Abstract
The rotavirus (RV) genome is comprised of eleven segments of double-stranded RNA (dsRNA) and is contained within a non-enveloped, icosahedral particle. During assembly, a highly-coordinated selective packaging mechanism ensures that progeny RV virions contain one of each genome segment. Cis-acting signals thought to mediate assortment and packaging are associated with putative panhandle structures formed by base-pairing of the ends of RV plus-strand RNAs (+RNAs). Viral polymerases within assembling core particles convert the eleven distinct +RNAs to dsRNA genome segments. It remains unclear whether RV +RNAs are assorted prior to or during encapsidation, and the functions of viral proteins during these processes are not resolved. However, as reviewed here, recent insights gained from the study of RV and two other segmented RNA viruses, influenza A virus and bacteriophage Φ6, reveal potential mechanisms of RV assortment and packaging.
Random versus selective genome segment packaging
Viruses that maintain their genomes as separate RNA molecules are faced with a daunting challenge during assembly—how to package a full complement of genome segments. Some RNA viruses utilize a non-selective packaging mechanism in which segments are randomly encapsidated into virions. This random packaging mechanism creates a large number of particles that lack complete genomes and are thus unable to mediate subsequent infections [1]. Other RNA viruses, particularly those with three or more genome segments, have evolved a more sophisticated packaging mechanism whereby each viral RNA is explicitly recognized. Members of the Reoviridae family are thought to utilize this gene-specific approach to package their genomes of 9, 10, 11, or 12 segments of double-stranded RNA (dsRNA). The strongest evidence in support of selective packaging, rather than random packaging, for the Reovirdae comes from the observation that the particle-to-plaque forming unit ratio can be quite low [2, 3]. Moreover, a Reoviridae member has never been identified that contains more than one copy of each gene, suggesting a precise equimolar process of assortment [4]. While the exact manner by which the Reoviridae achieve assortment is unknown, these viruses share several features with the eight-segmented, negative-strand RNA viruses of the Orthomyxoviridae family (e.g. influenza A virus) and the three-segmented, dsRNA bacteriophages of the Cystoviridae family (e.g. Φ6) [5, 6]. In particular, for all these viruses, cis-acting elements in single-stranded viral RNA are thought to determine segment selection, and viral proteins play critical roles in orchestrating the assortment and packaging processes. One of the remaining mysteries for Reoviridae is whether they (i) assort their genome segments prior to packaging, similar to influenza A virus, or (ii) package each segment individually into a pre-formed particle, similar to Φ6. In this review, we discuss the evidence supporting each of these assortment and packaging models for rotavirus (RV), a Reoviridae family member and significant pediatric gastrointestinal pathogen [7].
RV virion architecture and replication cycle
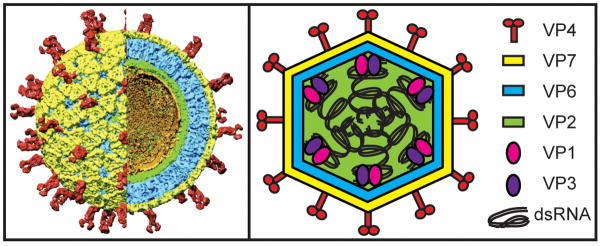
The mature RV virion is a triple-layered particle that encases eleven dsRNA genome segments (Figure 1) [8–12]. The outermost layer of the virion has T=13 icosahedral symmetry and is composed of the VP7 glycoprotein with several embedded copies of the VP4 spike attachment protein [13, 14]. The intermediate layer, also exhibiting T=13 symmetry, is made up of VP6 and surrounds a thin T=1 VP2 core shell [14]. Aqueous channels penetrate the VP6 and VP2 layers, allowing divalent cations and nucleotides to access the particle interior [14]. Viral polymerase complexes (PCs) consisting of a single subunit each of the viral RNA-dependent RNA polymerase (VP1) and RNA capping enzyme (VP3) are attached to the inner surface of the VP2 shell, proximal to most if not all of the twelve fivefold axes [10, 14, 15]. These enzymes are linked to the core shell through interactions requiring the amino-terminal residues of VP2, which form inwardly protruding fivefold hubs [14–18]. The RV dsRNA genome encodes six structural (VP1-4, VP6-7) and five or six nonstructural (NSP1-5/6) proteins and is predicted to be arranged as tubules that spool around the PCs [14, 15].
Figure 1. Architecture and protein composition of the RV virion.
The left panel shows a cryo-electron micrograph image reconstruction of a mature, RV triple-layered particle (TLP) at 9.5 Å resolution and was used with permission from B.V.V. Prasad (Baylor University). A portion of the particle has been computationally removed to reveal the internal virion layers. The smooth external surface is made up of the VP7 glycoprotein (yellow) and is embedded with the VP4 spike attachment protein (red). The intermediate VP6 layer is shown in blue and the thin VP2 core shell is shown in green. Ordered portions of viral dsRNA that line the VP2 shell are shown in gold. Polymerase complex (PC) components, VP1 (the viral polymerase) and VP3 (the viral capping enzyme), are not visualized in this reconstruction, but are predicted to be tethered to the inner surface of VP2 near each fivefold axis. The right panel shows a cartoon schematic of a RV TLP with proteins and dsRNA colored according to the legend.
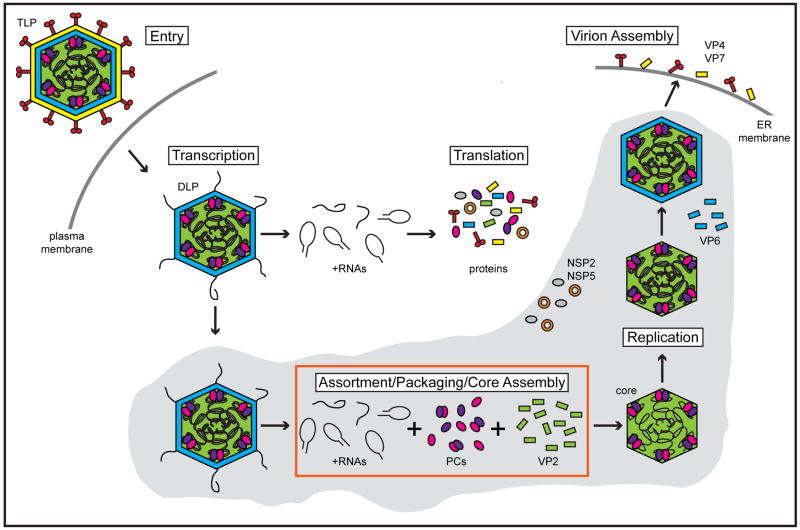
The primary site of RV infection is the small intestinal villi, where the virus replicates in the cytoplasm of mature enterocytes (Figure 2) [13]. Trypsin-like proteases of the gastrointestinal tract cleave the VP4 spike protein into VP5* and VP8*, an event that primes RV for entry into the cell [12, 19, 20]. During internalization, the outermost virion layer is lost, yielding a VP2-VP6 double-layered particle (DLP). Loss of the outer capsid triggers viral PCs within the DLP to become transcriptionally active and produce numerous copies of capped, non-poly (A) plus-strand RNAs (+RNAs) using the minus-strands of dsRNAs as templates [21, 22]. Each tethered PC is dedicated to transcribing an individual genome segment, but acts in synchrony with the others to simultaneously create eleven species of +RNA [16, 17]. Because the PCs operate independently, transcription is not equimolar; some species of +RNA are produced more abundantly than others [22–25]. RV DLPs are capable of robustly synthesizing transcripts for several hours in vitro, suggesting that PCs efficiently re-engage the 3′ ends of the minus-strand templates [26–28]. Newly synthesized RV +RNAs acquire a 5′ cap structure (m7GpppG) by the activity of the PC component VP3 prior to their extrusion from the DLP via channels near the fivefold axes [29, 30].
Figure 2. Schematic of the RV replication cycle.
During entry of a RV triple-layered particle (TLP) into a host cell, VP4 and VP7 are lost, resulting in the release of a double-layered particle (DLP). The viral PCs, composed of VP1 (pink spheres) and VP3 (purple spheres), within the DLP interior are transcriptionally active and synthesize multiple copies of eleven species of capped, non-poly (A) +RNAs (black lines). The nascent +RNAs are extruded out of the DLP and deposited into the cytosol where they serve as templates for translation of viral proteins. Newly made non-structural proteins NSP2 (orange donut) and NSP5 (gray sphere) form inclusions (viroplasms; gray shaded area) around DLPs, thereby trapping +RNAs that will be used for assortment, packaging, and genome replication. Two models are proposed for how the eleven species of +RNA associate with each other, PCs, and VP2 (green) to form a fully-packaged RV core (red box; see Figure 4). During or following their encapsidation, +RNAs are used as templates for replication by the core-associated PCs to recreate eleven dsRNA segments within a pre-virion particle. A VP6 layer (blue) is acquired, and then DLPs bud into the endoplasmic reticulum (ER) during which the outer capsid proteins (VP4 and VP7) are acquired. Mature RV TLPs exit non-polarized cells predominantly by lysis.
Nascent RV +RNAs serve dual roles during the replication cycle, acting as mRNAs for protein synthesis and as templates for genome replication. The intracellular localization of a viral transcript is predicted to largely determine its use in the infected cell [31]. The +RNA products of transcription from incoming DLPs accumulate in the cytosol and are available for translation by host ribosomes into viral proteins. Two viral nonstructural proteins (NSP2 and NSP5) are thought to co-localize around transcribing DLPs, forming dense inclusion bodies termed viroplasms [32, 33]. Viroplasm-associated RV +RNAs are selectively packaged into assembled or assembling VP2 cores (detailed below) [31, 34]. Following or during encapsidation of the eleven +RNA species, core-associated PCs perform minus-strand synthesis, thereby reconstituting the dsRNA genome inside of a pre-virion particle [34]. In contrast to viral transcription, which occurs multiple times on each segment, genome replication is equimolar and produces exactly one of each of the eleven dsRNAs per virion [4]. The timing of genome packaging and replication is regulated in part by interactions between the viral polymerase VP1 and core shell protein VP2 [16, 17]. Engagement of VP1 by VP2 triggers conformational changes in the enzyme that allow for the initiation of RNA synthesis [35–37]. This core shell requirement ensures that the polymerase does not aberrantly replicate the RV genome outside of particles. Following genome replication, progeny cores acquire a VP6 layer; such DLPs can amplify the replication cycle by supporting secondary rounds of transcription. DLPs eventually bud into the lumen of the endoplasmic reticulum, during which the outer capsid proteins VP4 and VP7 are added [38]. Mature, triple-layered particles are predominantly released from non-polarized cells via lysis, but may be released from polarized cells by both lytic and a non-lytic mechanisms [39].
Cis-acting functional elements of RV +RNAs
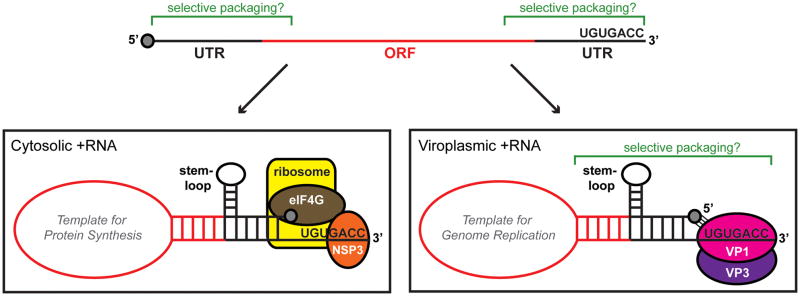
RV +RNA transcripts contain important sequence and structural elements that promote their (i) recognition by host ribosomes, (ii) assortment and packaging, and (iii) use as templates for genome replication. The eleven RV +RNAs range in size from 0.7 to 3.1 kb and generally contain a single open-reading frame (ORF) that is flanked at the 5′ and 3′ ends by untranslated regions (UTRs) (Figure 3) [40]. These molecules contain 5′ cap structures but lack 3′ poly (A) tails and instead end with a highly conserved seven-nucleotide sequence [34]. For group A RV strains, those that cause the majority of disease in humans and animals, the 33′ consensus sequence (33′CS) is UGUGACC. This stretch of nucleotides is an important functional element of viral +RNAs that are located in both the cytosol and the viroplasm. In particular, the 33′CSs of cytosolic +RNAs are bound by NSP3, a viral nonstructural protein that is proposed to serve as a surrogate of cellular poly (A) binding protein and enhance the translation of RV transcripts in the infected host [41–46]. For viral +RNAs being packaged and used as templates for genome replication in the viroplasm, the 3′CS serves as a critical polymerase recognition element [31, 36, 37, 47]. Binding of the 3′CS by VP1 in context of a PC presumably facilitates the incorporation of +RNAs into core particles [17]. This interaction is also predicted to temporally regulate initiation of minus-strand synthesis [17, 37, 48]. Specifically, VP1 engages the 3′CS in a manner that holds the terminal end of the +RNA out-of-register with the enzyme’s active site, thereby producing a stable, catalytically-inactive complex [37]. It has been hypothesized that the auto-inhibited polymerase is subsequently activated when engaged by VP2 during packaging of the VP1-bound +RNAs [17, 37, 48]. Following VP1 activation, the 3′CS functions as a minimal promoter, supporting the de novo initiation of minus-strand synthesis within the confines of a core particle [36, 49].
Figure 3. Cis-acting elements of RV +RNAs.
The top cartoon schematic represents a linear RV +RNA molecule. The central open-reading frame (ORF) is shown in red, and the 5′ and 3′ untranslated regions (UTRs) are shown in black. A cap structure (gray) is at the 5′ end of the molecule, and the consensus sequence (UGUGACC) is at the 3′ end. Regions of the +RNA thought to be important for selective packaging are indicated using green brackets. The lower left cartoon schematic represents a hypothetical cytosolic +RNA that would be used as template for protein synthesis. A panhandle structure is formed by base-pairing of the 5′ and 3′ ends, and RNA-specific stem-loop(s) are thought to project from these regions. The 3′ terminus is predicted to be bound by the nonstructural protein NSP3, which itself interacts with eukaryotic initiation factor eIF4G. The NSP3-eIF4G interaction, along with 5′-3′ complementarily, is thought to cause the +RNA to be held in a circular conformation, which might be important for efficient translation by host ribosomes. The lower right cartoon schematic represents a hypothetical viroplasmic +RNA that is selectively packaged into cores and used as a template for genome replication. Similar to the cytoplasmic +RNA, a panhandle structure is formed by base-pairing of the 5′ and 3′ ends, and RNA-specific stem-loop(s) project from these regions. The extreme 3′ end of the template is accessible to the polymerase VP1 (pink sphere) as a single-stranded tail. The 5′ cap of the template is presumed to interact with a cap-binding site on VP1. The VP3 capping enzyme is shown as a purple sphere interacting with VP1. Regions of the folded, viroplasmic +RNA thought to be important for selective packaging are indicated using green brackets.
RV +RNAs are predicted to form complex secondary and tertiary structures in infected cells (Figure 3). RV +RNAs used for protein synthesis are likely held in a circular conformation by the interaction of NSP3 (bound to the 3′CS) with the 5′ cap-associated eukaryotic initiation factor (eIF4G) [43, 44, 46]. Furthermore, computer modeling and RNase mapping experiments suggest that viral +RNAs fold into panhandles via 5′ and 3′ base-pairing [50–52]. In the tertiary +RNA structure, the 3′CS extends from the panhandle as a single-stranded tail [50–52]. Mutations made to the 3′CS that induce base-pairing with the 5′ end of the +RNA template diminish its capacity to be replicated in vitro [50]. Therefore, one function of the 5′-3′ panhandle could be to stabilize the 3′CS in a manner that is sterically accessible to the polymerase. The stable association of the 3′CS tail with VP1 might be further enhanced by recruitment of the 5′ cap of the folded +RNA to a cap-binding site on the polymerase [37].
In addition to roles in genome replication, the predicted panhandle structures of RV +RNAs are likely to contain important functional elements for assortment and packaging. With the exception of the extreme terminal nucleotides, the eleven species of viral +RNA share no homology to each other. Nonetheless, when comparing the same RNA segment of different group A RV strains, the 5′ and 3′ UTRs and neighboring ORF regions show high levels of nucleotide sequence identity. These conserved nucleotides comprise the 5′-3′ panhandles of folded RV +RNAs. Stable stem-loops are predicted within these conserved regions (i.e. within the panhandle) and can be formed by sequences at either the 5′ or 3′ end [50–52]. The observation that the putative stem-loops differ between the eleven +RNAs suggests that they might function as assortment signals. RV variants have been identified whose genomes include segments that are missing part or nearly all of the ORF [53]. The mutant RNAs can still undergo assortment, packaging, and replication, demonstrating that internal ORF nucleotides are not necessary for these processes. Moreover, when passed at a high multiplicity of infection in cell culture, RVs containing genome segments with large duplications tend to appear [53]. This phenomenon indicates that the duplicated +RNAs have advantages over wild-type +RNAs during packaging, possibly due to repetition of cis-acting elements [54]. Still, the functional identification and validation of +RNA elements required for RV assortment awaits efficient complementary (c)DNA-based reverse genetic and/or in vitro packaging systems.
For the closely related Reoviridae family member, mammalian orthoreovirus (MRV), an infectious +RNA system has been used to identify important determinants of genome assortment and packaging [55]. In this system, the ORF of an individual MRV +RNA is replaced with that of the reporter chloramphenicol acetyltransferase (CAT). Thus, the CAT ORF is flanked by the 5′ and 3′ UTRs of the parental viral +RNA. When transfected into cells expressing the protein encoded by the deleted ORF (along with the nine other MRV +RNAs), the chimeric CAT RNA undergoes assortment, packaging, and replication. These recombinant MRVs are capable of mediating subsequent infections and expressing the reporter protein in complementing cells. With this approach, it has been shown that the 5′ UTR contains the specific packaging signals for at least three different MRV +RNAs (m1, s2, and l1) [56, 57]. Like those of RV, conserved 5′ and 3′ terminal sequences of MRV +RNAs are predicted to form 5′-3′ panhandle structures. It is possible that the 5′ packaging signals identified for MRV +RNAs are associated with unique stem-loops that protrude from their panhandles [57]. A similar strategy analyzing the assortment and packaging signals of the +RNAs of the Reoviridae member, bluetongue virus, indicates that they likewise are located at the ends of the RNA and include sequences of the 5′ and 3′ UTRs and adjoining regions of the ORF [58].
The cis-acting RV and MRV +RNA elements important for assortment and packaging may be analogous to those identified for influenza A virus [5]. The eight segments of influenza viral RNA (vRNA) share a common organization, with a long central coding region (in antisense) and relatively short 5′ and 3′ UTRs. The vRNA segments are separately coated by multiple copies of the viral nucleoprotein (NP) and a single copy of the heterotrimeric viral PC to create a ribonucleoprotein (RNP) [59]. Similar to RV +RNAs, the termini of influenza vRNAs are partially base-paired to form panhandle structures, which in turn form corkscrew-like shapes in the context of RNPs [5, 59]. RNA-specific packaging signals are predicted to reside in putative stem-loops that are located adjacent to, rather than within, the panhandle of each vRNA and formed by both UTR and terminal ORF nucleotides. The exact manner in which influenza assorts its eight RNPs remains unclear; however, the available data is most consistent with the notion that interactions among the RNA molecules of RNPs mediate this process [60]. It has been proposed that influenza A virus utilizes a concerted packaging approach, whereby the eight RNPs are first selectively assorted into a ‘genome complex’ prior to their incorporation into a budding virion [61].
The dsRNA bacteriophage Φ6 specifically recognizes its three polycistronic +RNAs (s, m, and l) via pac sequences located near the 5′ ends [62]. These pac sequences are contained within distinctive stem-loop structures and are required for assortment and packaging [6, 63–65]. In contrast to influenza and possibly RV +RNAs, the segment-specific pac sequences of Φ6 do not overlap with the ORFs, but instead reside entirely with the 5′ UTRs. Φ6 +RNAs are also thought not to directly interact with each other prior to encapsidation. Instead, the three +RNAs are each introduced, one-by-one, into a pre-formed core particle [64, 66]. The reaction is mediated by a hexameric, viral motor protein (P4) and is sequential in that the s segment is packaged first, followed by m and then l [64, 67, 68]. An empty core initially displays a binding site for s, leading to its recruitment and packaging. Thereafter, a conformational change occurs in the core that reveals a binding site for m. Again, only after packaging of the m segment is the binding site for l uncovered. Once the Φ6 +RNAs are packaged inside the core, internal polymerases convert them into the full-length dsRNA genome segments (S, M, and L) [6]. Forceful expansion of the core shell as a result of the complete packaging of all three +RNA segments triggers Φ6 dsRNA synthesis [65]. Thus, segmented RNA viruses all seem to utilize cis-acting RNA elements as assortment signals, although their overall assortment processes differ markedly.
RV proteins involved in assortment and packaging
The nature of the complexes within which RV +RNAs are assorted and packaged remains poorly understood. Fractionation or immunoprecipitation of RV-infected cells has allowed for the physical isolation of core replication intermediates (core RIs) that have the capacity to replicate endogenous +RNAs, recreating the eleven-segmented dsRNA genome in vitro [69–72]. Importantly, exogenously added RV +RNAs cannot be replicated by the isolated complexes, suggesting that they are captured post-assortment. Core RIs may represent a step in assembly just prior to the final maturation of a T=1 shell, as the +RNAs are accessible to degradation by single-strand specific RNases [73]. These post-assortment, partially-packaged +RNA complexes contain the viral proteins VP1, VP2, VP3, NSP2, and NSP5, any or all of which could play important roles in selective packaging for RV.
VP1, VP2, and VP3 are critical for core assembly and genome replication and are undoubtedly important for +RNA packaging as well. The VP2 core shell is hypothesized to form by the knitting together of twelve decamer units, which themselves are organized from five asymmetric VP2 dimers [14]. Protruding inward from each fivefold axis are hubs composed of VP2 amino-terminal residues [14]. Deletion of the fivefold hub does not prevent recombinant VP2 from forming core-like particles, but does abrogate VP1 and VP3 encapsidation into those assemblies [18]. Likewise, VP2 lacking amino-terminal residues is not capable of supporting efficient VP1-mediated minus-strand synthesis in vitro, suggesting that the fivefold hub plays a role in polymerase activation [35]. Indeed, these biochemical data are consistent with the idea that VP2 fivefold hubs may provide scaffolds upon which PCs function. Still, if VP1 binding of +RNA precedes interactions between the PC and the core shell hub, it is also possible that this VP2 structure plays an indirect role in genome packaging. VP1 is the only known viroplasm-associated protein capable of specifically recognizing RV +RNAs, supporting the notion that this interaction may contribute to selective packaging [36, 37, 47].
Because NSP2 and NSP5 together induce and maintain viroplasms, they are critical for genome assortment, packaging, and replication. However, these nonstructural proteins might also play more direct roles, facilitating or even mediating the selective packaging of the eleven RV +RNAs [32]. NSP2 is an octameric protein with strong affinity for single-stranded RNA [74, 75]. This protein has helix unwinding and NTPase activities that are not essential for viroplasm formation, but are critical for virus replication [32, 76, 77]. The helix unwinding activity of NSP2 might help organize +RNAs for packaging and replication by removing interfering secondary structures. The NTPase activity of NSP2 suggests that this protein could function as a molecular motor, facilitating the insertion of +RNAs into assembling cores in a manner similar to the P4 protein of Φ6 [74]. Compared to NSP2, less is known about NSP5 and its putative role in selective packaging. NSP5 is a dimeric, serine/threonine-rich protein that, due to varying degrees of phosphorylation, exists as multiple isoforms in infected cells [78]. The functional significance of NSP5 phosphorylation remains speculative, as it is unconnected to viroplasm formation [79]. Like NSP2, NSP5 interacts with RNA in a nonspecific manner [80]. These proteins also bind to each other; NSP5 competes with RNA for binding to the tetramer-tetramer grooves on the NSP2 octamer [81]. Additionally, there is biochemical evidence suggesting that NSP2 and NSP5 bind VP1 and VP2 [82, 83]. Such interactions likely aid in the recruitment or retention of core proteins in the viroplasm, but could also serve to orchestrate the sequence of events required to assemble progeny cores. For example, during infection, the self-assembly tendency of VP2 must be suppressed, possibly via NSP2/NSP5 interactions, until appropriate packaging and replication events have occurred [84–86].
Models of RV +RNA assortment and packaging
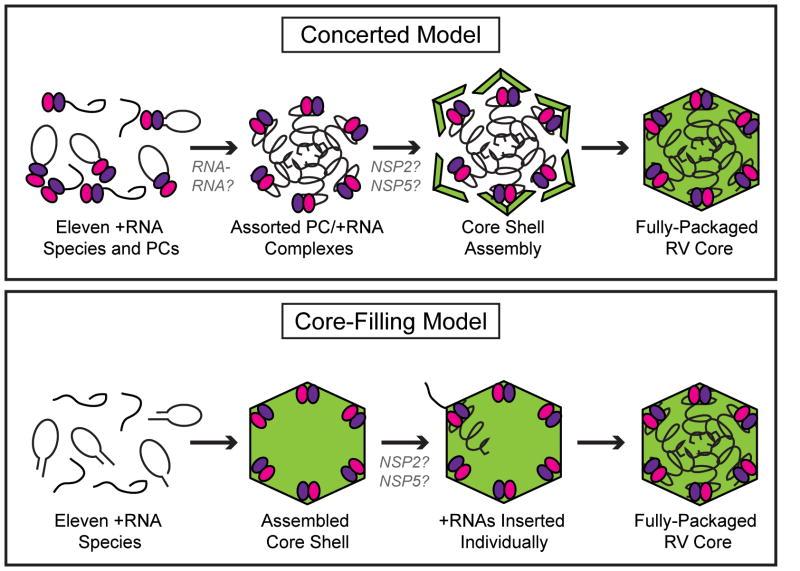
We propose two models of selective +RNA packaging by RV, concerted or core-filling, based on the strategies used by influenza A virus or Φ6, respectively (Figure 4). In the concerted packaging model, assortment is mediated by RNA-RNA interactions and happens prior to genome packaging. For influenza, vRNAs in the context of RNPs undergo assortment before they acquire an envelope by budding from the plasma membrane. Interactions among the eight vRNA molecules via cis-acting stem-loop structures mediate RNA specificity. In the concerted packaging model for RV, each +RNA species made by DLPs within the viroplasm is first bound by a single PC as a result of VP1’s affinity for the 5′ cap and 3′CS. Aided by interactions with NSP2 and NSP5, the eleven unique PC/+RNAs then undergo assortment in the absence of VP2. In support of this model, complexes have been isolated from RV-infected cells that contain VP1, VP3, NSP2, NSP5 and single-stranded RNA molecules [69, 71]. The existence of such pre-core RIs, and their capacity to be chased into more complex viral structures, suggests that +RNA recognition by PCs precedes VP2 association [69]. Even more, the affinity of VP1 for RV +RNA, and the manner in which this interaction creates a catalytically-inactive complex, also supports the concerted packaging model [37]. It is possible that the auto-inhibited binding of +RNAs by VP1 allows time for the eleven replication templates to find each other prior to core shell encapsidation. RV RNA specificity in the concerted packaging model, similar to that for influenza, is mediated by RNA-RNA interactions involving stem-loop structures within the 5′-3′ terminal panhandle of the +RNA molecules. Following assortment, the VP2 core shell assembles around the PC/+RNAs, in turn creating a loosely-packaged core RI [69, 71, 73]. A precise, but as of yet unidentified, interaction occurs between VP1 and VP2 that leads to enzymatic activation of the polymerase [35]. Minus-strand synthesis creates the eleven dsRNA genome segments inside of a pre-virion particle. However, it is not known whether core RIs become fully mature T=1 cores prior to or during genome replication. In vitro studies have shown that core RIs decrease in size and become RNase resistant as genome replication proceeds [73]. These data suggest that the condensation of VP2 into a tightly closed T=1 core shell occurs during minus-strand synthesis. Portions of the +RNA template may extend from the outer surface of the assembling VP2 core and be drawn into the particle as they are replicated.
Figure 4. Models of RV +RNA assortment and packaging.
Two models of selective +RNA packaging, concerted or core-filling, are proposed for RV based on the strategies used by influenza A virus or Φ6, respectively. In the concerted packaging model (top), eleven species of RV +RNAs (black lines) are bound by PC components VP1 (pink spheres) and VP3 (purple spheres). These PC/+RNAs undergo assortment via gene-specific interactions among the RNA molecules. A VP2 shell (green) then assembles around the assorted PC/+RNAs to create a fully packaged, and replication-competent core. NSP2 and NSP5 may function to regulate the timing of core assembly. In the core-filling model (bottom), a VP2 shell (green) containing internally tethered PCs (pink and purple spheres), but lacking nucleic acid, first assembles. Each of the eleven +RNAs (black lines) are then individually inserted into the core, possibly by the functions of NSP2 and/or NSP5. Complete packaging triggers core expansion and initiation of genome replication. The cartoons are meant to illustrate the order of events and not the nature of protein and RNA interactions.
In contrast to concerted packaging, the core-filling model dictates that the eleven +RNAs undergo ordered insertion into a pre-formed protein shell, similar to the strategy used by some bacteriophages. For Φ6, core particles containing viral polymerases are able to self-assemble in vitro. When incubated in the presence of the packaging motor protein P4, Φ6 +RNAs are introduced into the core through a portal located at a specific fivefold axis. Following the introduction of each of the three +RNAs in a sequential manner, an expansion of the viral capsid takes place; this conformational change in the core shell induces activation of the internal polymerases. Φ6 minus-strand synthesis occurs inside of a particle that is the morphogenic precursor to a mature virion. In the core-filling model for RV, intact but empty T=1 core shells with internally-tethered PCs, assemble de novo in viroplasms. Through the NTPase activity of NSP2, each of the eleven +RNAs are individually translocated into the core shells. In this manner, RNA selection is mediated not by interactions among the +RNAs, but by the specific binding of viral protein(s). For example, packaging of one +RNA could expose a binding site for the specific packaging of the next +RNA. Following introduction of the eleven +RNAs, core expansion possibly triggers the PCs to initiate minus-strand synthesis to generate the dsRNA genome.
There are several problems in suggesting a core-filling packaging model for RV. For instance, although empty virus particles can be recovered from infected cells [25], no data has been reported indicating that such particles are precursors of infectious virions, nor have particles been recovered that contain partially packaged or replicated genomes. Moreover, although empty core-like particles can be formed by co-expression of VP1, VP2, and VP3, there is no experimental data suggesting that such particles can be packaged by +RNA in vitro, even in the presence of NSP2 [18, 84]. In addition, RNA interference (RNAi) experiments have revealed that knockdown of VP1 expression in infected cells leads to the accumulation of empty particles [25], a result that is not consistent with the core-filling model, in which VP1 has no anticipated role in +RNA translocation into the core. It also seems unreasonable to propose that the NSP2 octamer is the equivalent of the Φ6 P4 packaging motor. In particular, while +RNA moves into the Φ6 core through a central hole located in the P4 hexamer [67, 68], the RNA-binding sites are located on the surface of the NSP2 octamer, suggesting that NSP2 is not a functional homolog of P4 [81]. Finally, in this packaging model, it is difficult to image how efficient segment recognition could be achieved by the sequential exposure of eleven separate +RNA interaction sites on the core. Notably, +RNAs of partially-packaged RV cores (i.e. core RIs) are RNase-sensitive, but those of partially-packaged Φ6 cores are RNase-resistant [86]. This biochemical observation suggests that even if RV utilizes a core-filling model, significant differences exist when compared with Φ6.
Concluding remarks and future directions
One of the most interesting topics of RV biology is the mechanism by which the virus assorts, packages, and replicates its segmented genome. The evidence to date is most consistent with a concerted packaging model, whereby the eleven +RNAs interact with each other using cis-acting sequence and structural elements prior to encapsidation within a core. Regardless of the mechanism, packaging a segmented genome complicates the assembly process for RV relative to other viruses whose genomes consist of a single piece of nucleic acid. Even so, genome segmentation confers distinct evolutionary advantages, as it affords the opportunity for reassortment during co-infection. In particular, segment exchanges could allow RVs to acquire advantageous genes and thus to rapidly adapt to selective pressures. Yet, reassortment between divergent strains requires that critical protein-protein interactions be maintained during viral replication (Box 1). Recent large-scale genomics studies suggest that co-circulating human RVs do not freely reassort genome segments, but instead have preferred RNA sets (i.e. genome constellations) [87]. It is possible that the functional constraints imposed by viral protein and RNA interactions during replication represent important determinants of viral fitness, thereby affecting whether or not a reassortant will appear in the human population [88, 89]. Future research is necessary, not only to define the RNA and protein interactions critical for RV assortment and packaging, but also to elucidate the limitations on segment exchange between genetically-divergent strains (Box 2). Efficient single-gene reverse genetic methods that are now being developed for RVs will undoubtedly provide a scientific platform for such studies.
Box 1. Restrictions on RV segment reassortment.
The capacity of RVs to reassort their genome segments during co-infection is predicted to be a major driving force in the evolution of this pathogen. Yet, there exist certain RV strain combinations that appear incapable of reassortment under experimental conditions and in nature. In particular, there has been no demonstration of gene reassortment between RV strains belonging to different serogroups (A–G). Although the reason for the observed restriction is not known, it is possible that multiple determinants, both direct and indirect, prevent intergroup genetic exchange. Direct determinants refer to the capacity of ‘foreign’ RV +RNA to be packaged and replicated by another group’s proteins. For example, co-infection of a host cell with a group A and C RV must provide an opportunity for physical mixing of their gene segments in common viroplasms, the stable interaction of their +RNA and core proteins during assortment and packaging, and an ability of their polymerases to replicate each other template RNAs. Indirect determinants of gene reassortment restriction relate to the capacity of the exchanged genes to function together in the new strain. Emerging evidence suggests that, while group A RVs readily undergo reassortment events, there are selection pressures for the maintenance of certain sets of genes (i.e. preferred genome constellations) [87, 88]. One of these pressures relates to how well the proteins encoded by the new RV interact during subsequent rounds of replication. For group A RV reassortants, the pressures are subtle and might be seen during the evolution of circulating viruses. In contrast, because RV groups encode very divergent proteins, intergroup reassortants that break preferred constellations might not be capable of carrying on a productive infection [89]. Given that gene reassortment has the potential to create novel and possibly more pathogenic RV strains, future studies in this area are warranted.
Box 2. Questions for future research.
Do RVs utilize a concerted or core-filling approach to assortment and packaging?
Where are the packaging signals located in RV +RNAs?
What are the functions of viral proteins (VP1, VP2, VP3, NSP2, and NSP5) during the assortment, packaging, and replication of the eleven-segmented RV genome?
Do RNA-RNA or protein-RNA interactions drive RV +RNA assortment?
What are the molecular determinants of RV gene reassortment restriction?
How does gene reassortment contribute to RV diversity and evolution?
Acknowledgments
We would like to thank Michelle Arnold, Kristen Ogden, Aitor Navarro, and Shane Trask for editorial advice and critical reading of the manuscript. SMM and JTP are supported by the Intramural Research Program of the NIH National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luque D, et al. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci USA. 2009;106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hundley F, et al. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology. 1985;143:88–103. doi: 10.1016/0042-6822(85)90099-6. [DOI] [PubMed] [Google Scholar]
- 3.Joklik WK, Roner MR. What reassorts when reovirus genome segments reassort? J Bio Chem. 1995;270:4181–4184. doi: 10.1074/jbc.270.9.4181. [DOI] [PubMed] [Google Scholar]
- 4.Patton JT. Evidence for equimolar synthesis of double-stranded RNA and minus- strand RNA in rotavirus-infected cells. Virus Res. 1990;17:199–208. doi: 10.1016/0168-1702(90)90065-j. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson EC, et al. Genome packaging in influenza A virus. J Gen Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 6.Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage phi6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Parashar UD, et al. Global mortality associated with rotavirus disease among children in 2004. J Infect Dis. 2009;200:S9–S15. doi: 10.1086/605025. [DOI] [PubMed] [Google Scholar]
- 8.Prasad BV, et al. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 9.Yeager M, et al. Three-dimensional structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, et al. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pesavento JB, et al. Rotavirus proteins: structure and assembly. Curr Top Microbiol Immunol. 2006;309:189–219. doi: 10.1007/3-540-30773-7_7. [DOI] [PubMed] [Google Scholar]
- 13.Estes MK, Kapikian AZ. Fields Virology. 5. Lippincott Williams and Wilkens; Philadelphia: 2007. Rotaviruses; pp. 1917–1974. [Google Scholar]
- 14.McClain B, et al. X-ray crystal structure of the rotavirus inner capsid particle at 3. A resolution. J Mol Biol. 2010;397:587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad BV, et al. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 16.McDonald SM, Patton JT. Viral Genome Replication. Springer Science+Business Media; New York: 2009. Core-associated genome replication mechanisms of dsRNA viruses; pp. 201–224. [Google Scholar]
- 17.Guglielmi KM, et al. Mechanism of intra-particle synthesis of the rotavirus double-stranded RNA genome. J Biol Chem. 2010;24:18123–18128. doi: 10.1074/jbc.R110.117671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng CQ, et al. The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. J Virol. 1998;72:201–208. doi: 10.1128/jvi.72.1.201-208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker M, Prasad BV. Rotavirus cell entry. Curr Top Microbiol Immunol. 2010;343:121–148. doi: 10.1007/82_2010_34. [DOI] [PubMed] [Google Scholar]
- 20.Lopez S, Arias CF. Early steps in rotavirus cell entry. Curr Top Microbiol Immunol. 2006;309:39–66. doi: 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]
- 21.Patton JT, et al. Replication and transcription of the rotavirus genome. Curr Pharm Des. 2004;10:3769–3777. doi: 10.2174/1381612043382620. [DOI] [PubMed] [Google Scholar]
- 22.Lawton JA, et al. Mechanism of genome transcription in segmented dsRNA viruses. Adv Virus Res. 2000;55:185–229. doi: 10.1016/S0065-3527(00)55004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stacy-Phipps S, Patton JT. Synthesis of plus- and minus-strand RNA in rotavirus-infected cells. J Virol. 1985;61:3479–3484. doi: 10.1128/jvi.61.11.3479-3484.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson MA, McCrae MA. Molecular biology of rotaviruses: quantitative analysis of regulation of gene expression during virus replication. J Virol. 1989;63:2048–2055. doi: 10.1128/jvi.63.5.2048-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayala-Breton C, et al. Analysis of the kinetics of transcription and replication of the rotavirus genome by RNA interference. J Virol. 2009;83:8819–8831. doi: 10.1128/JVI.02308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen J. Ribonucleic acid polymerase activity associated with purified calf rotavirus. J Gen Virol. 1977;36:395–402. doi: 10.1099/0022-1317-36-3-395. [DOI] [PubMed] [Google Scholar]
- 27.Cohen J, Dobos P. Cell free transcription and translation of rotavirus RNA. Biochem Biophys Res Commun. 1979;88:791–796. doi: 10.1016/0006-291x(79)91477-3. [DOI] [PubMed] [Google Scholar]
- 28.Spencer E, Arias ML. In vitro transcription catalyzed by heat-treated human rotavirus. J Virol. 1981;40:1–10. doi: 10.1128/jvi.40.1.1-10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawton JA, et al. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struc Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 30.Liu M, et al. Rotavirus VP3 expressed in insect cells possesses guanylyltransferase activity. Virology. 1992;188:77–84. doi: 10.1016/0042-6822(92)90736-9. [DOI] [PubMed] [Google Scholar]
- 31.Silvestri LS, et al. Rotavirus replication: plus-sense templates for double- stranded RNA synthesis are made in viroplasms. J Virol. 2004;78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taraporewala ZF, Patton JT. Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae. Virus Res. 2004;101:57–66. doi: 10.1016/j.virusres.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Patton JT, et al. Rotavirus genome replication and morphogenesis: role of the viroplasm. Curr Top Microbiol Immunol. 2006;309:169–187. doi: 10.1007/3-540-30773-7_6. [DOI] [PubMed] [Google Scholar]
- 34.Patton JT, et al. Coupling of rotavirus genome replication and capsid assembly. Adv Virus Res. 2007;69:167–201. doi: 10.1016/S0065-3527(06)69004-0. [DOI] [PubMed] [Google Scholar]
- 35.Patton JT, et al. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tortorici MA, et al. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J Biol Chem. 2003;278:32673–32682. doi: 10.1074/jbc.M305358200. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez RA, et al. Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch Virol. 2000;145:1963–1973. doi: 10.1007/s007050070069. [DOI] [PubMed] [Google Scholar]
- 39.Jourdan N, et al. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J Virol. 1997;71:8268–8278. doi: 10.1128/jvi.71.11.8268-8278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell DB, Both GW. Completion of the genomic sequence of the simian rotavirus SA11: nucleotide sequences of segments 1, 2, and 3. Virology. 1990;177:324–331. doi: 10.1016/0042-6822(90)90487-c. [DOI] [PubMed] [Google Scholar]
- 41.Poncet D, et al. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO. 1994;13:4165–4173. doi: 10.1002/j.1460-2075.1994.tb06734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poncet D, et al. Structure and function of rotavirus nonstructural protein NSP3. Arch Virol. 1996;12:29–35. doi: 10.1007/978-3-7091-6553-9_4. [DOI] [PubMed] [Google Scholar]
- 43.Piron M, et al. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vende P, et al. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J Virol. 2000;74:7064–7071. doi: 10.1128/jvi.74.15.7064-7071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deo RC, et al. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell. 2002;108:71–81. doi: 10.1016/s0092-8674(01)00632-8. [DOI] [PubMed] [Google Scholar]
- 46.Groft CM, Burley SK. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- 47.Patton JT. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald SM, et al. The ins and outs of four-tunneled Reoviridae RNA- dependent RNA polymerases. Curr Opin Struct Biol. 2009;19:775–782. doi: 10.1016/j.sbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wentz MJ, et al. The 3−-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D, Patton JT. Rotavirus RNA replication requires a single-stranded 3− end for efficient minus-strand synthesis. J Virol. 1998;72:7387–7396. doi: 10.1128/jvi.72.9.7387-7396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, et al. Genomic analysis of codon, sequence and structural conservation with selective biochemical-structure mapping reveals highly conserved and dynamic structures in rotavirus RNAs with potential cis-acting functions. Nuc Acids Res. 2010 doi: 10.1093/nar/gkq663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tortorici MA, et al. A base-specific recognition signal in the 5− consensus sequence of rotavirus plus-strand RNAs promotes replication of the double-stranded RNA genome segments. RNA. 2005;12:133–146. doi: 10.1261/rna.2122606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desselberger U. Genome rearrangements of rotaviruses. Arch Virol Suppl. 1996;12:37–51. doi: 10.1007/978-3-7091-6553-9_5. [DOI] [PubMed] [Google Scholar]
- 54.Troupin C, et al. Rearranged genomic RNA segments offer a new approach to the reverse genetics of rotaviruses. J Virol. 2010;84:6711–6719. doi: 10.1128/JVI.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roner MR, Joklik WK. Reovirus reverse genetics: Incorporation of the CAT gene into the reovirus genome. Proc Natl Acad Sci USA. 2001;98:8036–8041. doi: 10.1073/pnas.131203198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roner MR, Steele BG. Localizing the reovirus packaging signals using an engineered m1 and s2 ssRNA. Virology. 2007;358:89–97. doi: 10.1016/j.virol.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 57.Roner MR, Steele BG. Features of the mammalian orthoreovirus Dearing l1 single-stranded RNA that direct packaging and serotype restriction. J Gen Virol. 2007;88:3401–3412. doi: 10.1099/vir.0.83209-0. [DOI] [PubMed] [Google Scholar]
- 58.Matsuo E, Roy P. Bluetongue virus VP6 acts early in the replication cycle and can form the basis of chimeric virus formation. J Virol. 2009;83:8842–8848. doi: 10.1128/JVI.00465-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheung TK, Poon LL. Biology of influenza A virus. Ann NY Acad Sci. 2007;1102:1–25. doi: 10.1196/annals.1408.001. [DOI] [PubMed] [Google Scholar]
- 60.Fujii Y, et al. Selective incorporation of influenza virus RNA segments into virions. Proc Natl Acad Sci USA. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noda T, et al. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 62.Gottlieb P, et al. Identification of the packaging regions within the genomic RNA segments of bacteriophage phi6. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- 63.Pirttimaa MJ, Bamford DH. RNA secondary structures of the bacteriophage phi6 packaging regions. RNA. 2000;6:880–889. doi: 10.1017/s1355838200992598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mindich L. Precise packaging of the three genomic segments of the double- stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huiskonen JT, et al. Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure. 2006;14:1039–1048. doi: 10.1016/j.str.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 66.Frilander M, Bamford DH. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage phi6: the three segments modulate each other’s packaging efficiency. J Mol Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- 67.Kainov DE, et al. RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J Biol Chem. 2003;278:48084–48091. doi: 10.1074/jbc.M306928200. [DOI] [PubMed] [Google Scholar]
- 68.Pirttimaa MJ, et al. Nonspecific nucleoside triphosphatase P4 of double- stranded RNA bacteriophage phi6 is required for single-stranded RNA packaging and transcription. J Virol. 2002;76:10122–10127. doi: 10.1128/JVI.76.20.10122-10127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallegos CO, Patton JT. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–627. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 70.Patton JT, Gallegos CO. Structure and protein composition of the rotavirus replicase particle. Virology. 1988;166:358–365. doi: 10.1016/0042-6822(88)90506-5. [DOI] [PubMed] [Google Scholar]
- 71.Helmberger-Jones M, Patton JT. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology. 1986;155:655–665. doi: 10.1016/0042-6822(86)90225-4. [DOI] [PubMed] [Google Scholar]
- 72.Aponte C, et al. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2. J Virol. 1996;70:985–991. doi: 10.1128/jvi.70.2.985-991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patton JT, Gallegos CO. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J Gen Virol. 1990;71:1087–1094. doi: 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- 74.Jayaram H, et al. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature. 2002;417:311–315. doi: 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- 75.Schuck P, et al. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J Biol Chem. 2001;276:9679–9687. doi: 10.1074/jbc.M009398200. [DOI] [PubMed] [Google Scholar]
- 76.Taraporewala ZF, Patton JT. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J Virol. 2001;75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taraporewala Z, et al. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Afrikanova I, et al. Phosphorylation generates different forms of rotavirus NSP5. J Gen Virol. 1996;77:2059–2065. doi: 10.1099/0022-1317-77-9-2059. [DOI] [PubMed] [Google Scholar]
- 79.Carpio RV, et al. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J Biol Chem. 2004;279:10624–10633. doi: 10.1074/jbc.M311563200. [DOI] [PubMed] [Google Scholar]
- 80.Vende P, et al. RNA-binding activity of the rotavirus phosphoprotein NSP5 includes affinity for double-stranded RNA. J Virol. 2002;76:5291–5299. doi: 10.1128/JVI.76.10.5291-5299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang X, et al. Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: implications for genome replication. J Virol. 2006;80:10829–10835. doi: 10.1128/JVI.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arnoldi F, et al. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J Virol. 2007;81:2128–2137. doi: 10.1128/JVI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berois M, et al. Rotavirus nonstructural protein NSP5 interacts with major core protein VP2. J Virol. 2003;77:1757–1763. doi: 10.1128/JVI.77.3.1757-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeng CQ, et al. Characterization of rotavirus VP2 particles. Virology. 1994;201:55–65. doi: 10.1006/viro.1994.1265. [DOI] [PubMed] [Google Scholar]
- 85.Vende P, et al. Rotavirus NSP2 interferes with the core lattice protein VP2 in initiation of minus-strand synthesis. Virology. 2003;313:261–273. doi: 10.1016/s0042-6822(03)00302-7. [DOI] [PubMed] [Google Scholar]
- 86.Patton JT, Spencer E. Genome replication and packaging of segmented double-stranded RNA viruses. Virology. 2000;277:217–225. doi: 10.1006/viro.2000.0645. [DOI] [PubMed] [Google Scholar]
- 87.McDonald SM, et al. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog. 2009;5:e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heiman EM, et al. Group A human rotavirus genomics: evidence that gene constellations are influenced by viral protein interactions. J Virol. 2008;82:11106–11116. doi: 10.1128/JVI.01402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McDonald SM, et al. Shared and group-specific features of the rotavirus RNA polymerase reveal potential determinants of gene reassortment restriction. J Virol. 2009;83:6135–6148. doi: 10.1128/JVI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]