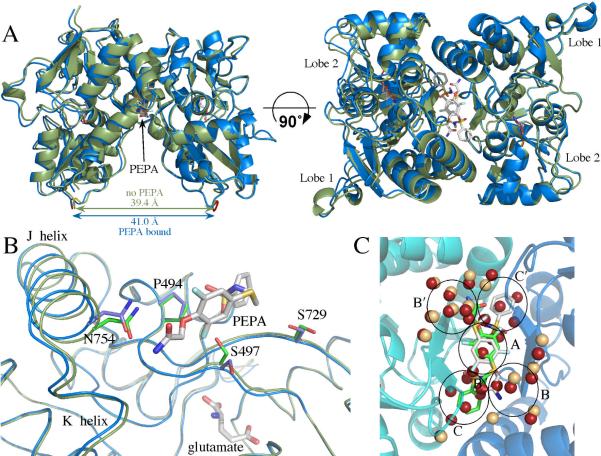
(A) Comparison of glutamate-bound GluA2o S1S2 in the presence (blue) and the absence of PEPA (green) in two orientations. Both orientations of PEPA are shown. Note that the binding of PEPA results in a separation of the two components of the dimer (distance between the Cα atoms of the threonine in the linker) by approximately 1.5 Å. (B) One monomer of GluA2o S1S2 in the presence (blue) and the absence of PEPA (green) with one orientation of PEPA shown. Both the J/K helices and the strand near S497 are displaced upon binding PEPA. Also, the sidechains of S497 and S729 change rotameric states. (C) Comparison of the water molecules at the dimer interface in the presence (tan spheres) and the absence of PEPA (red spheres). PEPA is shown in both orientations. Despite the greater separation of the dimer interface, a number of the ordered water molecules found in the absence of PEPA are displaced by PEPA. The black circles delineate subsites of the allosteric modulator binding site as described previously (31).
Experimental Procedures
Materials
PEPA was purchased from Tocris (Ellisville, MO). The GluA2 S1S2J construct was obtained from Eric Gouaux (Vollum Institute; 12).
Protein Preparation and Purification
GluA2 S1S2 consists of residues N392 - K506 and P632 - S775 of the full rat GluA2o subunit (40), a `GA' segment at the N-terminus, and a `GT' linker connecting K506 and P632 (12). A similar construct of GluA3 S1S2 was prepared as described previously (41). pET-22b(+) plasmids were transformed in E. coli strain Origami B (DE3) cells and were grown at 37°C to OD600 of 0.9 to 1.0 in LB medium supplemented with the antibiotics (ampicillin and kanamycin). The cultures were cooled to 20°C for 20 min. and isopropyl-β-D-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM. Cultures were allowed to grow at 20°C for 20 h. The cells were then pelleted and the S1S2 protein purified using a Ni-NTA column, followed by a sizing column (Superose 12, XK 26/100), and finally an HT-SP-ion exchange-Sepharose column (Amersham Pharmacia). Glutamate (1 mM) was maintained in all buffers throughout purification. After the last column, the protein was concentrated and stored in 20 mM sodium acetate, 1 mM sodium azide, and 10 mM glutamate at pH 5.5.
Crystallography
For crystallization trials, the protein was concentrated to 0.2 – 0.5 mM in 10 mM glutamate using a Centricon 10 centrifugal filter (Millipore, Bedford, MA). For the PEPA-bound structures, PEPA was added to 5 mM. The final protein concentration was 0.2 to 0.3 mM. Crystals were grown at 4°C using the hanging drop technique, and the drops contained a 1:1 (v/v) ratio of protein solution to reservoir solution. The reservoir solution contained 14–15% PEG 8K, 0.1 M sodium cacodylate, 0.1–0.15 M zinc acetate, and 0.25 M ammonium sulfate, pH 6.5.
Data were collected at the Cornell High Energy Synchrotron Source beam line A1 using a Quantum-210 Area Detector Systems charge-coupled device detector. Data sets were indexed and scaled with HKL-2000 (42). Structures were solved with molecular replacement using Phenix (43). Refinement was performed with Phenix (43), and Coot 0.5 (44) was used for model building.
Results
Structure of PEPA bound to GluA2 S1S2 flop
The structure of glutamate bound to GluA2o S1S2 (3dp6; 41) was used as the initial search probe for the molecular replacement solution of PEPA bound to GluA2o S1S2 with glutamate in the agonist-binding site. PEPA was then modeled into two symmetrical positions within the density found at the dimer interface, and the structure was optimized using Phenix (43). The refinement statistics are given in Table 1. The resolution is 1.85 Å, and three unique copies are found in the unit cell. The overall structure of the S1S2 domain is very similar to the structure in the absence of PEPA, with contacts between glutamate and the protein unchanged. However, PEPA clearly binds within the dimer interface, making contacts with both monomers within the dimer. As shown in Figure 1, one PEPA molecule binds per dimer interface. However because the dimer interface is symmetrical, two equivalent orientations (related by a 180° rotation) are possible. Electron density for both is seen in the crystal structure, although the intensity of one orientation is greater than the other.
Table 1.
Structural Statistics
| GluA2o (PEPA) | GluA3o (PEPA) | GluA3o | |
|---|---|---|---|
| Space Group | P22121 | P222 | P222 |
| Unit Cell (Å) | a=47.13 b=113.92 c=164.81 | a=46.95 b=52.26 c=115.98 | a=46.03 b=110.33 c=161.192 |
| X-ray source | CHESS (A1) | CHESS (A1) | CHESS (A1) |
| Wavelength (Å) | 0.977 | 0.977 | 0.977 |
| Resolution (Å) | 50–2.0 (2.03–2.00) | 50–2.5 (2.54–2.00) | 50–1.85 (1.88–1.85) |
| Measured reflections (#) | 817961 | 62549 | 344340 |
| Unique reflections (#) | 70175 | 9141 | 69379 |
| Data redundancy | 6.9 (7.1) | 6.0 (4.2) | 4.7 (3.0) |
| Completeness (%) | 99.9 (100.0) | 99.5 (89.8) | 96.5 (73.5) |
| Rsym (%) | 11.4 (34.2) | 13.0 (45.5) | 7.5 (24.6) |
| I/σi | 33.3 (7.1) | 19.2 (2.5) | 34.3 (3.3) |
| PDB ID | * | * | * |
| Current Model Refinement Statistics | |||
| Phasing | MR | MR | MR |
| Molecules/AU | 3 (no NCS applied) | 1 | 3 (no NCS applied) |
| Rwork/Rfree (%) | 18.7/24.2 | 18.9/28.5 | 20.1/23.4 |
| Free R test set size (#/%) | 2000 (2.85) | 914 (10.0) | 2000 (2.88) |
| Number of protein atoms | 5979 | 2030 | 6091 |
| Number of heteroatoms | 111 | 62 | 30 |
| Rmsd bond length (Å) | 0.011 | 0.015 | 0.009 |
| Rmsd bond angles (°) | 1.3 | 1.8 | 1.3 |
to be submitted to RCSB Protein Data Bank
The binding of PEPA to the dimer interface increases the distance between the two monomers that form the dimer by approximately 1.5 Å. This allows the relatively large PEPA molecule to fit within the interface, but also increases the separation between the linkers to the ion channel (the distance increases from 39.4 Å to 41 Å; Figure 1A). Relative to the core of Lobe 1, both the J/K helices and one β strand (P105-G110) connecting the two lobes are displaced slightly away from the dimer interface (Figure 1B). In addition, Lobe 2 is slightly twisted relative to glutamate-bound S1S2 in the absence of PEPA (3dp6; 41). PEPA binds at the bottom of a water-filled, inverted U-shaped cleft with five subsites (A, B/B′, and C/C′; 31). Upon binding, crystallographic waters are displaced from the central A subsite and more buried C/C′ sites, with the waters in the B/B′ subsite remaining (Figure 1C). This displacement of presumably ordered water would be likely to contribute a favorable entropy component to binding.
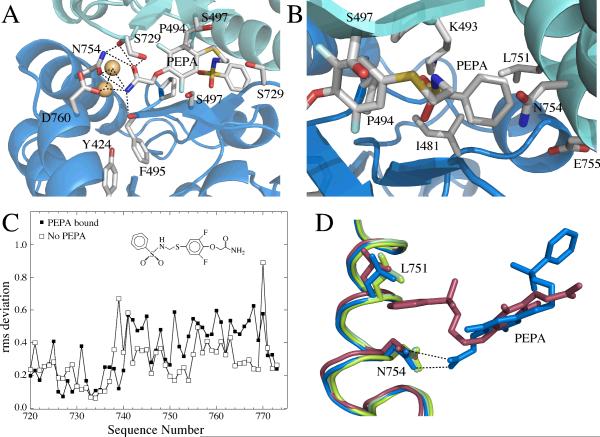
The sidechains of P494 are at the center of the interface and the edge of the two proline rings from each monomer form the base of the binding site in which the difluorophenyl ring resides (Figure 2A). This is close to the position of the methoxybenzoyl ring of aniracetam in its structure bound to GluA2-S1S2(FW) (29). The other side of the ring is exposed to S497 and S729. The sidechain hydroxyl of S497 is oriented toward the dimer interface in the absence of PEPA, but rotates out toward the solvent to accommodate the difluorophenyl ring of PEPA (Figure 1B). The amide of PEPA is involved in a network of hydrogen bonds with sidechain hydroxyl of Y424, the backbone carbonyl of F495, the sidechain carboxyl of D760, the sidechain amide of N754, and two water molecules (Figure 2A). The most striking of these hydrogen bond pairs is with N754. This represents the only difference between the flip (S754) and flop (N754) isoforms in the PEPA binding site and is almost certainly a major source of the preference for the flop isoform. The phenyl-sulfonylamide side of PEPA inserts into a hydrophobic pocket formed by sidechain methyls of I481 and L751 as well as methylene groups contributed by K493, N754, and E755 (Figure 2B). It is possible that the contribution by methylene group of N754 provides a more hydrophobic pocket than S754 in the flip form, further contributing to the preference for the flop form.
Figure 2.
The PEPA binding site, emphasizing the important interactions, shown in two orientations. (A) A view of the amide side of PEPA bound to GluA2 S1S2. The hydrogen bonding network with the amide of PEPA is shown as dotted lines. The H-bond with the sidechain of S729 is difficult to display in the orientation used in the figure. (B) A view of the phenyl group of PEPA inserted into a hydrophobic pocket in GluA2 S1S2. (C) RMS plot showing more variability in the J/K helices for the PEPA-bound structure than the unbound structure. (D) J/K helix showing where differences in the two orientations were analyzed. The amide of PEPA-N754 interaction (blue) maintains the position of the J helix in the absence of PEPA (green) The J helix is displaced on the phenyl side of PEPA (red).
Because the dimer interface is symmetrical, PEPA can bind in two orientations and both are observed in the crystal. For this reason, changes in the protein due to a specific interaction with PEPA can be partially masked because each monomer is a weighted average of two orientations of bound PEPA. However, one orientation has a stronger density than the other, providing some insight into the extent of changes in the dimer interface that are produced by PEPA binding. As shown in Figure 2C, the two monomers comprising the dimer differ more within the PEPA binding site than the corresponding monomers in the absence of PEPA. One turn of helix J (L751 to N754) contains important determinants for both orientations of PEPA. In one orientation the amide group of PEPA interacts with the sidechain of N754, and in the other, the aromatic ring of PEPA inserts between a hydrophobic pocket formed by the sidechain of L751 and the methylene group of N754. In the orientation for which the density of the amide of PEPA is stronger, N754 is better positioned to form an H-bond (Figure 2D); whereas, in the other side of the interface, N754 is oriented to form an H-bond with the carbonyl of S729. This change in orientation facilitates the insertion of the aromatic ring of PEPA into the hydrophobic pocket, which is accompanied by a small shift in the sidechain of L751 to accommodate the aromatic ring (Figure 2D). Since these structures are weighted averages, it is possible that the actual positions of these sidechains involve an even greater movement than is seen from the asymmetry of the crystal.
Structure of PEPA bound to GluA3 S1S2 flop
In studies of the physiological effects of PEPA, a significant difference between subtypes has been observed, with GluA3 being most susceptible to modulation (33). The structure of GluA3i S1S2 bound (flip form) to glutamate has been reported previously (41). Since PEPA preferentially binds to the flop form, the GluA3o structure was determined bound to glutamate with and without PEPA (Figure 3A). Like GluA2o, in the absence of PEPA, GluA3o has three copies in the asymmetric unit. Comparing lobe closure between GluA3i and GluA3o, the flop form is slightly more closed (1.6° ± 0.7°).
Figure 3.
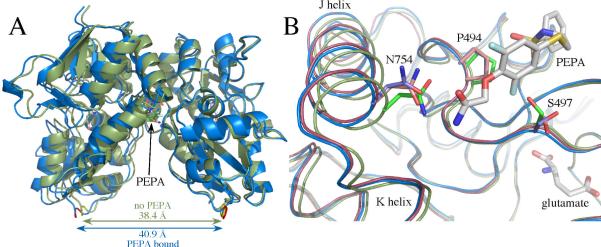
(A) Comparison of glutamate-bound GluA3o S1S2 in the presence (blue) and the absence of PEPA (green) in two orientations. Both orientations of PEPA are shown. Note that the binding of PEPA results in a separation of the two components of the dimer (distance between the Cα atoms of the threonine in the linker) by approximately 2.5 Å. (B) One monomer of GluA3o S1S2 in the presence (blue) and the absence of PEPA (green) with one orientation of PEPA shown. Shown for comparison is the PEPA-bound form of GluA2o (red). Both the J/K helices and the strand near S497 are displaced upon binding PEPA for both GluA2o and GluA3o. Also, the sidechains of S497 and S729 are in different rotameric states for GluA3o bound to PEPA compared with GluA3o in the absence of PEPA and GluA2o bound to PEPA. Also, N754 is displaced in PEPA-bound GluA3o, such that only one H-bond is possible with the amide of PEPA.
In the presence of PEPA, GluA3o was present in one copy in the asymmetric unit, and PEPA was observed with the same density in two symmetrical orientations. Like GluA2o bound to PEPA, the dimer interface (assessed using the symmetrical molecule in the crystal) was displaced relative to the unbound from (Figure 3A) by approximately 2.5 Å at the position of the linker replacing the ion channel domain. Within the binding site, three sidechains exhibited different rotamers compared with the GluA2o structure bound to PEPA (Figure 3B). For PEPA-bound GluA3o, both S497 and S729 assumed rotameric states that differed both from GluA2o bound to PEPA and from GluA2o and GluA3o in the absence of PEPA. In the case of S729, the rotameric state in combination with a slight movement of the amide of PEPA (relative to the GluA2o structure) would make an H-bond with the sidechain of S729 (shown in Figure 2A for GluA2o) unlikely. In the case of N754, the sidechain is displaced relative to the GluA2o-PEPA structure so that only one H-bond is made to the amide of PEPA. This may be a result of averaging of the two orientations of PEPA only one of which forms a bidentate H-bond with N754.
Discussion
The goal of allosteric modulation, like orthosteric modulation, is often to stabilize a conformational state of a dynamic protein (45). The activated state of iGluRs is naturally unstable allowing the channel to desensitize (46). Disruption of the symmetrical dimer interface between LBDs is thought to initiate desensitization-mediated channel closure (47). By maintaining the LBD dimer, positive allosteric modulators can prevent desensitization and prolong activation (18). Currently, 15 crystal structures of the GluA2 LBD with bound allosteric modulators are deposited in the Protein Data Bank (48). All of these modulators bind to a large crevice with 2-fold symmetry along the symmetric dimer interface (18). The large variation in structure among allosteric modulators results in significant variations in binding orientations and interactions. At least four distinct binding modes have been identified: (1) A-subsite class (aniracetam, CX614 (29)), (2) classical thiazide (cyclothiazide (18), TCMZ, ALTZ (31)), (3) the shifted thiazide class (IDRA-21, HCMZ, HFMZ; (31)), and (4) the full spanning class (PEPA (this paper), dimeric biarylpropylsulfonamide (30), LY404187 (8)). Overlaying modulators from these structural classes has led to the proposal that the allosteric modulator site is comprised of a series of subsites (Figure 1C; 31). Positioned at the center of the binding-site, the symmetric A subsite is narrow and allows entrance to only one molecule. Two subsites (B and C) lie at each end of the A subsite with the hydrophobic C subsite located more deeply in the pocket effectively defining five subsites (A, B, B′, C, and C′).
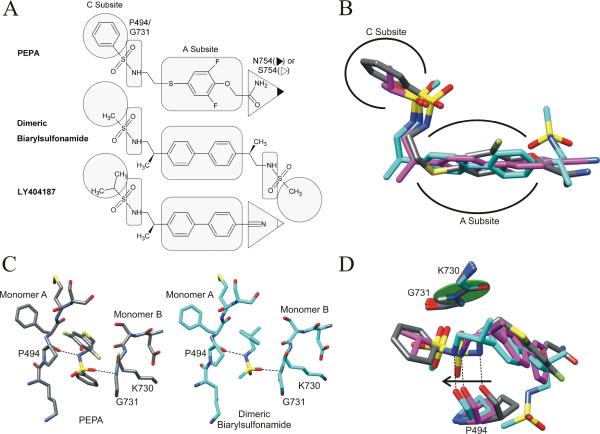
In the open state, the subsites are filled with water, which may act to weakly stabilize the dimer. Allosteric modulators generate stronger interactions across the subsites thereby increasing the linkages between the monomers. The simplest modulator class, including aniracetam and CX614, fills the A subsite with one molecule but does not enter the peripheral B and C subsites (29). The two classes of thiazide-based modulators account for 10 of the 15 solved allosteric modulator-GluA2 crystal structure complexes (18, 28, 31). The classical thiazide (CTZ-like) binding class and the shifted thiazide (IDRA-21-like) binding class are positioned respectively in the B and C subsite or mainly the C subsite. Most of the thiazide modulators do not extend across the A subsite and therefore can bind two molecules per dimer. However, a few of the newly described shifted thiazides (HFMZ, HCTZ; 31) enter the A subsite but only enough to impair binding of a second modulator. The dimeric biarylpropylsulfonamide compound ((R,R)-N,N-(2,2'-[Biphenyl-4-4'-Diyl]Bis[Propane-2,1-Diyl]) Dimethanesulfonamide) described by Kaae et al. (30) was the first allosteric modulator shown by crystallography to extend along the entire length of the inner dimer cavity from C to C′ subsites. PEPA also interacts with J helices from both monomers, which cap the ends of the modulator-binding pocket. The density occupied by both symmetrical copies of PEPA overlays the dimeric biarylsulfonamide compound as both modulators represent the full spanning class (Figure 4B).
Figure 4.
(A) Members of the full spanning class of allosteric modulators. The shape-highlighted regions of the modulators illuminate key contact points to the specific binding pocket residues and subsites (as labeled for PEPA). (B) Overlay of the full spanning modulator structures. The structures were aligned at both sets of P494 and G731 residues. PEPA (gray) occupies a similar arrangement of subsites as the dimeric biarylsulfonamide (PDB entry 3bbr, cyan, 30) and LY404187 (PDB entry 3kgc, magenta, 8). (C) The sulfonamide bridges the two monomers in both PEPA and the dimeric biarylsulfonamide with the same interactions to P494 and G731. (D) The hydrogen bond between the carbonyl of P494 and the sulfonamide is maintained when the modulator is in a shifted position relative to the peptide plane of K730 and G731 (green disk) located on the opposite monomer.
The GluA flip and flop splice variants differ by only a few residues along the J helix in the LBD; however, residue 754 (Asn in flop and Ser in flip) is positioned between the B and C subsites. For thiazides, a clear preference in binding to the flip-form is mediated by a hydrogen bond between the hydrobenzothiadiazide ring and S754 (28). In contrast, PEPA is flop-selective and the PEPA-bound structure provides the first structure containing a direct interaction between a modulator and the flop form's N754. The amide of PEPA extends straight out from the A subsite and across the B and C subsite interface to make an amide-amide hydrogen bond with N754 (Figure 2A). Unlike most other AMPA modulators, PEPA fills neither the B nor the C subsites but interacts directly with the J helix. A similar interaction is seen with LY404187 (49) bound to GluA2i (8). Strong hydrogen bonding can occur between two amides (50) and has been shown to be responsible for driving oligomerization of transmembrane leucine zippers (51). The distances between the interacting amides in the PEPA-bound structure support a bidentate hydrogen-bonding pattern, which is much stronger and more specific than a typical hydrogen bond. While PEPA is selective for the AMPA receptor's flop form, a weaker but still existent potentiation of the flip form has been observed (33, 52). Replacing N754 (flop) with S754 (flip) would not prevent PEPA from binding; however, serine would provide only one hydrogen-bonding partner for PEPA's amide with an extended interaction distance. In contrast, LY404187 displays a preference for the flip isoform (53), and its cyano group extends out to interact directly with S754. The cyano-S754 interaction is a clear flip analog of the flop-selective PEPA amide-N754 interaction (Figure 4A).
Opposite to the amide on the PEPA molecule, a sulfonamide is tethered to the difluorophenyl ring (Figure 4A). Within the dimer interface, the sulfonamide is positioned so the nitrogen can hydrogen bond directly with the carbonyl of P494 (Figure 4C). A sulfonamide oxygen points toward the amide nitrogen of G731. The angle of the peptide plane is perpendicular to the sulfonamide oxygen, making a hydrogen bonding interaction unlikely (Figure 4D). Instead, a dipole-dipole or charge-dipole interaction may occur. The amide nitrogen of a polypeptide supports at least a partial positive charge (54), which would interact with the strongly electronegative sulfonamide oxygen (55). Interestingly, both the dimeric biarylsulfonamide (30) and LY404187 (8), other members of the full spanning modulator class, also have a sulfonamide that interacts with the same backbone atoms of P494 and G731 as PEPA (Figure 4C).
A large number of biarylsulfonamides have been identified that modulate AMPA receptors and are being evaluated for therapeutic use in the treatment of depression and Parkinson's disease (56). The conserved sulfonamide reveals a previously unidentified relationship between PEPA and the biarylsulfonamide modulators. When the perpendicular peptide bond plane including G731 is fixed, the sulfonamide on three overlayed modulators varies by 1.2 Å along the length of the interface with the PEPA sulfonamide being positioned closer to the A subsite (Figure 4D). A shift of the sulfonamide also results in a shift in the corresponding P494 across the interface presumably to maintain the hydrogen bond with the modulator's amine. The sulfonamide forms an important bridge between the two dimer halves. For PEPA, a phenyl-sulfonamide replaces the methyl-sulfonamide in the dimeric biarylsulfonamide and fits snuggly against L751. Based on the orientation-induced asymmetry within the GluA2-complex structure, the phenyl pushes the J helix away from PEPA thereby affecting the C subsite (Figure 2C and D). Residues lining the C subsite are on the same beta strand as G731, which must shift if the C subsite is to remain together and presumably explain the 1.2 Å shift relative to the dimeric biarylsulfonamide. In fact, the same phenyl-sulfonamide group substitution in a biarylpropylsulfonamide decreases the modulatory effect of the derivative in SAR studies (57). For biarylpropylsulfonamides, the optimal sulfonamide substitution was found to be either an ethyl or an iso-propyl group, which should both fit without significantly disrupting the J helix or C subsite (57).
The PEPA-bound crystal structure from AMPA receptor subtypes, GluA2 and GluA3, do not display major differences in binding interactions even though PEPA exhibits a stronger effect on GluA3 (33). For GluA2, an asymmetry in the receptor-binding pocket was observed while no significant difference in PEPA density was seen for the each orientation within the GluA3 crystal structure. In addition, a number of side chains exhibit different rotameric states between the two structures, although it is unlikely that these small changes significantly impact the differential effects on the two subtypes. Although no structural differences have been identified between GluA2 and GluA3 that would obviously impact PEPA affinity, the possibility exists that subtle differences arising from the sequence differences peripheral to the binding site may be important as has been described in the case of the agonist binding site of GluA4 (58).
We have explored how PEPA (this paper) and other allosteric modulators (31) interact with the GluA interface in the context of drug design. Together the identification of a conserved group between PEPA (this paper) and biarylpropylsulfonamides (8, 30) and the regional nature of various subsite-functional group interactions provide a backdrop to extend biarylpropylsulfonamide SAR studies (57) to include PEPA and biarylpropylsulfonamide chimeras. Although optimizing the stability of the dimer interface provides a starting point for SAR studies, additional constraints should be considered including the ability of the modulator to enter the cavity, the dynamic structure of the dimer interface during closed, open, and desensitized state transitions, and the ability of the modulator to cross the blood-brain barrier before being metabolized. This definition of the allosteric modulator binding-site should provide guidance in glutamate receptor allosteric modulator pharmacology.
Acknowledgement
We thank Prof. Eric Gouaux (Vollum Institute) for the GluA2 S1S2J construct, and Prof. Linda Nowak (Cornell) for the full-length GluA3 construct.
This work was supported by a grants from the National Institutes of Health (R01-GM068935, R01 NS049223, and R21 NS067562). This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR 0225180, using the Macromolecular Diffraction at the CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources.
Abbreviations
- ALTZ
althiazide
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid
- CLTZ
chlorothiazide
- CX614
pyrrolidino-1,3-oxazino benzo-1,4-dioxan-10-one
- CTZ
cyclothiazide
- FW
(S)-5-fluorowillardiine
- flip and flop
alternatively spliced versions of AMPA receptors that vary in rates of desensitization and sensitivity to allosteric modulators
- iGluR
ionotropic glutamate receptor
- GluA1-4
four subtypes of AMPA receptor
- HCTZ
hydrochlorothiazide
- HFMZ
hydroflumethiazide
- IDRA-21
7-chloro-3-methyl-3,4-dihydro-2H-benzo[e][1,2,4]thiadiazine 1,1-dioxide
- IPTG
isopropyl-β-D-thiogalactoside
- LY404187
N-[2-(4′-cyanobiphenyl-4-yl)propyl]propane-2-sulfamide
- PEPA
4-[2-(phenylsulphonylamino)ethylthio]-2,6,-difluorophenoxy acetamide
- NMDA
N-methyl-D-aspartic acid
- S1S2
extracellular ligand-binding domain of GluA2 and GluA3
- SAR
structure-activity relationships
- TCMZ
trichlormethiazide
References
- 1.Christopoulos A. Allosteric binding sites on cell-surface receptors: novel targets for drug discovery. Nat Rev Drug Discov. 2002;1:198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 2.Changeux JP, Taly A. Nicotinic receptors, allosteric proteins and medicine. Trends Mol Med. 2008;14:93–102. doi: 10.1016/j.molmed.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Bowie D. Ionotropic glutamate receptors & CNS disorders. CNS Neurol Disord Drug Targets. 2008;7:129–143. doi: 10.2174/187152708784083821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingledine R, Borges K, Bowie D, Traynelis S. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 5.Oswald RE, Ahmed A, Fenwick MK, Loh AP. Structure of glutamate receptors. Current drug targets. 2007;8:573–582. doi: 10.2174/138945007780618526. [DOI] [PubMed] [Google Scholar]
- 6.Grigoriev VV, Proshin AN, Kinzirsky AS, Bachurin SO. Modern approaches to the design of memory and cognitive enhancers based on AMPA receptor ligands. Russian Chemical Reviews. 2009;78:485–494. [Google Scholar]
- 7.Black MD. Therapeutic potential of positive AMPA modulators and their relationship to AMPA receptor subunits. A review of preclinical data. Psychopharmacology (Berl) 2005;179:154–163. doi: 10.1007/s00213-004-2065-6. [DOI] [PubMed] [Google Scholar]
- 8.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosciences. 1995;18:161–168. doi: 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- 10.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greger IH, Ziff EB, Penn AC. Molecular determinants of AMPA receptor subunit assembly. Trends Neurosci. 2007;30:407–416. doi: 10.1016/j.tins.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 13.Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- 14.Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle DA, Cabral JM, Pfuetzner RA, Kuo AL, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of a potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 17.Balannik V, Menniti FS, Paternain AV, Lerma J, Stern-Bach Y. Molecular mechanism of AMPA receptor noncompetitive antagonism. Neuron. 2005;48:279–288. doi: 10.1016/j.neuron.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed A, Thompson M, Fenwick M, Romero B, Loh A, Jane D, Sondermann H, Oswald R. Mechanisms of antagonism of the GluR2 AMPA receptor: Structure and dynamics of the complex of two willardiine antagonists with the glutamate binding domain. Biochemistry. 2009;48:3894–3903. doi: 10.1021/bi900107m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: Molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 23.Partin KM, Fleck MW, Mayer ML. AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci. 1996;16:6634–6647. doi: 10.1523/JNEUROSCI.16-21-06634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin JR, Cumin R, Aschwanden W, Moreau JL, Jenck F, Haefely WE. Aniracetam improves radial maze performance in rats. Neuroreport. 1992;3:81–83. doi: 10.1097/00001756-199201000-00021. [DOI] [PubMed] [Google Scholar]
- 25.Naur P, Vestergaard B, Skov LK, Egebjerg J, Gajhede M, Kastrup JS. Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate. FEBS Lett. 2005;579:1154–1160. doi: 10.1016/j.febslet.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 27.Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hald H, Ahring PK, Timmermann DB, Liljefors T, Gajhede M, Kastrup JS. Distinct Structural Features of Cyclothiazide are Responsible for Effects on Peak Current Amplitude and Desensitization Kinetics at iGluR2. J Mol Biol. 2009;391:906–917. doi: 10.1016/j.jmb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaae BH, Harpsoe K, Kastrup JS, Sanz AC, Pickering DS, Metzler B, Clausen RP, Gajhede M, Sauerberg P, Liljefors T, Madsen U. Structural proof of a dimeric positive modulator bridging two identical AMPA receptor-binding sites. Chemistry & biology. 2007;14:1294–1303. doi: 10.1016/j.chembiol.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Ptak CP, Ahmed AH, Oswald RE. Probing the allosteric modulator binding site of GluR2 with thiazide derivatives. Biochemistry. 2009;48:8594–8602. doi: 10.1021/bi901127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Partin KM, Bowie D, Mayer ML. Structural determinants of allosteric regulation in alternatively spliced AMPA receptors. Neuron. 1995;14:833–843. doi: 10.1016/0896-6273(95)90227-9. [DOI] [PubMed] [Google Scholar]
- 33.Sekiguchi M, Fleck MW, Mayer ML, Takeo J, Chiba Y, Yamashita S, Wada K. A novel allosteric potentiator of AMPA receptors: 4-[2-(phenylsulfonylamino)ethylthio]-2,6-difluoro-phenoxyacetamide. J Neurosci. 1997;17:5760–5771. doi: 10.1523/JNEUROSCI.17-15-05760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakmann G, Seeburg PH. Flip and flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1584. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 35.Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 36.Pei W, Huang Z, Niu L. GluR3 flip and flop: differences in channel opening kinetics. Biochemistry. 2007;46:2027–2036. doi: 10.1021/bi062213s. [DOI] [PubMed] [Google Scholar]
- 37.Sekiguchi M, Takeo J, Harada T, Morimoto T, Kudo Y, Yamashita S, Kohsaka S, Wada K. Pharmacological detection of AMPA receptor heterogeneity by use of two allosteric potentiators in rat hippocampal cultures. Br J Pharmacol. 1998;123:1294–1303. doi: 10.1038/sj.bjp.0701707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiguchi M, Yamada K, Jin J, Hachitanda M, Murata Y, Namura S, Kamichi S, Kimura I, Wada K. The AMPA receptor allosteric potentiator PEPA ameliorates post-ischemic memory impairment. Neuroreport. 2001;12:2947–2950. doi: 10.1097/00001756-200109170-00038. [DOI] [PubMed] [Google Scholar]
- 39.Zushida K, Sakurai M, Wada K, Sekiguchi M. Facilitation of extinction learning for contextual fear memory by PEPA: a potentiator of AMPA receptors. J Neurosci. 2007;27:158–166. doi: 10.1523/JNEUROSCI.3842-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed AH, Wang Q, Sondermann H, Oswald RE. Structure of the S1S2 glutamate binding domain of GluR3. Proteins: Structure, Function, and Bioinformatics. 2009;75:628–637. doi: 10.1002/prot.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology, Vol. 276, Macromolecular Crystallography, part A. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 46.Trussell LO, Fischbach GD. Glutamate receptor desensitization and its role in synaptic transmission. Neuron. 1989;3:209–218. doi: 10.1016/0896-6273(89)90034-2. [DOI] [PubMed] [Google Scholar]
- 47.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 48.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miu P, Jarvie KR, Radhakrishnan V, Gates MR, Ogden A, Ornstein PL, Zarrinmayeh H, Ho K, Peters D, Grabell J, Gupta A, Zimmerman DM, Bleakman D. Novel AMPA receptor potentiators LY392098 and LY404187: effects on recombinant human AMPA receptors in vitro. Neuropharmacology. 2001;40:976–983. doi: 10.1016/s0028-3908(01)00027-2. [DOI] [PubMed] [Google Scholar]
- 50.Shimoni L, Glusker JP. Hydrogen bonding motifs of protein side chains: descriptions of binding of arginine and amide groups. Protein Sci. 1995;4:65–74. doi: 10.1002/pro.5560040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- 52.Sekiguchi M, Nishikawa K, Aoki S, Wada K. A desensitization-selective potentiator of AMPA-type glutamate receptors. Br J Pharmacol. 2002;136:1033–1041. doi: 10.1038/sj.bjp.0704804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quirk JC, Nisenbaum ES. Multiple molecular determinants for allosteric modulation of alternatively spliced AMPA receptors. J Neurosci. 2003;23:10953–10962. doi: 10.1523/JNEUROSCI.23-34-10953.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kemnitz CR, Loewen MJ. “Amide resonance” correlates with a breadth of C-N rotation barriers. J Am Chem Soc. 2007;129:2521–2528. doi: 10.1021/ja0663024. [DOI] [PubMed] [Google Scholar]
- 55.Cazenave-Gassiot A, Boughtflower R, Caldwell J, Coxhead R, Hitzel L, Lane S, Oakley P, Holyoak C, Pullen F, Langley GJ. Prediction of retention for sulfonamides in supercritical fluid chromatography. J Chromatogr A. 2008;1189:254–265. doi: 10.1016/j.chroma.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 56.O'Neill MJ, Witkin JM. AMPA receptor potentiators: application for depression and Parkinson's disease. Current drug targets. 2007;8:603–620. doi: 10.2174/138945007780618517. [DOI] [PubMed] [Google Scholar]
- 57.Ornstein PL, Zimmerman DM, Arnold MB, Bleisch TJ, Cantrell B, Simon R, Zarrinmayeh H, Baker SR, Gates M, Tizzano JP, Bleakman D, Mandelzys A, Jarvie KR, Ho K, Deverill M, Kamboj RK. Biarylpropylsulfonamides as novel, potent potentiators of 2-amino-3- (5-methyl-3-hydroxyisoxazol-4-yl)- propanoic acid (AMPA) receptors. J Med Chem. 2000;43:4354–4358. doi: 10.1021/jm0002836. [DOI] [PubMed] [Google Scholar]
- 58.Gill A, Birdsey-Benson A, Jones BL, Henderson LP, Madden DR. Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists. Biochemistry. 2008;47:13831–13841. doi: 10.1021/bi8013196. [DOI] [PMC free article] [PubMed] [Google Scholar]