Abstract
Historically, consumption of Green tea (Camellia sinensis) has been associated with health benefits against multiple diseases including cancer, atherosclerosis and cardiovascular disorders. Emerging evidence has suggested a pathogenic role for HMGB1, a newly identified “late” mediator of lethal systemic inflammation, in the aforementioned diseases. Here we demonstrated that a major ingredient of Green tea, EGCG, was internalized into HMGB1-containing LC3-positive cytoplasmic vesicles (likely autophagosomes) in macrophages, and induced HMGB1 aggregation in a time-dependent manner. Furthermore, EGCG stimulated LC3-II production and autophagosome formation, and inhibited LPS-induced HMGB1 up-regulation and extracellular release. The EGCG-mediated HMGB1 inhibitory effects were diminished by inhibition of class III phosphatidylinositol-3 kinase (with 3-methyladenine) or knockdown of an essential autophagy-regulating protein, beclin-1. Moreover, the EGCG-mediated protection against lethal sepsis was partly impaired by co-administration of an autophagy inhibitor, chloroquine. Taken together, the present study has suggested a possibility that EGCG inhibits HMGB1 release by stimulating its autophagic degradation.
1. Introduction
As an evolutionarily conserved degradation process to maintain cellular homeostasis, macrophages degrade endogenous cytoplasmic macromolecules via autophagy (“self-eating”), and degrade exogenous pathogens using a special form of autophagy distinctly termed “xenophagy” [1,2]. Autophagy begins with the formation of double-membraned structures called phagophores, which elongate and engulf portions of the cytoplasm to form autophagosomes. Subsequently, autophagosomes fuse with lysosomes to form degradative autophagolysosomes, where the engulfed contents are degraded by acidic lysosomal hydrolases [2]. Autophagy normally occurs at basal levels in most tissues, but it can be further stimulated by starvation [3], pathogen-associated molecular patterns (PAMPs, such as endotoxin) [4], or cytokines (such as IFN-γ) [3]. Although it has been shown that xenophagy may be strategically employed by macrophages to destroy intracellular bacteria [5–7], the potential role of autophagy in human diseases or in the regulation of any specific disease mediators remains poorly understood [2,8].
Sepsis refers to a systemic inflammatory response syndrome resulting from a microbial infection, and is partly mediated by various PAMPs, which stimulate macrophages to sequentially release early (e.g., TNF) and late (e.g., HMGB1) proinflammatory mediators [9,10]. Although an appropriate early cytokine response may be essential against infection, excessive accumulation of late mediators (such as HMGB1) may adversely disrupt homeostasis, and contribute to the pathogenesis of lethal endotoxemia [10] and sepsis [11]. Indeed, substantial evidence has supported HMGB1 as an alarmin signal that serves to alert, recruit, and activate innate immune cells [12–14], thereby sustaining an injurious inflammatory response in sepsis [15,16]. Consistently, HMGB1-specific neutralizing antibodies [10,11] or inhibitors (e.g., tanshinones, ethyl pyruvate, nicotine, stearoyl lysophosphatidylcholine) [10,17–23] confer significant protection against lethal endotoxemia and sepsis, supporting HMGB1 as a critically important late mediator of lethal sepsis. In addition, HMGB1 has been implicated in the pathogenesis of a variety of other diseases including oxidative stress [24], atherosclerosis [25], cancer [26], and cardiovascular diseases [27].
Historically, herbal medicine has formed the basis of folk remedies for various ailments, but their mechanisms of action remain elusive. Brewed from the leaves of the plant, Camellia sinensis, Green tea has been associated with health benefits against multiple diseases including oxidative stress [28], atherosclerosis [28], cancer [29], and cardiovascular diseases [30]. These healing properties are attributable to its abundant polyphenolic catechins, such as epigallocatechin-3-gallate or EGCG (up to 50 mg in a single cup of tea) [31]. Recently, we discovered that Green tea [32] and its major component, EGCG [33], dose-dependently abrogated endotoxin-induced HMGB1 release, and consequently protected animals against lethal endotoxemia and sepsis [33]. However, the mechanism underlying EGCG-mediated HMGB1 inhibition was previously unknown. In the present study, we uncovered a possible mechanism by which EGCG effectively inhibits HMGB1 release by inducing its aggregation and autophagic degradation in macrophages.
2. Material and Methods
2.1. Cell culture
Murine macrophage-like RAW 264.7 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). GFP-LC3-transfected RAW 264.7 cells were established as previously described [4], and maintained in RPMI 1640/10%FBS/2 mM glutamine supplemented with puromycin (2 μg/ml; Sigma-Aldrich, St. Louis, MO, USA) to retain clonal homogeneity. Primary peritoneal macrophages were isolated from Balb/C mice (male, 7–8 weeks, 20–25 grams) at 3 days after intraperitoneal injection of 2 ml thioglycollate broth (4%) as previously described [18,19,34,35].
2.2. LPS stimulation
Adherent macrophages were gently washed with, and cultured in, serum-free OPTI-MEM I medium 2 h before stimulation with endotoxin (lipopolysaccharide, LPS, E. coli 0111:B4, Sigma-Aldrich, St. Louis, MO, USA). At 16 hours after LPS stimulation, intra- and extra-cellular HMGB1 levels were determined by Western blotting analysis as previously described [18,34]. To assess the involvement of autophagy in EGCG-mediated HMGB1 inhibition, autophagy was inhibited genetically by siRNA against beclin-1 or pharmacologically by 3-methyladenine. Epigallocatechin gallate (EGCG, C22H18O11), catechin gallate (CG, C22H18O10), catechin (C, C15H14O6), epigallocatechin (C15H14O7), ethyl gallate (C9H10O5), bafilomycin A1, and 3-methyladenine were obtained from Sigma-Aldrich (St. Louis, MO, USA).
2.3. EGCG intracellular trafficking
EGCG (10 mM) was oxidized by sodium meta-periodate (Thermo Scientific, Waltham, MA, USA; 10 mM in 1 × PBS pH 7.2, 10 min, room temperature), and subsequently incubated with EZ-Link biotin-LC-hydrazide (Thermo Scientific, Waltham, MA, USA; 20 mM, 2 h, 4° C). The resulting products were fractionated by HPLC using a Nova-pak C18 column (3.9 × 150 mm) and 0.065% trifluoroacetic acid (TFA, v/v, in water) as the mobile phase. The sample was eluted by a linear gradient of 0–59% acetonitrile (v/v, in 0.065% TFA) over 12 minutes at a flow rate of 1.0 ml/min, and monitored at a wavelength of 254 nm. Each HPLC peak was screened for activity in inhibiting LPS-induced HMGB1 release as previously described [33], and biologically active biotin-EGCG was used for intracellular trafficking studies. Briefly, RAW 264.7 cells were incubated with biotin-EGCG (10 μM) for various time periods, fixed with 4% paraformaldehyde, and respectively stained with streptavidin-conjugated Alexa fluor 594 (Invitrogen, Carlsbad, CA, USA) for EGCG, and DAPI (Vector Lab, Burlingame, CA, USA) for cell nuclei. Images were captured using a fluorescence microscope (Carl Zeiss Microimaging).
2.4. Visualization of autophagosomes
Murine macrophage-like RAW 264.7 cells stably transfected with GFP-LC3 were stimulated with LPS in the absence or presence of EGCG for 16 h, and cells were examined for the presence of GFP-LC3 punctate structures under a fluorescence microscope as previously described [4]. The ratio between the 18-kD cytosolic LC3-I and 16-kD lipidated autophagosome-bound LC3-II was determined by Western blotting analysis as previously described [4]. The autophagic flux was measured by evaluating the effects of EGCG on LC3-II turnover in the absence or presence of an autophagy inhibitor, bafilomycin A1. Specifically, macrophage cultures were stimulated with EGCG for 12 h, and bafilomycin A1 was added at various concentrations (0, 5, 25, 100, 200, 250 nM). At 4 h post bafilomycin A1 addition, cells were harvested and assayed for LC3 concentrations by Western blotting analysis. For human breast adenocarcinoma MDA-MB-361 and MCF-7 cancer cell lines, autophagic vacuoles were detected by staining with acidotropic dyes such as monodansylcadaverine (MDC) as previously described [36]. Briefly, cells were incubated with 0.05 mM MDC for 1 h (at 37°C), and fixed in 4% paraformaldehyde for 15 min. After extensive washing with 1×PBS, cells were observed under Nikon Mikrophot-FXA microscopy (with a 356-nm excitation filter and a 545-nm barrier filter).
2.5. Fluorescence Immunostaining
RAW 264.7 cells were stimulated with LPS (200 ng/ml) in the absence or presence of biotin-labeled EGCG (10 μM) for 16 h. Subsequently, cells were fixed with 2% formalin for 10 min, and permeabilized with 0.1% Triton X-100 in PBS (1 min, room temperature). After extensive washing with PBS, cells were stained with LAMP2-specific monoclonal antibody (Santa Cruz Biotech, Santa Cruz, CA, USA), or HMGB1-specific antigen-affinity purified polyclonal rabbit antibodies. Afterwards, cells were incubated with Alexa-488-conjugated donkey anti-mouse antibody (Invitrogen, Carlsbad, CA, USA), and Alexa-594-conjugated donkey anti-rabbit antibody (Invitrogen, Carlsbad, CA, USA), respectively.
2.6. Transmission Electron Microscopy
At 24 h post EGCG stimulation, cells were fixed in 1.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.0) for 2 h, postfixed in 2% osmium tetroxide for 2 h, dehydrated with increasing concentrations of ethanol, and gradually infiltrated with Araldite resin. Ultrathin sections (80 nm) were obtained using an ultramicrotome (RMC MT6000-XL). Sections were stained with uranyl acetate and lead citrate and examined using a Hitachi H-300 transmission electron microscope.
2.7. Preparation of recombinant HMGB1
The cDNA encoding for rat HMGB1 was cloned onto a pCAL-n vector, and recombinant HMGB1 was expressed in E. coli BL21 (DE3) pLysS cells as previously described [10]. Contaminating endotoxin was removed from the HMGB1 preparation by Triton X-114 extraction as previously described [12]. To determine whether EGCG binds to HMGB1 in aqueous solution, HMGB1 (100 μg/ml) was incubated with EGCG (10 μM) in 1 × PBS (pH 7.4, 37° C) in the absence or presence of DTT (0.65 mM), and subsequently assayed for protein aggregation by SDS-PAGE, NBT redox-cycling staining, or Western blot analysis.
2.8. Nitroblue tetrazolium staining
Following SDS-PAGE gel electrophoresis, proteins were transblotted onto a PVDF membrane, and stained with NBT (240 μM) in potassium glycinate (2.0 M, pH 10) as described previously [37].
2.9. HMGB1 Western blotting analysis
The levels of HMGB1 in whole-cell lysate, various cellular fractions, and the culture medium were determined by Western blotting analysis as previously described [10,18,19,34,35]. Cytoplasmic and nuclear fractions were isolated after selective lysis of the plasma membrane in low salt buffer (10 mM HEPES, pH 7.9; 10 mM KCl; 0.1 mM EDTA; 0.1 mM EGTA; 1 mM DTT; 0.5 mM PMSF, 1% NP-40) as previously described [18]. Each fraction was assayed for HMGB1 levels by Western blotting analysis with reference to a cytoplasmic protein (β-actin, Sigma-Aldrich, St. Louis, MO, USA) or a nuclear protein (PCNA, BD Biosciences, San Jose, CA, USA). HMGB1-specific polyclonal antibodies were generated in rabbits as previously described [10].
2.10. Cytokine antibody array
Murine cytokine antibody array (Cat. No. M0308003, RayBiotech Inc., Norcross, GA, USA) was used to determine the relative levels of 62 different cytokines in macrophage lysate as previously described [18].
2.11. RNA interference
siRNAs against beclin-1 (sc-29798) were obtained from the Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), and used according to the manufacturer’s instructions. Briefly, murine peritoneal macrophages were transiently transfected with beclin-1 siRNA (50 pmols/well) using siRNA Transfection Reagent (sc-29528, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 48 h. After transfection, Transfection Medium was replaced with tissue culture medium, and incubated for 12 h before stimulating with LPS in the absence or presence of EGCG. Beclin-1 expression levels were determined by Western blotting analysis using beclin-1-specific antibodies (sc-11427).
2.12. Animal models of endotoxemia and sepsis
This study was approved and performed in accordance with the guidelines for the care and use of laboratory animals at the Feinstein Institute for Medical Research, Manhasset, New York. Endotoxemia was induced in Balb/C mice (male, 7–8 weeks) by intraperitoneal injection of bacterial endotoxin (LPS, 10 mg/kg) as previously described [10,18,19,21]. Sepsis was induced in male Balb/C mice (7–8 weeks, 20–25 g) by cecal ligation and puncture (CLP) as previously described [18,19,38]. EGCG was orally administered at indicated doses and time points, and mice were monitored for survival for up to two weeks.
2.13. Statistical Analysis
Data are expressed as mean ± SD of at least 2–3 independent experiments (n = 2–3). One-way ANOVA was used for comparison among all different groups. When the ANOVA was significant, post-hoc testing of differences between groups was performed using Tukey’s test. The Kaplan-Meier method was used to compare the differences in mortality rates between groups. A P value < 0.05 was considered statistically significant.
3. Results
3.1. EGCG was internalized into LC3-, LAMP2-, and HMGB1-containing cytoplasmic vesicles
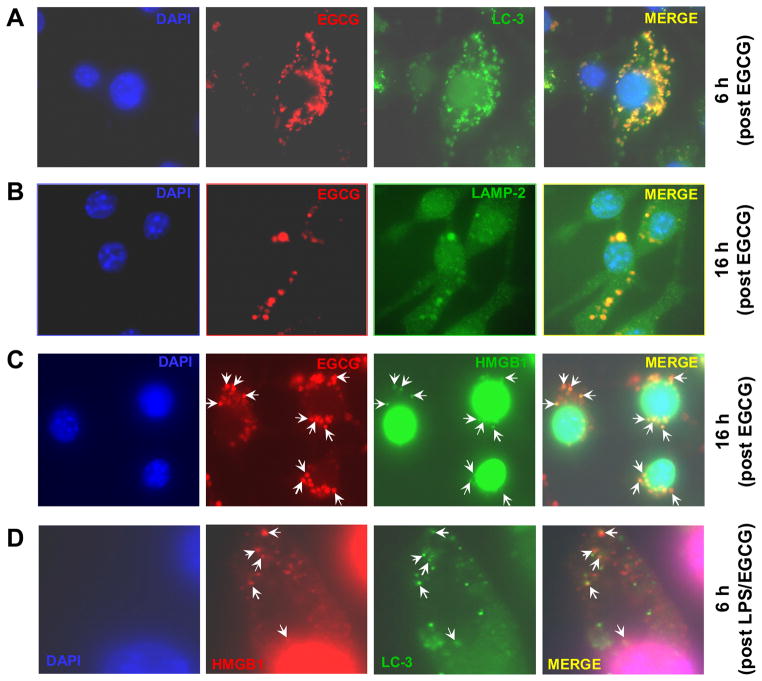
Previous studies have suggested divergent subcellular localizations for exogenous EGCG that include the cell membrane [39] and cytoplasmic vesicles [40]. To elucidate the mechanisms by which EGCG blocks HMGB1 nuclear-cytoplasmic translocation and extracellular release, we performed the trafficking experiment to determine the functional localization of EGCG in macrophage cultures. Consistent with previous reports [40], we found that EGCG was rapidly internalized into cytoplasmic vesicles within 6 h (Fig. 1A). Surprisingly, these EGCG-containing vesicles almost completely co-localized with LC3 punctate structures (likely autophagosomes, Fig. 1A). Subsequently, these EGCG-containing vesicles increased in size (within 12–16 h), and co-localized with the lysosomal-associated membrane protein 2 (LAMP2)-containing cytoplasmic compartments (Fig. 1B). These large LAMP2-positive compartments were likely autophagolysosomes – the fusion product of autophagosomes and lysosomes [41]. The observation that EGCG co-localized with two distinct cytoplasmic vesicles (the LC3-positive autophagosomes and LAMP2-positive autophagolysosomes) at two different time points (6 h and 16 h post EGCG treatment) suggests a persistent involvement of EGCG in the induction of autophagy.
Figure 1. EGCG was internalized into cytoplasmic vesicles containing LC3, LAMP2, and HMGB1.
A). EGCG was co-localized with LC3-containining vesicles. GFP-LC3-transfected RAW 264.7 cells were treated with biotin-labeled EGCG for 6 h, subsequently stained with streptavidin-conjugated Alexa-594 to visualize intracellular EGCG, and stained with DAPI to visualize the nuclei (blue). B). EGCG was co-localized with LAMP2-positive vesicles. RAW 264.7 cells were treated with biotin-labeled EGCG for 16 h, and stained with LAMP2-specific primary antibody and Alexa-488-conjugated secondary antibody. C). EGCG co-localized with HMGB1-containing cytoplasmic vesicles. RAW 264.7 cells were stimulated with LPS in the presence of biotin-EGCG for 16 h, and stained HMGB1-specific antibodies, and Alexa-488-conjugated donkey anti-rabbit antibody. D). Cytoplasmic HMGB1 “puncta” were partly co-localized with LC3-containining vesicles. GFP-LC3-transfected RAW 264.7 cells were stimulated with LPS and EGCG for 6 h, and sequentially incubated with HMGB1-specific and Alexa-594-conjugated donkey anti-rabbit antibodies. Note that cytoplasmic HMGB1 “puncta” (Red) partly co-localized with LC3-containing cytoplasmic vesicles (Green).
To determine whether cytoplasmic EGCG co-localized with HMGB1, we stained LPS-stimulated macrophages with HMGB1-specific antibodies. Consistent with a previous report [18], cytoplasmic HMGB1-positive “puncta” were observed in LPS-stimulated macrophage cultures (Fig. 1C). Interestingly, these HMGB1-containing “puncta” appeared to co-localize with some EGCG-containing vesicles (Fig. 1C). Although co-localization of HMGB1 “puncta” and GFP-LC3 vesicles was not found in LPS-stimulated macrophages (in the absence of EGCG, data not shown), some cytoplasmic HMGB1 “puncta” were co-localized with GFP-LC3 vesicles at 6 h post co-stimulation with LPS and EGCG (Fig. 1D). Taken together, these experimental data support a possibility that EGCG interacts with HMGB1 within autophagosomes of LPS-stimulated macrophages.
3.2. EGCG induced HMGB1 aggregation in vitro
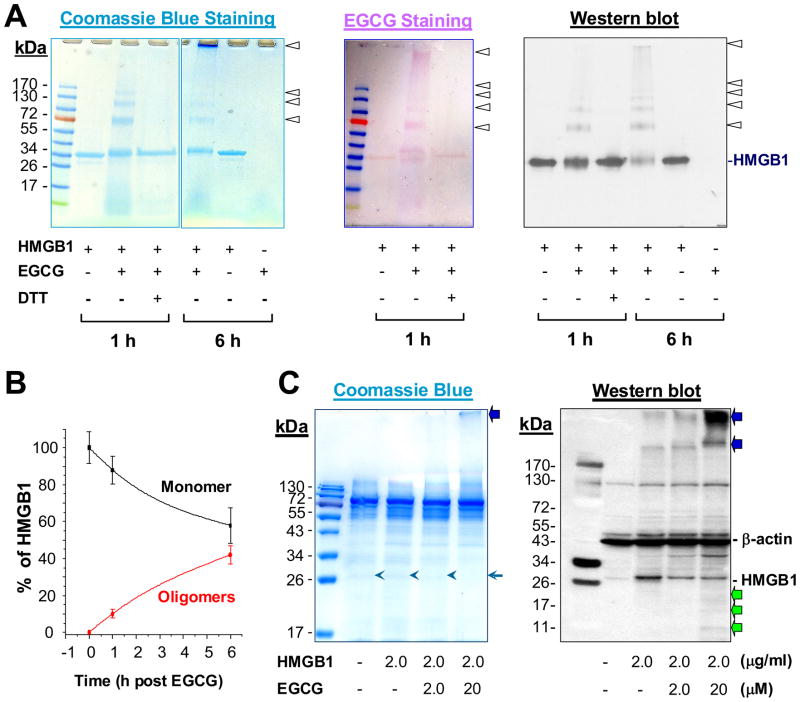
To test this possibility, highly purified HMGB1 protein was incubated with EGCG for various time periods, and subjected to SDS-PAGE analysis. In the absence of EGCG, recombinant HMGB1 migrated as a ~ 33 kDa band on SDS-PAGE gels (Fig. 2A, left panel). Following incubation with EGCG, however, additional bands with higher molecular weights (MW, 66 kDa, 99 kDa, 132 kDa, etc) were observed (Fig. 2A, left panel), indicating that EGCG induced the formation of SDS-resistant HMGB1 aggregates.
Figure 2. EGCG induced oxidative HMGB1 aggregation and potential degradation.
A). EGCG induced HMGB1 aggregation in vitro. Highly purified HMGB1 protein was incubated with EGCG in the absence or presence of DTT for indicated time periods, resolved on SDS-PAGE gel, and stained by Coomassie blue (Left Panel), NBT (Middle Panel), or anti-HMGB1 antibodies (Western blotting) (Right Panel). B). Quantitative analysis of EGCG-induced HMGB1 aggregation. The relative level of HMGB1 monomer and oligomers were determined by densitometry, and expressed as % of total HMGB1 protein. C). EGCG induced HMGB1 aggregation and degradation in vivo. RAW 264.7 cells were incubated with highly purified HMGB1 in the absence or presence of EGCG for 16 h, and cellular protein was subjected to SDS-PAGE (Left Panel), or Western blotting analysis (Right Panel). Note that EGCG caused formation of higher MW aggregation (blue arrows) and lower MW degradation products (green arrows).
To elucidate the nature of EGCG-HMGB1 interaction, we adopted a redox-cycling nitroblue tetrazolium (NBT) staining method to detect protein-bound EGCG molecules [37]. Superimposable with the Coomassie blue staining profile (Fig. 2A, left), NBT staining revealed multiple purple bands with identical MW (66 kDa, 99 kDa, 132 kDa) (Fig. 2A, middle panel). Notably, the EGCG-induced protein aggregation was completely prevented by a reducing agent, 1, 4- dithiothreitol (DTT) (Fig. 2A, left and middle panel), indicating that EGCG induces HMGB1 aggregation likely via an oxidative reaction. The identities of these HMGB1-EGCG complexes were confirmed by Western blotting analysis (Fig. 2A, right panel), which revealed multiple superimposable bands as shown by Coomassie blue or NBT staining. Densitometric analysis revealed that EGCG converted 10–40% of the HMGB1 monomer into oligomers within 1–6 h of incubation (Fig. 2B). Taken together, these data showed that EGCG can induce HMGB1 aggregation in a time-dependent manner.
To gain insight into the structure-function relationship, we also examined several other catechin derivatives [such as catechin (C), epigallocatechin (EGC), catechin gallate (CG), and ethyl gallate (G)] for similar activities. Consistent with previous observations that C or G failed to inhibit LPS-induced HMGB1 release [33], they similarly failed to induce HMGB1 aggregation (data not shown). In agreement with its capacities to inhibit LPS-induced HMGB1 release, CG also dramatically induced HMGB1 aggregation (data not shown). These data suggest that functional groups of both catechins and gallate are needed for EGCG or CG-mediated HMGB1 aggregation.
3.3. EGCG induced HMGB1 degradation in vivo
To determine whether EGCG similarly induces HMGB1 aggregation in vivo, highly purified HMGB1 was supplemented to macrophage cultures in the absence or presence of EGCG, and cell lysate was subsequently assayed for HMGB1 aggregation by Western blotting analysis. In vivo, stimulation with HMGB1 (2 μg/ml) led to a dramatic increase in the intensity of a 30 kDa band (Fig. 2C), which was recognized by HMGB1-specific antibodies (Fig. 2C). In the presence of EGCG, however, the intensity of this 30 kDa band was dramatically reduced (Fig. 2C), which was accompanied by the appearance of a number of larger (> 170 kDa) and smaller (< 26 kDa) bands reactive with HMGB1-specific antibodies (Fig. 2C). These experimental data suggest that EGCG facilitates HMGB1 aggregation and subsequent degradation in macrophage cultures.
3.4. EGCG enhanced LPS-induced autophagy in macrophages
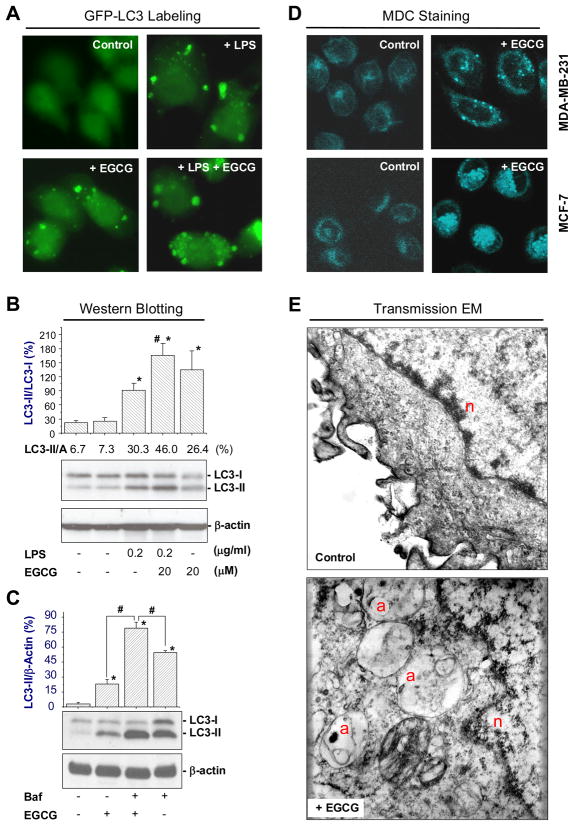
To gain insight into the mechanism of HMGB1 degradation, we determined whether EGCG enhanced LPS-induced autophagic activities. Consistent with a previous report [4], LPS induced formation of LC3-containing cytoplasmic vesicles in GFP-LC3-transfected macrophages (autophagosomes, Fig. 3A), and elevated LC3-I to LC3-II conversion in non-transfected macrophages (Fig. 3B). At the concentrations effective for inhibiting LPS-induced HMGB1 release, EGCG dramatically enhanced LPS-induced autophagosome formation (Fig. 3A) and LC3-II production (Fig. 3B). Even in the absence of LPS, EGCG was still able to induce formation of LC3-containing punctate structures (Fig. 3A), and significantly elevate LC3-II concentration in macrophage cultures (Fig. 3B). To determine whether EGCG elevated LC3-II production or decreased LC3-II degradation (i.e., autophagic flux), we determined the effect of a proton pump inhibitor, bafilomycin A1, on EGCG-induced elevation of LC3-II levels. Even in the presence of bafilomycin A1 at saturating concentrations for LC3-II accumulation (100 nM), EGCG still significantly increased LC3-II levels (Fig. 3C), suggesting that EGCG increases autophagosome synthesis, rather than merely inhibiting LC3-II degradation (by reducing autophagosomes trafficking to, and fusion with, lysosomes). Similarly, EGCG (100 μM) induced formation of autophagic vacuoles in non-immune cells, such as human breast adenocarcinoma (MDA-MB-361) and cancer (MCF-7) cell lines (Fig. 3D). Employing transmission electron microscopy (EM), double-membraned vacuoles typical of autophagosomes were observed in EGCG-treated cells (Fig. 3E), confirming that EGCG can indeed stimulate autophagy in both immune and non-immune cells.
Figure 3. EGCG stimulated autophagy in immune or non-immune cells.
A) EGCG enhanced LPS-induced formation of LC3 punctates in macrophage cultures. GFP-LC3-transfected RAW 264.7 cells were stimulated with LPS (200 ng/ml) in the absence or presence of EGCG (10 μM) for 16 h, and the formation of LC3 punctates were examined under fluorescent microscopy. B). EGCG enhanced LPS-induced LC3-II formation. Macrophage cultures were stimulated with LPS in the absence, or presence of EGCG for 16 h, and cellular levels of LC3-I and LC3-II were determined by Western blotting analysis with reference to β-actin. C). EGCG enhanced LC3-II levels in the presence of bafilomycin A1. Macrophage cultures were stimulated with EGCG (15 μM) for 12 h, bafilomycin A1 was added at a saturating concentration (100 nM), and further incubated for additional 4 h. Cellular levels of LC3-I and LC3-II were determined by Western blotting analysis with reference to β-actin. D). EGCG induced formation of autophagosomes in non-immune cells. Human breast adenocarcinoma (MDA-MB-361) and cancer (MCF-7) cells were stimulated with EGCG for 24 h, and stained with acidotropic monodansylcadaverine (MDC) dye to visualize autophagosomes. E). EGCG induced formation of double-membraned autophagic vacuoles. Ultrastructural analysis of EGCG-induced autophagy by transmission electron microscopy. a, autophagic vacuole (autophagosome); n, nucleus; m, mitochondria.
3.5. EGCG reduced cellular HMGB1 levels in LPS-stimulated macrophages
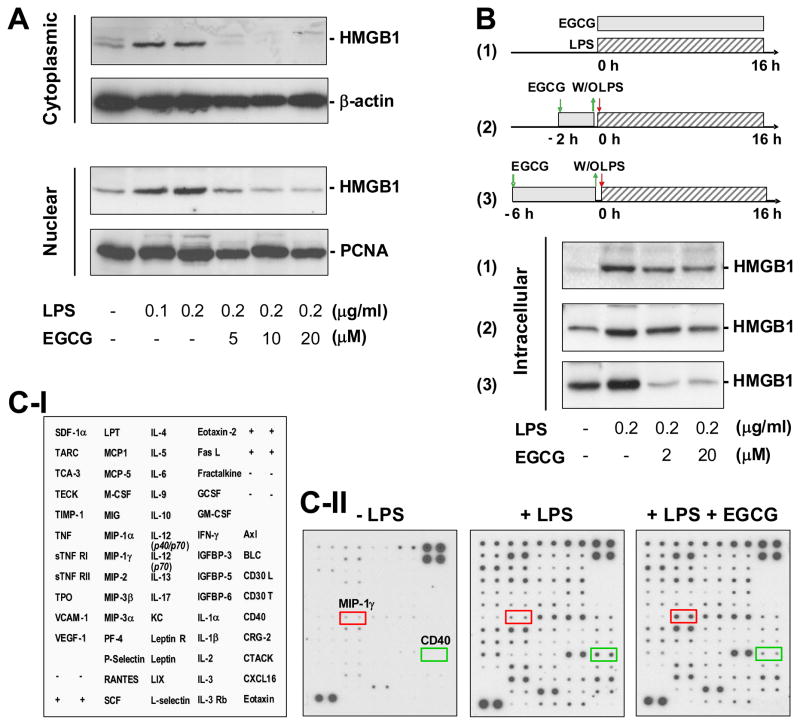
To understand its potential consequences, we determined the effects of EGCG on cytoplasmic and nuclear HMGB1 levels in LPS-stimulated macrophages. In sharp contrast to murine macrophage-like RAW 264.7 cells [10], LPS dose-dependently increased both nuclear and cytoplasmic HMGB1 protein levels in primary macrophage cultures (Fig. 4A). At the concentration (10 μM) that abrogated HMGB1 release [33], EGCG effectively prevented LPS-induced elevation of both cytoplasmic and nuclear HMGB1 levels (Fig. 4A), but did not affect basal HMGB1 levels in the nucleus of quiescent macrophages. Nevertheless, at these low concentrations (10–20 μM), EGCG effectively prevented occasional elevation of cytoplasmic HMGB1 levels in control cell cultures (data not shown). Taken together, our experimental data suggest a possibility that EGCG prevents LPS-induced HMGB1 release by strategically destroying it in the cytoplasm via a cellular degradation process – autophagy.
Figure 4. EGCG selectively prevented LPS-induced elevation of cellular HMGB1 levels.
Primary murine peritoneal macrophages were stimulated with LPS in the absence or presence of EGCG at indicated regimens, and assayed for cellular levels of HMGB1 (Panel A, B) or other 62 cytokines (Panel C) by Western blotting analysis or Cytokine Antibody Arrays, respectively. A, B). EGCG dose-dependently prevented LPS-induced increase of cellular HMGB1 levels. C). EGCG did not affect LPS-induced elevation of most other cytokines. Panel C-I denotes names of cytokines on the Cytokine Antibody Arrays shown in Panel C-II. Shown in Panel A, B, and C-II were representative Western blots or Cytokine Arrays of three experiments with similar results.
We previously found that EGCG was still able to inhibit LPS-induced HMGB1 release when it was added 2 to 6 h after LPS [33]. To elucidate the underlying mechanism, we performed parallel wash-out experiments to determine whether a brief prior exposure was sufficient for EGCG to prevent LPS-induced HMGB1 up-regulation. As predicted, prolonged incubation with EGCG dramatically suppressed LPS-induced increase of cellular HMGB1 levels (Fig. 4B, panel 1). However, even a brief pre-exposure of EGCG for 6 h appeared to be sufficient to confer a dramatic inhibition of LPS-induced elevation of cellular HMGB1 levels (Fig. 4B, panel 3).
To assess its selectivity, we determined the effects of EGCG on cellular levels of 62 other cytokines using cytokine antibody array. Consistent with a previous report [33], LPS elevated cellular levels of many cytokines and chemokines including CD40, CXCL16, Eotaxin-2, G-CSF, GM-CSF, IGFBP-3, IL-1a, IL-4, IL-9, IL-12, LIX, LPT, MCP-1, MCP-5, MIP-2, PF-4, P-selection, TIMP-1 and TPO. However, at the concentrations that effectively inhibited LPS-induced elevation of cellular HMGB1 levels, EGCG (10 μM) did not affect levels of most other cytokines (Fig. 4C), indicating that EGCG selectively regulates cellular HMGB1 levels. Intriguingly, EGCG dramatically inhibited LPS-induced upregulation of CD40 (Fig. 4C), a trigger of the fusion between pathogen-containing vacuoles and lysosomes during “xenophagy” [1,42]. Although CD40 may be important in destroying exogenous pathogens during xenophagy, its regulatory role in the degradation of endogenous macromolecules (such as EGCG/HMGB1 aggregates) during autophagy is not yet known.
3.6. Suppression of autophagy impaired EGCG-mediated HMGB1 inhibition
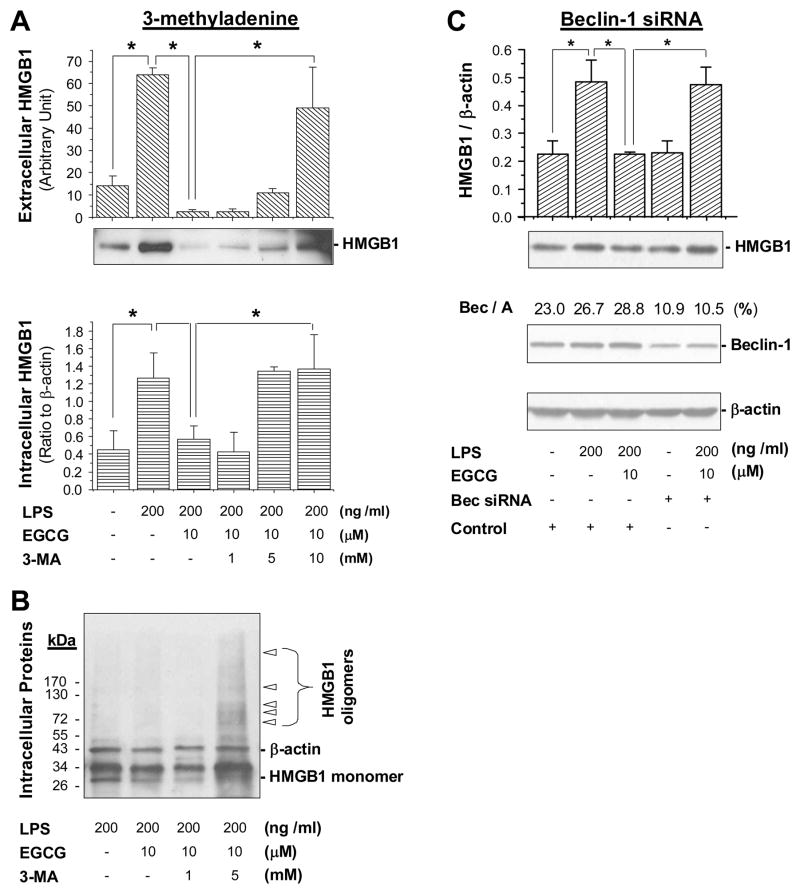
To test the possibility that EGCG inhibits HMGB1 by stimulating its autophagic degradation, we determined whether blockage of autophagy impaired EGCG-mediated HMGB1 inhibition in macrophage cultures. A specific inhibitor of autophagy-regulating signaling molecule (PI3K-class III), 3-methyladenine (3-MA), was shown to effectively block LPS-induced autophagy in macrophage cultures [4]. We found that 3-MA dose-dependently impaired EGCG-mediated inhibition of intracellular HMGB1 elevation (Fig. 5A, bottom panel) and extracellular HMGB1 release (Fig. 5A, top panel) in primary macrophage cultures. Although aggregation of endogenous HMGB1 was barely detectable in macrophages that were treated with LPS and EGCG for 12–16 h (Fig. 5B), co-treatment with 3-MA (5 mM) led to dramatic accumulation of HMGB1 aggregates in macrophage cultures (Fig. 5B). It suggests that the EGCG-mediated HMGB1 inhibition may be partly dependent on its autophagy-inducing properties.
Figure 5. PI3K inhibitor and beclin-1 siRNA impaired EGCG-mediated inhibition of HMGB1 expression or release.
A) An autophagy inhibitor, 3-methyladenine, impaired EGCG-mediated inhibition of HMGB1 expression and release. Primary macrophages were incubated with LPS in the absence or presence of EGCG or 3-methyladenine for 16 hr, and intra- (Top Panel) or extracellular (Bottom Panel) HMGB1 levels were determined by immunoblotting analysis. Data represents mean ± SD of three experiments. *P < 0.05. B). Co-treatment with 3-MA led to accumulation of HMGB1 oligomers in primary macrophage cultures. Primary macrophages were incubated with LPS in the absence or presence of EGCG or 3-MA for 16 h, and cell lysates were assayed for HMGB1 aggregation by Western blotting analysis. Note that EGCG treatment decreased cellular levels of HMGB1 monomer, but co-treatment with 3-MA led to dramatic accumulation of HMGB1 oligomers. C). Knock-down of beclin-1 impaired EGCG-mediated inhibition of HMGB1 expression. Thioglycollate-elicited peritoneal macrophages were transfected with Beclin-1-specific siRNA for 48 h, and subsequently stimulated with LPS in the absence or presence of EGCG for additional 16 h. Cellular levels of beclin-1, HMGB1 and β-actin were determined by Western blotting analysis. Bar graphs: quantitative analysis of HMGB1/β-actin ratio.
To further test this possibility, we determined whether knock-down of an essential autophagy gene [43], beclin-1, similarly affects EGCG-mediated inhibition of HMGB1 expression. Treatment of thioglycollate-elicited peritoneal macrophages with siRNA specific to beclin-1 led to a >60% reduction of cellular beclin-1 levels (Fig. 5C). Similarly, knockdown of beclin-1 reduced cellular LC3-II levels by 50–60% (data not shown), and dramatically impaired EGCG-mediated inhibition of HMGB1 expression in LPS-stimulated macrophage cultures (Fig. 5C), supporting the possibility that EGCG inhibits LPS-induced HMGB1 expression by stimulating its autophagic degradation.
3.7. An autophagy inhibitor impaired EGCG-mediated protection against lethal sepsis
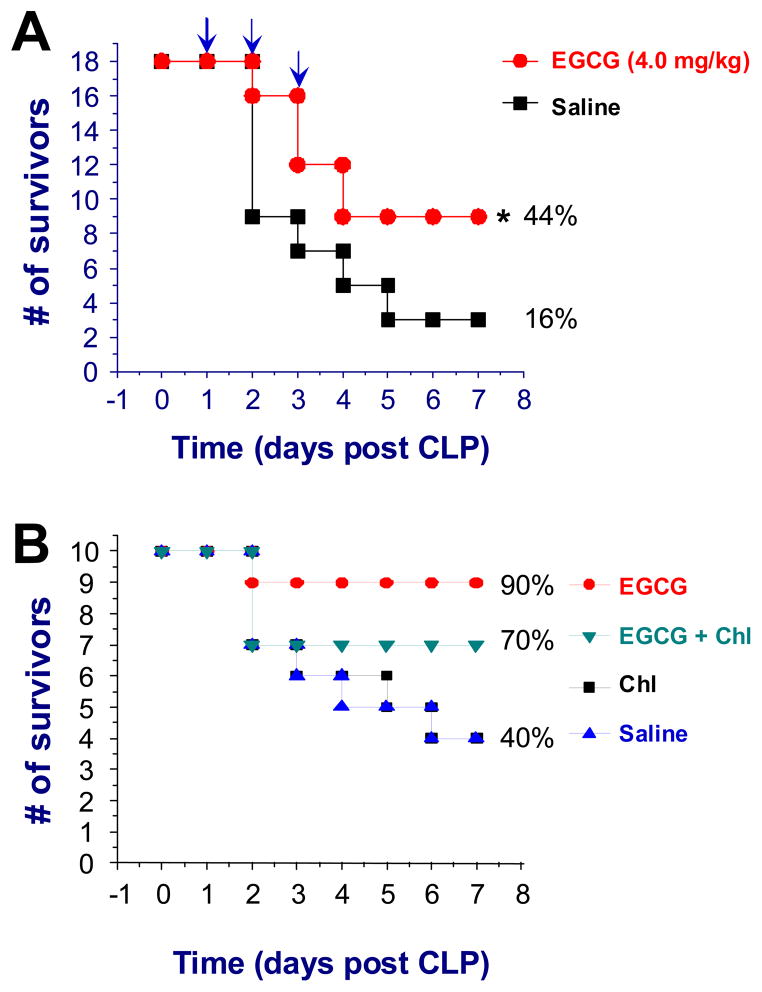
To appreciate the prophylactic and therapeutic benefits of tea consumption, we determined whether administration of EGCG via a clinically feasible route confers protection in animal models of lethal endotoxemia or sepsis. In animal models of lethal endotoxemia (LPS = 10 mg/kg), oral administration of EGCG (4 mg/kg, daily for three days) significantly increased survival rate from 25% (for control group receiving saline, N = 16 mice/group) to 62.5% (for experimental group receiving EGCG, N = 16 mice/group, P < 0.05). In a clinically relevant model of sepsis (induced by cecal ligation and puncture), delayed administration of EGCG conferred a significant protection against lethal sepsis (N = 18 mice per group, Fig. 6A), significantly increasing animal survival rate from 16% to 44% (P < 0.05), supporting a therapeutic potential for EGCG in the treatment of sepsis. Our previous study suggested that EGCG confers protection against lethal sepsis partly by attenuating systemic accumulation of HMGB1, a late mediator of lethal sepsis [33]. To explore additional protective mechanisms, we determined whether autophagy inhibitors, such as 3-MA or chloroquine, impaired EGCG-mediated protective effects. 3-MA has been widely used as an autophagy inhibitor in vitro, but its poor solubility in water affects its bioavailability in vivo, thereby limiting its use for animal studies. In contrast, chloroquine is readily water-soluble, and has been successfully employed by others to inhibit autophagy in vivo. It was previously shown that administration of chloroquine at a higher dose (50 mg/kg) conferred protection against CLP-induced sepsis, increasing the 4-day animal survival rate from 24% to 55% [44]. At a lower dose (30 mg/kg), however, chloroquine did not affect animal survival rates (Fig. 6B), but partly impaired EGCG-mediated protective effects (Fig. 6B). It will thus be critically important to further investigate whether EGCG confers protection against lethal sepsis partly by stimulating autophagy in future studies.
Figure 6. An autophagy inhibitor impaired EGCG-mediated protection against lethal sepsis.
A). Delayed oral administration of EGCG rescued mice from lethal sepsis. Balb/C mice were subjected to lethal sepsis (induced by CLP). At +24, +48, and +72 h post the onset of sepsis, animals were orally gavaged with saline (0.2 ml/mouse) or EGCG (4.0 mg/ml), and animal survival rates were monitored for up to two weeks. The Kaplan-Meier method was used to compare the differences in mortality rates between groups. *, P < 0.05 versus saline. B). Co-administration of an autophagy inhibitor, chloroquine, partly impaired EGCG-mediated protection. Balb/C mice were subjected to lethal sepsis (induced by CLP). At +2, +24, +48, and +72 h post CLP, animals were intraperitoneally administered with saline, EGCG (4.0 mg/kg), chloroquine (30 mg/kg, “Chl”), or EGCG (4.0 mg/kg) plus chroloquine (30 mg/kg), and animal survival rates were monitored for two weeks.
4. Discussion
Historically, consumption of Green tea has been associated with health benefits against multiple diseases including oxidative stress [28], atherosclerosis [28], cancer [29], and cardiovascular diseases [30]. Emerging evidence has suggested a common role for HMGB1 in the pathogenesis of lethal systemic inflammation [15] and the aforementioned diseases [24–27]. Recently, we discovered that green tea and its major ingredient, EGCG, effectively inhibited LPS-induced HMGB1 release in macrophage cultures [32,33]. In the present study, we uncovered a novel mechanism by which EGCG inhibits HMGB1 release by inducing protein aggregation and subsequent degradation possibly in an autophagy-dependent manner.
Consistent with a previous report [40], we found that EGCG was rapidly internalized into cytoplasmic vesicles within 4–6 h. Surprisingly, these EGCG-containing vesicles almost completely co-localized with punctate structures positive for LC3, a specific marker of autophagosomes [45]. Subsequently, these EGCG-containing vesicles co-localized with LAMP2- and HMGB1-containing cytoplasmic compartments within 12–16 h of exposure. In the presence of EGCG, cytoplasmic HMGB1 “puncta” intercepted with LC3-containing vesicles in LPS-stimulated macrophages, supporting a possibility that EGCG interacts with HMGB1 within cytoplasmic vesicles (e.g., autophagosomes). The mechanism by which EGCG is internalized into macrophage cytoplasmic vesicles remains elusive. On one hand, EGCG can be auto-oxidized to form dimers such as theasinensin (Fig. 7) [40], which binds to the cytoplasmic membrane, and gains entry into the cytosol via passive diffusion [40]. On the other hand, EGCG is capable of binding to cell surface receptors (e.g., the 67 kDa laminin receptor, 67LR receptor) [46] through which EGCG mediates its anticancer [40,46] and anti-inflammatory activities [47–49]. It is not yet known whether EGCG is internalized into macrophages via passive diffusion or active 67LR-receptor-mediated endocytosis.
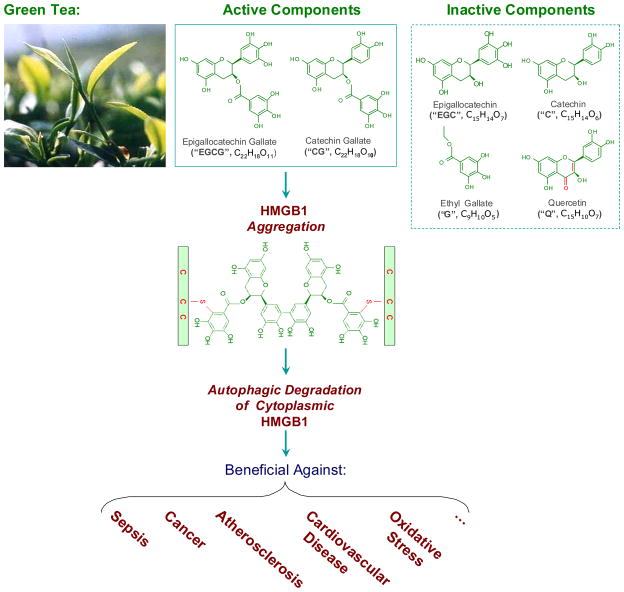
Figure 7. Proposed model for Green tea-associated health benefits.
Green tea contains ketone flavonoid (Q), non-ketone flavonoid (EGC and C), as well as catechin gallates (EGCG and CG). Structure-function relationship studies revealed that the active components capable of inducing HMGB1 aggregation (e.g., EGCG or CG) were able to induce autophagy and inhibit active HMGB1 release; the inactive components incapable of inducing HMGB1 aggregation (e.g., C, EGC, or G) all failed to induce autophagy and inhibit LPS-induced HMGB1 release. It is possible that EGCG conjugates to Cys23, Cys45, or Cys106 of HMGB1 via an oxidative reaction, thereby forming EGCG-HMGB1 aggregates. Given the potential role of HMGB1 in a variety of diseases, it is possible to consider that Green tea confers well-known health benefits by potentially modulating autophagic HMGB1 degradation in various cells.
The co-localization of EGCG with LC3-, LAMP2-, and HMGB1-containing cytoplasmic vesicles suggested a possibility that EGCG may modulate HMGB1 degradation through an autophagosome-lysosome-dependent pathway. Indeed, at the concentrations effective for inhibiting LPS-induced HMGB1 release [33], EGCG stimulated LC3 punctate (autophagosome) formation and LC3-I to LC3-II conversion, thereby effectively inducing autophagy in macrophage cultures. As a potential consequence, EGCG induces subsequent HMGB1 degradation via an autophagosome-lysosome-dependent pathway. This possibility was supported by our observation that co-treatment with an autophagy inhibitor, 3-MA, led to dramatic accumulation of HMGB1 aggregates in LPS/EGCG-treated macrophages. Intriguingly, EGCG can also conjugate to the proteasome complex, and consequently inhibit the ubiquitin-proteasome degradation pathway [50]. It is thus possible to selectively modulate autophagic degradation pathway using EGCG and other similar pharmacological agents.
The mechanism by which EGCG induces autophagy may relate to its capacity in inducing formation of EGCG-HMGB1 complexes (Fig. 7). On one hand, EGCG spontaneously forms aggregation products (e.g., the theasinensin dimer) via an oxidation reaction [40]. On the other hand, EGCG can conjugate to proteins either covalently (with the free thiol group of cysteine residues, Fig. 7) [37], or non-covalently (via hydrogen bonding, aromatic stacking or hydrophobic interactions) [51,52]. The direct interaction between EGCG aggregates and HMGB1 protein may underlie the formation of the SDS-resistant HMGB1/EGCG aggregates. Because large EGCG-HMGB1 complexes can not physically pass through the narrow pore of the proteasome barrel of the ubiquitin-proteasome pathway, it may function as a proximal trigger of autophagy [53].
In parallel with its autophagy-inducing capacities, EGCG prevented LPS-induced intracellular elevation and extracellular release of HMGB1 in primary macrophage cultures. Importantly, these parallel autophagy-stimulating and HMGB1-inhibiting effects were both achievable when cells were exposed to EGCG briefly (2–6 h), a time frame allowing for sufficient EGCG cytoplasmic trafficking and HMGB1 aggregation. Furthermore, knockdown of an autophagy-regulating protein (e.g., beclin-1) or inhibition of PI3K (with 3-methyladenine) impaired EGCG-mediated inhibition of HMGB1 expression and release. Intriguingly, it was recently suggested that cytoplasmic HMGB1 can interact with beclin-1, and function as an important regulator of autophagy [54,55]. It is thus important to investigate whether: 1) HMGB1 also occupies a critical role in EGCG-mediated autophagy; and 2) HMGB1 aggregation is absolutely necessary for its subsequent autophagic degradation. Nevertheless, our structure-function relationship studies support this possibility, because catechins (e.g., C or G) that failed to induce HMGB1 aggregation also failed to induce autophagy in macrophage cultures (Fig. 7). In contrast, catechins (e.g., EGCG and CG) that induced HMGB1 aggregation, also effectively induced autophagy (data not shown).
The potential role of autophagy in the regulation of HMGB1 release has just recently been investigated. Emerging evidence has suggested a potential link between autophagy and HMGB1 release, because agents capable of inducing (such as epidermal growth factor receptor-targeted diphtheria toxin) or inhibiting (such as a ketone-containing flavonoid, quercetin) autophagy similarly affected HMGB1 release in glioblastoma tumor cells [56] and murine-macrophage-like AW 264.7 cells [57]. In contrast, galloylated catechins such as EGCG and CG effectively induced autophagy, and yet still inhibited LPS-induced HMGB1 release (Fig. 7). These findings raised questions regarding an essential role for autophagy in the regulation of HMGB1 release, but suggested a pharmacological strategy to modulate HMGB1 release using autophagy-stimulating agents (such as EGCG) for the treatment of inflammatory diseases.
The relationships between autophagy (“self-eating”) and apoptosis (“self-killing”) are rather complex, and remain a subject of extensive on-going research [58]. Sharing some key molecular regulators in their machineries, autophagy and apoptosis may be triggered by common stimuli such as starvation, oxidative stress, and cytokines [58]. These stimuli may induce either combined autophagy and apoptosis, or exclusive switches between the two responses [8,58–60]. At the low concentrations (10–20 μM) effective for abrogating endotoxin-induced HMGB1 release, EGCG induced autophagy in macrophage cultures. At higher concentrations (e.g., 200 –1000 μM) however, ECGC also activates caspases, thereby inducing apoptotic cell death [61]. It remains elusive whether there is a reciprocal crosstalk between autophagy and apoptosis in EGCG-stimulated macrophages. Similarly, although the rise of intracellular [Ca2+] has been implicated as a common regulator of autophagy and apoptosis [62], it is presently unknown whether EGCG, particularly at low concentrations (10–20 μM), affects Ca2+ influx in macrophage cultures. Nevertheless, at high concentrations (e.g., 50–300 μM), EGCG can indeed elevate intracellular [Ca2+] possibly by altering cytoplasmic membrane permeability [63], thereby activating caspases and inducing apoptosis [61,64]. Paradoxically, EGCG could also attenuate Ca2+ influx induced by other stimuli, such as antigens (in mast cells) [65] or bacterial chemotactic peptides (in THP-1 monocytic cells) [66], implicating a complex relationship between EGCG and Ca2+ under various conditions.
Previously, we found that Green tea [32] and its major component, EGCG, inhibited bacterial endotoxin-induced HMGB1 release in vitro, and rescued mice from lethal sepsis when EGCG was given intraperitoneally [33,67]. In the present study, we further elucidated the intricate mechanisms by which EGCG inhibits endotoxin-induced HMGB1 release, and validated its therapeutic potential in animal models of sepsis by administering it via a clinically feasible route (oral gavage). Here we uncovered a novel mechanism by which EGCG effectively inhibits HMGB1 release by inducing its aggregation and autophagic degradation in macrophages. Furthermore, we demonstrated that EGCG still significantly rescued mice from lethal sepsis even when the first dose was given orally at 24 h after onset of the disease. We strategically administered EGCG in a delayed fashion to preserve a potentially beneficial early TNF response, but specifically attenuated late proinflammatory mediators such as HMGB1 [33]. Intriguingly, the EGCG-mediated protective effects were impaired by co-administration of an autophagy inhibitor, chloroquine. It will thus be critically important to investigate the potential role of autophagy in lethal systemic inflammatory diseases in future studies. The doses of EGCG orally given to septic mice (4 mg/kg, i.e., 10 μM) are comparable to those achievable in humans after ingestion of a few cups of green tea [68]. Given the multiple health benefits associated with Green tea consumption, as well as the shared role of HMGB1 in the pathogenesis of many diseases, it is tempting to consider that Green tea consumption may be beneficial to humans by selectively modulating autophagic HMGB1 degradation in targeted cells (Fig. 7). Further investigation in this area will improve our understanding of innate immune-modulating mechanisms of herbal medicine, and shed light on the development of novel autophagy-modulating therapeutic strategies for the treatment of human diseases.
Acknowledgments
We thank Dr. Mala Ashok for excellent technical assistance with animal experiments, and Dr. Yan Huang and Josh Gersten for critical reading of the manuscript. This work was supported by the National Center of Complementary and Alternative Medicine (NCCAM, R01AT005076) and the National Institute of General Medical Sciences (NIGMS, R01GM063075).
Abbreviations used in this paper
- CLP
cecal ligation and puncture
- EGCG
(−)-epigallocatechin-3-gallate
- HMGB1
high mobility group box 1
- LC3
microtubule-associated protein1 light chain 3
- LPS
lipopolysaccharide
Footnotes
AUTHOR CONTRIBUTIONS
W.L. and S.Z. performed most experiments and edited the manuscript; J.L. prepared recombinant HMGB1 protein and performed HPLC biotin-EGCG purification experiment; A.A. performed structure-function relationship experiment; A.J. performed the autophagic flux experiment; J.X. and S.F. performed autophagy experiments in non-immune cells; N.T.E. contributed key reagents; K.J.T and A.E.S. provided suggestions and project oversight; H.W. designed the study, analyzed the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–17. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Nara. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 7.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh YC, Athar M, Chaudry IH. When apoptosis meets autophagy: deciding cell fate after trauma and sepsis. Trends Mol Med. 2009;15:129–38. doi: 10.1016/j.molmed.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–51. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, Ochani M, Li J, Qiang X, Tanovic M, Harris HE, et al. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc Natl Acad Sci U S A. 2004;101:296–301. doi: 10.1073/pnas.2434651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu S, Ashok M, Li J, Li W, Yang H, Wang P, et al. Spermine protects mice against lethal sepsis partly by attenuating surrogate inflammatory markers. Mol Med. 2009;15:275–82. doi: 10.2119/molmed.2009.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson U, Wang H, Palmblad K, Aveberger AC, Bloom O, Erlandsson-Harris H, et al. High Mobility Group 1 Protein (HMG-1) Stimulates Proinflammatory Cytokine Synthesis in Human Monocytes. J Exp Med. 2000;192:565–70. doi: 10.1084/jem.192.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 selectively repress tissue damage- induced immune responses. Science. 2009;323:1722–5. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Zhu S, Zhou R, Li W, Sama AE. Therapeutic potential of HMGB1-targeting agents in sepsis. Expert Rev Mol Med. 2008;10:e32. doi: 10.1017/S1462399408000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Ward MF, Sama AE. Novel HMGB1-inhibiting therapeutic agents for experimental sepsis. Shock. 2009;32:348–57. doi: 10.1097/SHK.0b013e3181a551bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulloa L, Ochani M, Yang H, Tanovic M, Halperin D, Yang R, et al. Ethyl pyruvate prevents lethality in mice with established lethal sepsis and systemic inflammation. Proc Natl Acad Sci U S A. 2002;99:12351–6. doi: 10.1073/pnas.192222999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Li J, Ashok M, Wu R, Chen D, Yang L, et al. Yang A cardiovascular drug rescues mice from lethal sepsis by selectively attenuating a late-acting proinflammatory mediator, high mobility group box 1. J Immunol. 2007;178:3856–64. doi: 10.4049/jimmunol.178.6.3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Li W, Li J, Rendon-Mitchell B, Ochani M, Ashok M, et al. The Aqueous Extract of a Popular Herbal Nutrient Supplement, Angelica sinensis, Protects Mice against Lethal Endotoxemia and Sepsis. J Nutr. 2006;136:360–5. doi: 10.1093/jn/136.2.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Liao H, Ochani M, Justiniani M, Lin X, Yang L, et al. Cholinergic agonists inhibit HMGB1 release and improve survival in experimental sepsis. Nat Med. 2004;10:1216–21. doi: 10.1038/nm1124. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Li J, Qiang X, Czura CJ, Ochani M, Ochani K, et al. Suppression of HMGB1 release by stearoyl lysophosphatidylcholine:an additional mechanism for its therapeutic effects in experimental sepsis. J Lipid Res. 2005;46:623–7. doi: 10.1194/jlr.C400018-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Czura CJ, Tracey KJ. Lipid unites disparate syndromes of sepsis. Nat Med. 2004;10:124–5. doi: 10.1038/nm0204-124. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Zhu S, Li J, Huang Y, Zhou R, Fan X, et al. A hepatic protein, fetuin-A, occupies a protective role in lethal systemic inflammation. PLoS ONE. 2011;6:e16945. doi: 10.1371/journal.pone.0016945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsung A, Klune JR, Zhang X, Jeyabalan G, Cao Z, Peng X, et al. HMGB1 release induced by liver ischemia involves Toll-like receptor 4 dependent reactive oxygen species production and calcium-mediated signaling. J Exp Med. 2007;204:2913–23. doi: 10.1084/jem.20070247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harja E, Bu DX, Hudson BI, Chang JS, Shen X, Hallam K, et al. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE−/− mice. J Clin Invest. 2008;118:183–94. doi: 10.1172/JCI32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang D, Kang R, Zeh HJ, III, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–40. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrassy M, Volz HC, Igwe JC, Funke B, Eichberger SN, Kaya Z, et al. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation. 2008;117:3216–26. doi: 10.1161/CIRCULATIONAHA.108.769331. [DOI] [PubMed] [Google Scholar]
- 28.Frei B, Higdon JV. Antioxidant activity of tea polyphenols in vivo: evidence from animal studies. J Nutr. 2003;133:3275S–84S. doi: 10.1093/jn/133.10.3275S. [DOI] [PubMed] [Google Scholar]
- 29.Crespy V, Williamson G. A review of the health effects of green tea catechins in in vivo animal models. J Nutr. 2004;134:3431S–40S. doi: 10.1093/jn/134.12.3431S. [DOI] [PubMed] [Google Scholar]
- 30.Vita JA. Tea consumption and cardiovascular disease: effects on endothelial function. J Nutr. 2003;133:3293S–7S. doi: 10.1093/jn/133.10.3293S. [DOI] [PubMed] [Google Scholar]
- 31.Graham HN. Green tea composition, consumption, and polyphenol chemistry. Prev Med. 1992;21:334–50. doi: 10.1016/0091-7435(92)90041-f. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Li W, Wang H. More tea for septic patients? - Green tea may reduce endotoxin-induced release of high mobility group box 1 and other pro-inflammatory cytokines. Med Hypotheses. 2006;66:660–3. doi: 10.1016/j.mehy.2005.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Ashok M, Li J, Yang H, Sama AE, Wang H. A Major Ingredient of Green Tea Rescues Mice from Lethal Sepsis Partly by Inhibiting HMGB1. PLoS ONE. 2007;2:e1153. doi: 10.1371/journal.pone.0001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rendon-Mitchell B, Ochani M, Li J, Han J, Wang H, Yang H, et al. IFN-gamma Induces High Mobility Group Box 1 Protein Release Partly Through a TNF-Dependent Mechanism. J Immunol. 2003;170:3890–7. doi: 10.4049/jimmunol.170.7.3890. [DOI] [PubMed] [Google Scholar]
- 35.Chen G, Li J, Ochani M, Rendon-Mitchell B, Qiang X, Susarla S, et al. Bacterial endotoxin stimulates macrophages to release HMGB1 partly through CD14- and TNF-dependent mechanisms. J Leukoc Biol. 2004;76:994–1001. doi: 10.1189/jlb.0404242. [DOI] [PubMed] [Google Scholar]
- 36.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 37.Ishii T, Mori T, Tanaka T, Mizuno D, Yamaji R, Kumazawa S, et al. Covalent modification of proteins by green tea polyphenol (−)-epigallocatechin-3-gallate through autoxidation. Free Radic Biol Med. 2008;45:1384–94. doi: 10.1016/j.freeradbiomed.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Yang S, Zhou M, Chaudry IH, Wang P. Novel approach to prevent the transition from the hyperdynamic phase to the hypodynamic phase of sepsis: role of adrenomedullin and adrenomedullin binding protein-1. Ann Surg. 2002;236:625–33. doi: 10.1097/00000658-200211000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimura Y, Yamada K, Tachibana H. A lipid raft-associated 67kDa laminin receptor mediates suppressive effect of epigallocatechin-3-O-gallate on FcepsilonRI expression. Biochem Biophys Res Commun. 2005;336:674–81. doi: 10.1016/j.bbrc.2005.08.146. [DOI] [PubMed] [Google Scholar]
- 40.Hong J, Lu H, Meng X, Ryu JH, Hara Y, Yang CS. Stability, cellular uptake, biotransformation, and efflux of tea polyphenol (−)-epigallocatechin-3-gallate in HT-29 human colon adenocarcinoma cells. Cancer Res. 2002;62:7241–6. [PubMed] [Google Scholar]
- 41.Gonzalez-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquere S, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 42.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–77. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 44.Yasuda H, Leelahavanichkul A, Tsunoda S, Dear JW, Takahashi Y, Ito S, et al. Chloroquine and inhibition of Toll-like receptor 9 protect from sepsis-induced acute kidney injury. Am J Physiol Renal Physiol. 2008;294:F1050–8. doi: 10.1152/ajprenal.00461.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tachibana H, Koga K, Fujimura Y, Yamada K. A receptor for green tea polyphenol EGCG. Nat Struct Mol Biol. 2004;11:380–1. doi: 10.1038/nsmb743. [DOI] [PubMed] [Google Scholar]
- 47.Hong BE, Fujimura Y, Yamada K, Tachibana H. TLR4 signaling inhibitory pathway induced by green tea polyphenol epigallocatechin-3-gallate through 67-kDa laminin receptor. J Immunol. 2010;185:33–45. doi: 10.4049/jimmunol.0903742. [DOI] [PubMed] [Google Scholar]
- 48.Tachibana H, Fujimura Y, Yamada K. Tea polyphenol epigallocatechin-3-gallate associates with plasma membrane lipid rafts: lipid rafts mediate anti-allergic action of the catechin. Biofactors. 2004;21:383–5. doi: 10.1002/biof.552210174. [DOI] [PubMed] [Google Scholar]
- 49.Fujimura Y, Tachibana H, Yamada K. Lipid raft-associated catechin suppresses the FcepsilonRI expression by inhibiting phosphorylation of the extracellular signal-regulated kinase1/2. FEBS Lett. 2004;556:204–10. doi: 10.1016/s0014-5793(03)01432-7. [DOI] [PubMed] [Google Scholar]
- 50.Nam S, Smith DM, Dou QP. Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J Biol Chem. 2001;276:13322–30. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- 51.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, et al. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat Struct Mol Biol. 2008;15:558–66. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 52.Roberts BE, Shorter J. Escaping amyloid fate. Nat Struct Mol Biol. 2008;15:544–6. doi: 10.1038/nsmb0608-544. [DOI] [PubMed] [Google Scholar]
- 53.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–8. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang R, Livesey KM, Zeh HJ, Loze MT, Tang D. HMGB1: A novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6:1209–11. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- 56.Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ. 2009;16:175–83. doi: 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang D, Kang R, Xiao W, Zhang H, Lotze MT, Wang H, et al. Quercetin prevents LPS-induced high-mobility group box 1 release and proinflammatory function. Am J Respir Cell Mol Biol. 2009;41:651–60. doi: 10.1165/rcmb.2008-0119OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewaele M, Martinet W, Rubio N, Verfaillie T, de Witte PA, Piette J, et al. Autophagy pathways activated in response to PDT contribute to cell resistance against ROS damage. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01118.x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fimia GM, Piacentini M. Regulation of autophagy in mammals and its interplay with apoptosis. Cell Mol Life Sci. 2010;67:1581–8. doi: 10.1007/s00018-010-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 61.Hashimoto K, Sakagami H. Induction of apoptosis by epigallocatechin gallate and autophagy inhibitors in a mouse macrophage-like cell line. Anticancer Res. 2008;28:1713–8. [PubMed] [Google Scholar]
- 62.Giorgi C, Romagnoli A, Pinton P, Rizzuto R. Ca2+ signaling, mitochondria and cell death. Curr Mol Med. 2008;8:119–30. doi: 10.2174/156652408783769571. [DOI] [PubMed] [Google Scholar]
- 63.Feng W, Cherednichenko G, Ward CW, Padilla IT, Cabrales E, Lopez JR, et al. Green tea catechins are potent sensitizers of ryanodine receptor type 1 (RyR1) Biochem Pharmacol. 2010;80:512–21. doi: 10.1016/j.bcp.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das A, Banik NL, Ray SK. Mechanism of apoptosis with the involvement of calpain and caspase cascades in human malignant neuroblastoma SH-SY5Y cells exposed to flavonoids. Int J Cancer. 2006;119:2575–85. doi: 10.1002/ijc.22228. [DOI] [PubMed] [Google Scholar]
- 65.Inoue T, Suzuki Y, Ra C. Epigallocatechin-3-gallate inhibits mast cell degranulation, leukotriene C4 secretion, and calcium influx via mitochondrial calcium dysfunction. Free Radic Biol Med. 2010;49:632–40. doi: 10.1016/j.freeradbiomed.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Wang O, Ruan L, Hou X, Cui Y, Wang JM, et al. The green tea polyphenol (−)-epigallocatechin-3-gallate inhibits leukocyte activation by bacterial formylpeptide through the receptor FPR. Int Immunopharmacol. 2009;9:1126–30. doi: 10.1016/j.intimp.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang F, de Villiers WJ, McClain CJ, Varilek GW. Green tea polyphenols block endotoxin-induced tumor necrosis factor-production and lethality in a murine model. J Nutr. 1998;128:2334–40. doi: 10.1093/jn/128.12.2334. [DOI] [PubMed] [Google Scholar]
- 68.Yang CS, Chen L, Lee MJ, Balentine D, Kuo MC, Schantz SP. Blood and urine levels of tea catechins after ingestion of different amounts of green tea by human volunteers. Cancer Epidemiol Biomarkers Prev. 1998;7:351–54. [PubMed] [Google Scholar]