Abstract
Guanine nucleotide binding protein-like 3-like (GNL3L) is a nucleolar protein and the vertebrate paralogue of nucleostemin (NS). We previously reported that nucleoplasmic mobilization of NS stabilizes MDM2 (mouse double minute 2). Here, we investigated the role of GNL3L as a novel MDM2 regulator. We found that GNL3L binds MDM2 in vivo and displays the same function as NS in stabilizing MDM2 protein and preventing its ubiquitylation. The interaction between GNL3L and MDM2 also takes place in the nucleoplasm. However, the MDM2-regulatory activity of GNL3L occurs constitutively and does not so much depend on the nucleolar release mechanism as NS does. GNL3L depletion triggers G2/M arrest in the p53-wild-type HCT116 cells more than in the p53-null cells, and up-regulates specific p53 targets (i.e. Bax, 14-3-3σ, and p21) without affecting the ubiquitylation or stability of p53 proteins. The inhibitory activity of GNL3L on p53-mediated transcription correlates with the increased expression of GNL3L and reduced expression of 14-3-3σ and p21 in human gastrointestinal tumors. This work shows that, in contrast to most nucleolar proteins that negatively control MDM2, GNL3L and NS are the only two that are designed to stabilize MDM2 protein under basal or induced condition, respectively, and may act as tumor-promoting genes.
Keywords: Colorectal carcinoma, G2/M arrest, GNL3L, MDM2, nucleostemin, p53, ubiquitylation
Introduction
As proteins residing in the nucleolus are being cataloged systematically, so is the focus on this classic organelle turned from its ribosomal function to non-ribosomal role (Pederson & Tsai, 2009). Nucleostemin (NS) is one of the nucleolar proteins shown to be capable of exercising non-ribosomal activities. It is preferentially expressed by undifferentiated stem/progenitor cells undergoing active proliferation, and plays an essential role in early embryogenesis and self-renewal. The vertebrate NS family includes three members - NS, GNL3L, and Ngp-1, all of which contain a unique MMR1_HSR1 domain of five GTP-binding motifs arranged in a circularly permuted order (Tsai & Meng, 2009). NS and GNL3L are vertebrate paralogues that share a common invertebrate orthologue, whereas Ngp-1 exists as a single gene from yeast to human.
Mammalian NS was first cloned as a neural stem cell (NSC)-enriched gene (Tsai & McKay, 2002), and later found to be abundantly expressed by other stem cell types, cancer cells, and the adult testis (Baddoo et al., 2003; Maki et al., 2007; Ohmura et al., 2008; Siddiqi et al., 2008). In contrast, GNL3L is expressed lower in the undifferentiated NSCs than in their differentiated progeny during neural development, and is preferentially expressed by the cerebellum and forebrain in adult animals (Yasumoto et al., 2007). At the subcellular level, NS is highly concentrated in the nucleolus, whereas GNL3L displays a higher nucleoplasmic intensity and a much shorter nucleolar retention time than NS (Meng et al., 2007). Among the NS-interacting proteins, functional outcomes have been shown for p53 (Ma & Pederson, 2007; Tsai & McKay, 2002), MDM2 (mouse double minute 2) (Dai et al., 2008; Meng et al., 2008), TRF1 (telomeric repeat binding factor 1) (Zhu et al., 2006), ARF (alternative reading frame) (Ma & Pederson, 2007), and RSL1D1 (ribosomal L1 domain containing 1) (Meng et al., 2006). So far, only three proteins, i.e. the estrogen receptor-related protein (ERR), TERT (telomerase reverse transcriptase), and TRF1, have been shown to interact with GNL3L (Fu & Collins, 2007; Yasumoto et al., 2007; Zhu et al., 2009).
The biological importance of NS is best demonstrated by the early embryonic lethal phenotype of NS-null mice (Beekman et al., 2006; Zhu et al., 2006) but also by multiple studies showing its function in maintaining the continuous proliferation of cancer and stem cells. One of the proposed mechanisms for the NS activity is via MDM2 regulation. A study reported that NS depletion enhances MDM2 interaction with L5 and L11 and induces p53 activation (Dai et al., 2008). We found that nucleoplasmic mobilization of NS stabilizes MDM2 protein and inhibits the p53 transcription activity (Meng et al., 2008). Reciprocally, MDM2 is responsible for the degradation of NS protein induced by guanine nucleotide depletion (Huang et al., 2009). Compared to NS, the GNL3L function is much less explored. It has been shown to negatively regulate the telomere length (Fu & Collins, 2007) and to promote the metaphase-to-anaphase transition via TRF1 stabilization (Zhu et al., 2009). So far, it is not clear whether GNL3L regulates the MDM2/p53 pathway or how it relates to the MDM2-stabilizing activity of NS.
Given the paralogous relationship between NS and GNL3L, we began to investigate the possibility that GNL3L may be a novel MDM2 regulator that functions redundantly with or distinctively from NS. Here, we report that GNL3L can stabilize MDM2 protein by preventing its ubiquitylation, and rescue the NS-knockdown-induced MDM2 ubiquitylation. Unlike NS, the anti-MDM2-ubiquitylation activity of GNL3L occurs constitutively and is not as much controlled by the nucleolar release mechanism as NS is. Consistent with its MDM2-stabilizing ability, GNL3L knockdown triggers G2/M arrest and up-regulates specific p53 downstream targets, i.e. Bax, 14-3-3σ, and p21, more so in the p53-wild-type than in the p53-null HCT116 cells. In accordance, high percentages of human colorectal and gastric cancers express high levels of GNL3L and low levels of 14-3-3σ and p21. This work indicates a potential role of GNL3L as a tumor-promoting factor via MDM2 stabilization.
Results
GNL3L binds MDM2 via its G- or I-domain and the central domain of MDM2
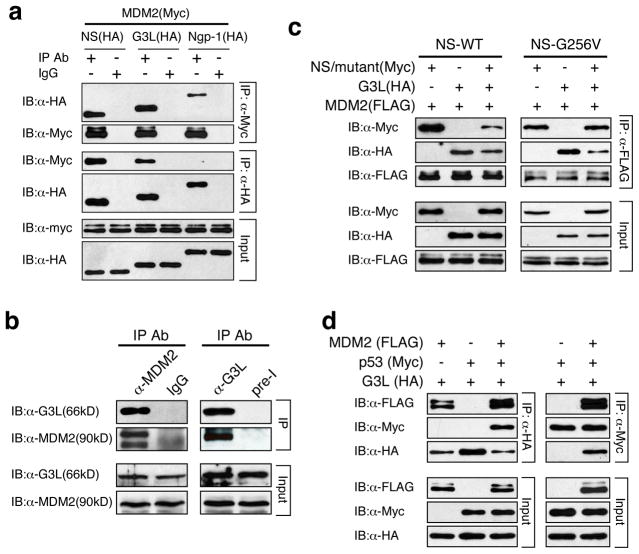
A connection between GNL3L and MDM2 was first revealed by coimmunoprecipitation (coIP) of MDM2 (Myc-tagged) and NS family proteins (HA-tagged), which showed that both NS and GNL3L can be coimmunoprecitated with MDM2 by anti-Myc (rows 1 and 2) or anti-HA antibody (rows 3 and 4) (Fig. 1a). In contrast, little or no Ngp-1 interacts with MDM2. Endogenous coIP further confirmed that GNL3L and MDM2 co-reside in the same protein complex immunoprecipitated by anti-MDM2 or anti-GNL3L antibody in HeLa cells (Fig. 1b). To investigate the MDM2-binding relationship between NS and GNL3L, anti-FLAG coIP of expressed MDM2 (FLAG), GNL3L (HA), and/or NS (Myc) was performed in HEK293 cells and showed that GNL3L appears to have a stronger activity in competing against NS for MDM2 binding than NS does against GNL3L (Fig. 1c, left panel). The better MDM2 binding of GNL3L than that of NS may be caused by their differential nucleoplasmic distribution (see Fig. S1). To examine this possibility, we conducted the same competitive coIP experiment using wild-type GNL3L and NS-G256V mutant, which is mislocalized to the nucleoplasm but still maintains its MDM2-binding activity. The results showed that NS-G256V displays a stronger MDM2-binding activity compared to GNL3L (Fig. 1c, right panel). Finally, triple coIP demonstrated that GNL3L does not bind p53 directly but does so in the presence of MDM2, suggesting that MDM2 may bridge between GNL3L and p53 (Fig. 1d).
Figure 1. GNL3L competes against NS for MDM2 binding.
(a) MDM2 (Myc) and NS family proteins (HA) were co-immunoprecipitated (coIP) and immunoblotted (IB) by anti-tag antibodies in HEK293 cells. The results showed that MDM2 preferentially interacts with NS and GNL3L. (b) Endogenous binding between MDM2 and GNL3L was demonstrated by anti-MDM2 (left panel) and anti-GNL3L (right panel) coIP experiments. IgG and preimmune serum (pre-I) precipitated samples were used as controls. (c) Anti-FLAG coIP of GNL3L (HA), NS (Myc), and MDM2 (FLAG) showed that GNL3L and NS compete against each other for MDM2 binding. By comparison, GNL3L shows more binding with MDM2 than wild-type NS does (left panel) and less binding with MDM2 than NS-G256V does (right panel). NS-G256V is a nucleoplasm-mislocalized mutant of NS (Fig. S1b3). (d) Triple coIP of GNL3L (HA), MDM2 (FLAG), and p53 (Myc) by anti-HA (left panel) or anti-Myc antibody (right panel) shows that GNL3L may bind p53 indirectly through MDM2.
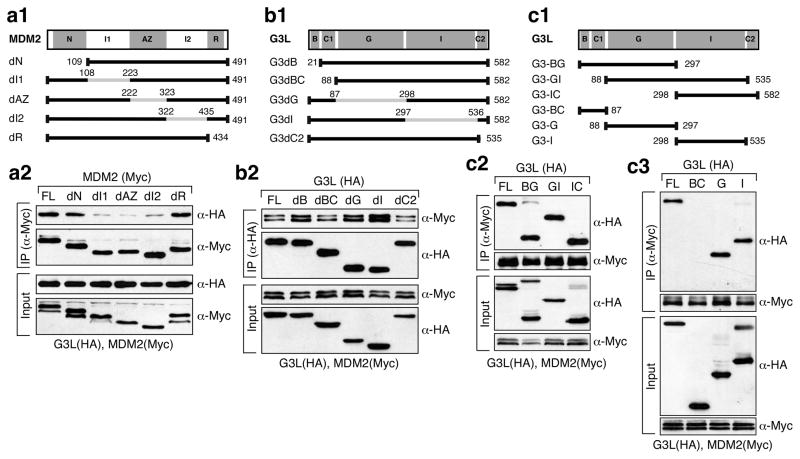
The binding interface between MDM2 and GNL3L was defined by non-overlapping deletion mutants of MDM2 on its p53-binding (N, a.a. 1–108), intermediate-1 (I1, a.a. 109–222), acidic-zinc finger (AZ, a.a. 223–322), intermediate-2 (I2, a.a. 323–434), and RING-finger domains (R, a.a. 435–491) (Fig. 2a1). Anti-Myc coIP of MDM2 mutants (Myc) and full-length GNL3L-HA showed that deleting the I1 (dI1), AZ (dAZ), or I2 domain (dI2) of MDM2 reduces its ability to bind GNL3L (Fig. 2a2). To map the MDM2-binding site(s) on GNL3L, GNL3L mutants lacking the basic (B, a.a. 1–20), basic-coiled-coil (BC, a.a. 1–87), GTP-binding (G, a.a. 88–297), intermediate (I, a.a. 298–535), or C2 (a.a. 536–582) domain were created (Fig. 2b1). Anti-HA coIP of full-length MDM2-Myc and mutant GNL3L (HA) showed that none of the single domain deletion mutants of GNL3L lose their ability to bind MDM2 and suggested the involvement of multiple GNL3L domains (Fig. 2b2). CoIP of complex deletion mutants of GNL3L (Fig. 2c1) confirmed that both the G and I-domains of GNL3L are sufficient to bind MDM2 (Fig. 2c2, c3). These data demonstrate that the interaction between GNL3L and MDM2 is mediated by the central region of MDM2 and the G or I domain of GNL3L.
Figure 2. GNL3L binds the central domain of MDM2 via its GTP-binding or intermediate domain.
Diagrams show MDM2 (a1) and GNL3L single (b1) or multi-domain (c1) deletion mutants. Grey lines and numbers indicate the deleted regions and amino acid positions. Abbreviations: N, p53-binding; I1 and I2, intermediate-1 and -2; AZ, acidic-zinc finger; R, RING-finger; B, basic; C, coiled-coil; G, GTP-binding; and I, intermediate. CoIP showed that binding between GNL3L and MDM2 requires the I1, AZ, and I2 domains of MDM2 (a2) and the G- or I-domain of GNL3L (b2, c2, and c3).
GNL3L stabilizes MDM2 protein by reducing its ubiquitylation
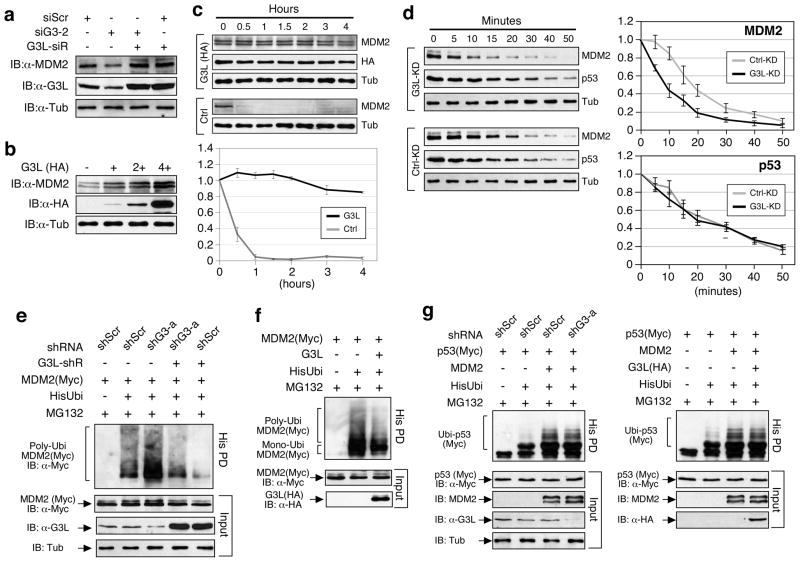
We investigated how GNL3L affects MDM2 and found that siRNA knockdown of GNL3L (siG3-2) reduces the amount of endogenous MDM2 protein in p53-inactive HeLa cells, which can be reversed by putting back a siG3-2-resistant GNL3L construct (G3L-siR) (Fig. 3a). In consistence, GNL3L overexpression increases the endogenous MDM2 protein level in a dose-dependent manner (Fig. 3b). We next determined the effect of GNL3L overexpression on MDM2 protein stability in cycloheximide-treated H1299 cells expressing recombinant MDM2 proteins. Western results showed that MDM2 proteins in the GNL3L-overexpressing cells are degraded much slower than that in the control cells (Fig. 3c, p < 0.0001 by Repeated Measures ANOVA). To confirm this finding, the endogenous MDM2 protein stability was measured in cycloheximide-treated HCT116-8 cells. GNL3L knockdown (G3L-KD) significantly reduces the stability of the endogenous MDM2 protein (protein half-life [T1/2] = 8.4 minutes) as compared to the control knockdown (T1/2 = 17.2 minutes, n=3, p < 0.005 by Repeated Measures ANOVA), but has no effect on the endogenous p53 protein stability (p=0.27) (Fig. 3d). To investigate whether GNL3L regulates MDM2 ubiquitylation, HEK293 cells were transfected with (His)6-tagged ubiquitin, MDM2, and GNL3L-targeting shRNAmir (shG3- a) constructs. Ubiquitylated proteins were captured by Ni2+-chelating sepharose and detected by anti-MDM2 western blots. The results showed that GNL3L knockdown significantly increases the ubiquitylation of MDM2, and that this effect can be rescued by the shG3-a-resistant GNL3L (G3L-shR) (Fig. 3e). Similarly, GNL3L overexpression decreases the amount of ubiquitylated MDM2 (Fig. 3f). In consistence with the lack of effect of GNL3L on p53 protein stability, neither knockdown nor overexpression of GNL3L changes the amount of MDM2-medidated p53 ubiquitylation (Fig. 3g).
Figure 3. GNL3L stabilizes MDM2 protein by preventing its ubiquitylation.
(a) Transient siRNA knockdown of GNL3L (siG3-2) in HeLa cells decreases the level of endogenous MDM2 proteins. This effect can be reversed by coexpression of a siG3-2-resistant GNL3L, G3L-siR. (b) Coexpression of GNL3L (HA) increases the protein level of endogenous MDM2 in a dose-dependent manner. (c) The effect of GNL3L on MDM2 protein stability was determined in H1299 cells transfected with MDM2 alone (Ctrl) or cotransfected with MDM2 and GNL3L (G3L). After cycloheximide treatment, the MDM2 protein amounts were measured, adjusted by their α-tubulin amounts, and expressed as percentages of the MDM2 protein amount at the 0h time-point (bottom). (d) The cycloheximide experiment showed that GNL3L knockdown in HCT116-8 cells significantly reduces the protein stability of endogenous MDM2 (n=3, p<0.005, Repeated Measures ANOVA) without affecting the stability of endogenous p53 (p=0.27). (e) HEK293 cells were transfected with the (His)6-tagged ubiquitin, MDM2 and a control (shScr) or GNL3L-specific (shG3-a) shRNAmir construct. Ubiquitylated MDM2 products were precipitated by Ni2+ sepharose (His PD) and detected by anti-MDM2 antibody. GNL3L depletion increases the ubiquitylation of MDM2, and this effect can be rescued by an shRNA-resistant GNL3L (G3L-shR). (f) Overexpression of wild-type GNL3L decreases MDM2 polyubiquitylation compared to the control sample. (g) GNL3L knockdown (left panel) or overexpression (right panel) has no effect on the MDM2-induced p53 ubiquitylation.
GNL3L constitutively interacts with MDM2 in the nucleoplasm
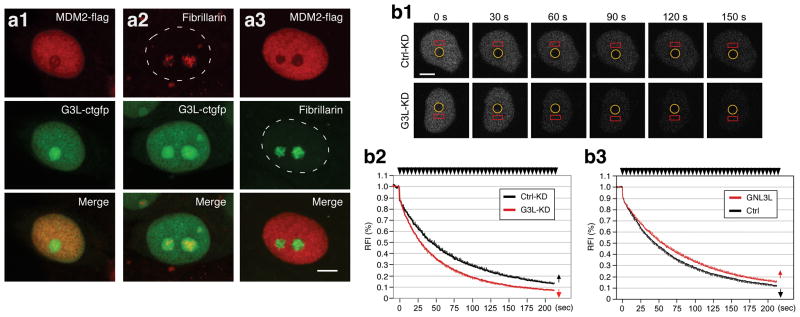
To determine where GNL3L and MDM2 interact within the cell, HCT116-8 cells were transfected with GFP-fused GNL3L and FLAG-tagged MDM2 individually or together. Confocal studies showed that GNL3L and MDM2 overlap considerably in the nucleoplasm but not in the nucleolus (Fig. 4a). To show their dynamic interaction in living cells, FLIP (fluorescence loss during photobleaching) was used to determine the nucleolar-nucleoplasmic exchange rate of GFP-fused MDM2, represented by the disappearance of its nucleoplasmic signal while bleaching the nucleolus (Meng et al., 2007). We reasoned that if GNL3L complexes with MDM2 in the nucleoplasm, the exchange rate of MDM2 will be increased by GNL3L knockdown. Indeed, the nucleolar-nucleoplasmic exchange rate of MDM2 is significantly increased by GNL3L knockdown (G3L-KD, mean decay half-time [T1/2] = 45.1 seconds) compared to the control-knockdown samples (Ctrl-KD, T1/2 = 28.7 seconds, p < 0.01 by Repeated Measures ANOVA, n=27) (Fig. 4b1, b2). By contrast, overexpression of GNL3L has only a small effect on reducing the exchange rate of MDM2 (p = 0.05 by Repeated Measures ANOVA, n=30) (Fig. 4b3), which may be due to the constitutive interaction between MDM2 and the endogenous GNL3L. This is in contrast to what we have found in NS, which shows a much clearer effect on the MDM2 FLIP rate when overexpressed than knocked down (Meng et al., 2008). These results demonstrate that GNL3L constitutively interacts with MDM2 in the nucleoplasm.
Figure 4. GNL3L colocalizes with MDM2 in the nucleoplasm. Knockdown of GNL3L increases the nucleolar-nucleoplasmic exchange rate of MDM2.
(a) Confocal studies showed that GFP-fused GNL3L and FLAG-tagged MDM2 colocalize in the nucleoplasm of HCT116-8 cells and that coexpression of these two proteins does not change each other's distributions. Anti-fibrillarin staining labels the nucleolus in singly transfected cells. Bars show 5um. (b1) The exchange rate between the nucleolar and nucleoplasmic pools of MDM2 was measured in HCT116-8 cells by FLIP (fluorescence loss in photobleaching). The nucleolar signal of GFP-fused MDM2 was bleached repeatedly (circle) and the nucleoplasmic signal was recorded (rectangle). Time-sequenced images and their time intervals (in seconds) to the first bleaching pulse were shown. (b2) Knockdown of GNL3L (G3L-KD, in red) significantly increases the nucleolar-nucleoplasmic exchange rate of MDM2 (p < 0.01). (b3) Overexperssion of GNL3L exerts only a small effect on decreasing the exchange rate of MDM2 (p = 0.05). Error bars represent s.e.m. shown in one direction (arrows in graph). Y-axis represents the relative fluorescence index (RFI). Top arrows indicate bleaching pulses.
The MDM2-regulatory activity of GNL3L is constitutively active and does not depend on the nucleolar release mechanism
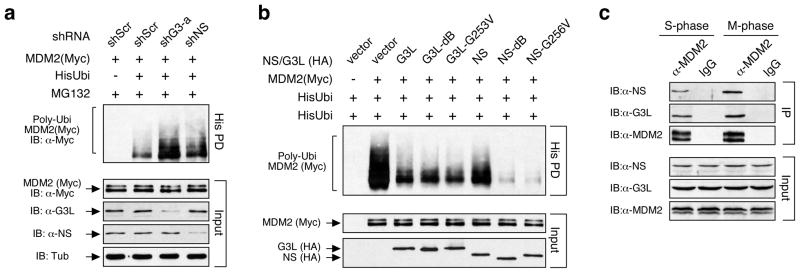
The FLIP results indicate that GNL3L binds MDM2 constitutively. We therefore hypothesize that the MDM2 ubiquitylation activity of GNL3L may also be constantly active and less regulated by the nucleolar release mechanism compared to that of NS. To test this idea, we first measured the effect of GNL3L and NS knockdown on MDM2 ubiquitylation and showed that GNL3L knockdown has a more profound effect in increasing MDM2 ubiquitylation than does NS knockdown (Fig. 5a). Next, we compared the MDM2 ubiquitylation effects of wild-type GNL3L and its nucleoplasm-mislocalized mutants. These mutations include deletion of its nucleolar localization signal (G3L-dB) or point mutation on the conserved GTP-binding residue (G3L-G253V) (Fig. S1). The results showed that these two nucleoplasmic forms of GNL3L show comparable activities in reducing MDM2 ubiquitylation to the wild-type GNL3L (Fig. 5b). The effect of NS overexpression in reducing MDM2 ubiquitylation is slightly less than that of GNL3L overexpression. Notably, the MDM2-regulatory activity of NS nucleoplasmic mutants is significantly stronger compared not only to wild-type NS but also to wild-type and mutant GNL3L, indicating that after being released from the nucleolus, NS may exert a stronger effect in reducing MDM2 ubiquitylation than GNL3L does. The release of nucleolar proteins from the nucleolus can be triggered by the mitotic entry. Since more NS is partitioned in the nucleolus than GNL3L, one may expect this event to increase the NS-MDM2 binding more than the GNL3L-MDM2 binding. To compare the effect of mitosis-induced nucleolar disassembly on the NS-MDM2 and GNL3L-MDM2 interactions, we measured the endogenous binding between MDM2 and GNL3L (or NS) in S-phase and M-phase synchronized cells, and found that the mitosis-induced increase of coIP efficiency is more evident in the NS-MDM2 binding than in the GNL3L-MDM2 binding (Fig. 5c). Together, these data demonstrate that despite the common activity of NS and GNL3L in preventing MDM2 ubiquitylation, they differ in that the GNL3L effect is constitutively active and the NS effect is triggered by the nucleolar release mechanism.
Figure 5. The activity of GNL3L in regulating MDM2 ubiquitylation is constitutively active and less controlled by the nucleolar release mechanism than NS is.
(a) For in vivo ubiquitylation assays, HEK293 cells were transfected with the (His)6- tagged ubiquitin, MDM2 (Myc), and GNL3L or NS-specific shRNA constructs. GNL3L knockdown shows a stronger effect in increasing MDM2 ubiquitylation than NS does. (b) The effects of GNL3L and NS overexpression on MDM2 ubiquitylation were compared using the same assay. Overexpression of wild-type and nucleoplasmic forms (G3L-dB and G3L-G253V) of GNL3L shows the same effect in decreasing the ubiquitylation of MDM2. On the other hand, nucleoplasmic forms of NS (NS-dB and NS-G256V) show much stronger effects compared not only to wild-type NS but also to wild-type and mutant GNL3L. (c) Endogenous coIP of MDM2 and GNL3L (or NS) showed that mitosis increases the coIP efficiency of NS and MDM2 more than it does on the coIP of GNL3L and MDM2.
GNL3L knockdown induced G2/M arrest and p53 activation
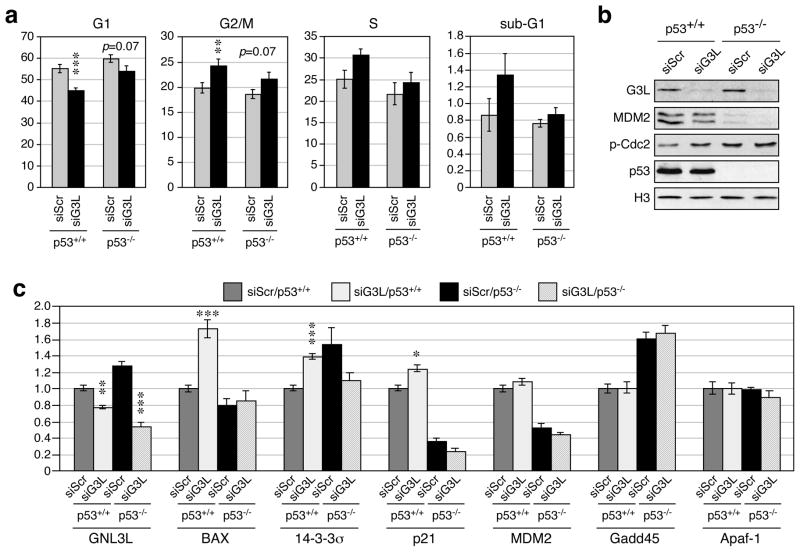
One frequently debated issue concerning the NS activity is whether its cell cycle effect is p53 dependent or not. To clarify this issue for GNL3L, we examined the cell cycle profiles of GNL3L knockdown in two isogenic cell lines that differ only in their p53 status. The parental HCT116-8 cells were derived from human colorectal carcinoma with normal p53 functions and a mutation in the ras proto-oncogene. The HCT116-2 line is a derivative of the HCT116-8 line, in which p53 is inactivated by homologous recombination (Bunz et al., 1998). In HCT116-8 (p53+/+) cells, GNL3L knockdown decreases the G1 cell percentage (p < 0.0001) and increases the percentage of G2/M cells (p = 0.001) (Fig. 6a). In HCT116-2 (p53−/−) cells, the G2/M arrest effect of GNL3L knockdown is less significant (p = 0.07). The differential sensitivity of p53+/+ and p53−/−cells to GNL3L knockdown is not caused by differences in their GNL3L knockdown efficiencies, as western blots confirmed a more effective GNL3L knockdown in the p53−/−cells than in the p53+/+ cells (Fig. 6b). Western blots also showed that GNL3L knockdown decreases the amount of MDM2 protein in both p53-wild-type and null cells and increases the amount of phospho-Cdc2 (Y15) (an inhibitor of mitotic entry) mainly in the p53-wild-type cells, but does not change the p53 protein level. To determine how GNL3L depletion affects the p53 activity, we measured the mRNA levels of several p53-regulated genes by real-time RT-PCR (qRT-PCR) (Fig.6c). The results first confirmed that the GNL3L knockdown efficiencies in p53-wild-type and null cells are 25% and 50%, respectively. Notably, GNL3L depletion induces a significant up-regulation of Bax (p<0.0001), 14-3-3-σ (p<0.0001), and p21 (p<0.01) in a p53-dependent manner, but does not affect the expression levels of MDM2, Gadd45, or Apaf-1.
Figure 6. GNL3L knockdown triggers G2/M arrest and p53 activation.
(a) The cell cycle phenotypes of control (siScr) and GNL3L (siG3L) knockdown were analyzed by propidium-iodide-labeled flow cytometry in HCT116-8 (p53+/+) and HCT116-2 (p53−/−) cells. GNL3L knockdown triggers G2/M arrest more in the p53+/+ cells than in the p53−/− cells. (b) Western blots confirm GNL3L knockdown efficiencies and reveal a slight decrease of MDM2 proteins in the p53-wild-type and null cells and an increase of phospho-Cdc2 (Y15) in the p53-wild-type cells by GNL3L knockdown. (c) The GNL3L knockdown effect on the p53 activity was determined by quantitative RT-PCR assays of several p53 downstream targets. GNL3L depletion up-regulates Bax, 14-3-3-σ, and p21 in a p53-dependent manner. *, p < 0.01; **, p < 0.001; ***, p < 0.0001.
NS depletion triggers a compensatory up-regulation of GNL3L
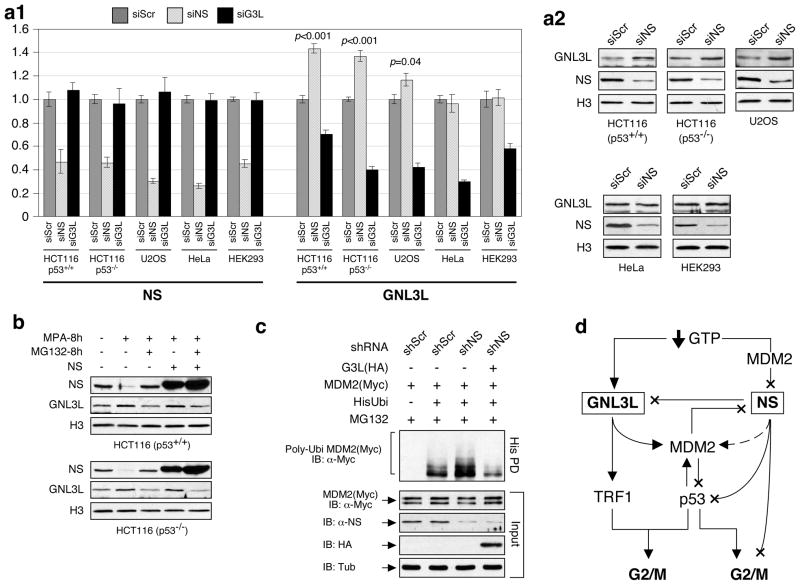
To determine whether the expressions of NS and GNL3L are co-regulated, we compared their mRNA and protein levels in control (siScr), NS-knockdown (siNS), and GNL3L-knockdown (siG3L) cells (Fig. 7a). qRT-PCR and western assays showed that NS depletion induces a significant up-regulation of GNL3L transcripts and proteins in both p53-wild-type and null HCT116 cells (p<0.001) and a moderate increase of GNL3L in U2OS cells (p=0.04). The increase of GNL3L following NS knockdown is cell type-specific, as NS-knockdown HeLa and HEK293 cells show no such an effect, and does not work reciprocally on NS when GNL3L is depleted. NS can also be decreased by guanine nucleotide depletion at the protein level (Huang et al., 2009). Interestingly, mycophenolic acid (MPA) treatment, which depletes the endogenous guanine nucleotide by blocking the de novo guanine nucleotide biosynthesis, concomitantly decreases the NS protein and increases the GNL3L protein (Fig. 7b). This increase of GNL3L protein can be reversed by adding a 26S proteasome inhibitor, MG132, but not by restoring the NS level, indicating that the MPA effect on GNL3L is not directly controlled by the level of NS protein. To further examine whether GNL3L can functionally compensate for the loss-of-NS effect on MDM2 ubiquitylation, we performed rescue experiments and showed that the NS-knockdown-induced MDM2 ubiquitylation increase can be fully reversed by coexpression of GNL3L (Fig. 7c). These results indicate that some cells may up-regulate GNL3L to functionally compensate for the loss of NS.
Figure 7. NS depletion triggers a compensatory up-regulation of GNL3L in HCT116 and U2OS cells.
(a1) qRT-PCR measurements revealed a compensatory up-regulation of GNL3L transcripts following NS knockdown in p53-wild-type and null HCT116 cells and U2OS cells but not in HeLa or HEK293 cells. Conversely, GNL3L knockdown shows no such effect on NS. (a2) The increase of GNL3L expression by NS knockdown was confirmed by western blots. (b) Mycophenolic acid (MPA) treatment triggers a reciprocal increase of GNL3L and decrease of NS in HCT116-8 and 2 cells. The MPA-induced GNL3L up-regulation can be reversed by MG132 treatment but not by NS overexpression. (c) GNL3L can functionally rescue the MDM2 ubiquitylation phenotype of NS-knockdown cells. (d) GNL3L and NS form a complex regulatory network on the MDM2-p53 pathway. Both proteins show similar activities in MDM2 stabilization but differ in their modes of regulation. Multiple points of feedback and cross-regulation exist. Arrows and X’s indicate excitatory or inhibitory functions, respectively, and the dashed line represents an event regulated by the nucleolar release mechanism.
Correlation between GNL3L expression and p53 activities in human cancers
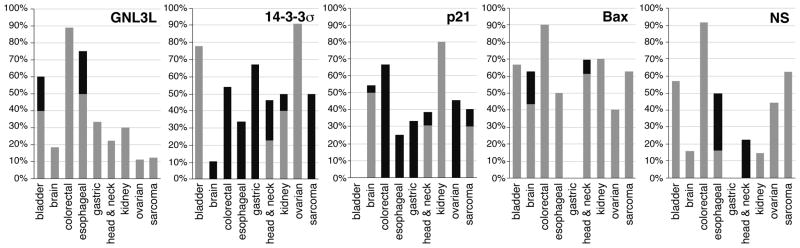
To establish the relevance of GNL3L and its p53 regulatory activity in human cancers, we first analyzed the expression levels of GNL3L in primary tumor samples using the Oncomine 4.3 (http://www.oncomine.org). We set the threshold of our analysis at p-value < 0.05 and fold-change > 1.5, and calculated the percentage of tumor samples showing up-regulation or down-regulation of the target genes (Fig. 8). We found that a significant portion of the bladder, brain, colorectal, esophageal, gastric, head and neck, kidney, ovarian, and sarcoma tumors show increased levels of GNL3L compared to the normal tissues. Particularly, the colorectal, esophageal, and gastric tumors display the highest percentages of samples with increased GNL3L expression. Notably, it is also in these three types of tumors that we found down-regulation of 14-3-3σ and p21. The decrease of 14-3-3σ and p21 in colorectal carcinomas indicates p53 suppression, which cannot be explained by the transcript changes in MDM2, p53, and other nucleolar proteins that stabilize p53, including ARF, B23, L5, L11, and L23 (Fig. S2). The expression of Bax, however, is increased in most tumor types. Together, these expression patterns support the idea that high levels of GNL3L may suppress p53 activities in human colorectal cancers.
Figure 8. Correlation between GNL3L expression and p53 activities in human cancers.
Nine types of human cancers show a significant percentage of samples with increased GNL3L expression. Among them, the colorectal, gastric, and esophageal carcinomas also show a down-regulation of 14-3-3σ and p21. Bax expression is increased in most tumors. Y-axis represents the percentage of tumor samples whose expression of the target gene is 1.5-fold higher (grey bars) or lower (black bars) than that of normal tissues.
Discussion
GNL3L and NS both stabilize MDM2 protein by preventing its ubiquitylation
This study reveals GNL3L as a novel MDM2 regulator as is its vertebrate paraglogue, NS (Fig. 7d). GNL3L and NS share similar MDM2-binding characteristics, i.e. both occur in the nucleoplasm and bind to the central domain of MDM2. The MDM2 interaction of GNL3L is mediated by its G- or I-domain, which is different from the MDM2-interactive C- and A-domains of NS. Functionally, GNL3L shows the same ability as NS in stabilizing MDM2 protein and preventing its ubiquitylation, and is capable of rescuing the MDM2 ubiquitylation effect caused by NS depletion. Like NS, MDM2 destabilization by GNL3L knockdown affects neither the ubiquitylation nor the stability of p53 proteins. Therefore, the GNL3L-stabilized MDM2 may inhibit the transcriptional activation of p53 through a direct binding mechanism without affecting the p53 protein level. Previous reports have shown that deletion of the acidic-zinc finger domain of MDM2 blocks its p53 degradation activity (Kubbutat et al., 1999) and that p53 ubiquitylation can be inhibited by peptide binding to the acidic domain of MDM2 (Wallace et al., 2006). Considering that GNL3L also binds to the central domain of MDM2, the lack of effect on p53 stability and ubiquitylation by GNL3L indicates that GNL3L binding to MDM2 may disrupt the E3 ligase activity of MDM2, which may also explain how NS and GNL3L prevent MDM2 ubiquitylation, as MDM2 also serves as its own E3 ligase (Fang et al., 2000).
Distinct features in GNL3L and NS-mediated MDM2 regulation
Despite their similar effects on MDM2 stabilization, GNL3L and NS are designed to operate under distinct biological contexts. The binding and ubiquitylation activity of GNL3L on MDM2 is constitutive and does not so much depend on the nucleolar release mechanism. In contrast, the MDM2-stabilizing effect of NS is only active when the nucleolar release mechanism is triggered and therefore is mainly used in the context of cell cycle counting or stress response. Such a difference in the GNL3L and NS-mediated MDM2 regulation may be due to their differential nucleolar-nucleoplasmic partitioning dynamics (Meng et al., 2007). Consistent with the idea that the MDM2-stabilizing effect of GNL3L is constitutively active and the NS effect is kept inactive in normal growing interphase cells, GNL3L knockdown shows a stronger effect on the FLIP kinetics and ubiquitylation of MDM2 than NS knockdown does. Even though wild-type NS is mostly inactive in MDM2 regulation in unstressed interphase cells, a significant amount of NS protein can still be seen in the nucleoplasm when overexpressed. This nucleoplasmic overflow of overexpressed NS and the constitutive binding of endogenous GNL3L and MDM2 may explain why NS and GNL3L overexpression show only a small difference in MDM2 ubiquitylation. It should be noted that the MDM2-regulatory activity of NS nucleoplasmic mutants (NS-dB and NS-G256V) is significantly stronger than that of wild-type and mutant GNL3L, and that NS-G256V shows a stronger MDM2-binding ability than GNL3L. These two results indicate that once released from the nucleolus, NS may exert a stronger effect in reducing MDM2 ubiquitylation than GNL3L does. Besides MDM2 regulation, NS and GNL3L also exercise distinct biological activities in TRF1 and ERRγ regulation, where they show opposite effects on TRF1 protein stability (Zhu et al., 2009), and only GNL3L can inhibit the transcription activity of ERRγ (Yasumoto et al., 2007).
Relationship between MDM2 destabilization and p53 activation in GNL3L-depleted cells
GNL3L knockdown produces a more prominent G2/M arrest phenotype in the p53-wild-type cells (p < 0.001) than in the p53-null cells (p = 0.07), showing that the p53 pathway clearly contributes to the cell cycle phenotype of GNL3L-knockdown cells. GNL3L knockdown also leads to increases in phospho-Cdc2 (Y15) and several p53 downstream targets, including Bax, 14-3-3σ, and p21. Up-regulation of 14-3-3σ or p21 can trigger G2/M arrest by retaining the Cdc2/cyclin B1 complex in the cytoplasm (Hermeking et al., 1997; Taylor & Stark, 2001) or by directly inhibiting Cdc2 (Bunz et al., 1998; Taylor & Stark, 2001), respectively. Notably, the expression profiles of GNL3L, 14-3-3σ, and p21 in primary tumor samples fit the idea that GNL3L plays a role in suppressing p53 activities in the colorectal, esophageal, and gastric cancers. However, most human tumors with increased GNL3L also show a paradoxical increase of Bax, even though GNL3L knockdown increases Bax more than 14-3-3σ or p21 in HCT116 cells. Considering the cell type complexity of human cancer samples, this result may not exclude the role of Bax in mediating the GNL3L effect in primary tumors. It is worthy of note that MDM2 can also be regulated by other nucleolar proteins, including ARF (alternative reading frame), B23, L5, L11, and L23 (Dai et al., 2008; Dai et al., 2004; Jin et al., 2004; Kurki et al., 2004; Meng et al., 2008; Tao & Levine, 1999; Zhang et al., 2003). Most of these proteins stabilize p53 by inhibiting or sequestering MDM2 in the nucleolus. In colorectal tumors, the levels of ARF, B23, L5, L11, and L23 genes are either up-regulated or not changed, which raises the possibility that GNL3L may be a key factor in suppressing the p53 activity in these tumors.
NS depletion is associated with a compensatory up-regulation of GNL3L
This new discovery was first made in the knockdown experiment. Besides siRNA knockdown, there are only two physiological events known so far that can reduce the expression of NS, i.e. cell differentiation (Tsai & McKay, 2002) and guanine nucleotide depletion (Huang et al., 2009). Indeed, the decrease of NS expression during neural differentiation is accompanied by a reciprocal increase of GNL3L expression (Tsai, unpublished data). Here, we also observe that guanine nucleotide depletion can trigger NS decrease and GNL3L increase simultaneously, and that this increase of GNL3L can be reversed by MG132 treatment but not by restoring the level of NS protein, which argues against a direct control over GNL3L expression by NS. Importantly, this phenomenon does not occur in all cancer cell lines. It appears in HCT116 and U2OS cells, but not in HeLa or HEK293 cells. Considering that HCT116 and U2OS cells are Rb1-wild-type and p16-inactive, HeLa cells are Rb1-inactive and p16-wild-type, and HEK293 cells are Rb1-wild-type and p16-wild-type, this cell type-dependent up-regulation of GNL3L by NS knockdown should not involve the p53 and Rb1 pathways and may explain some of the cell type-specific phenotypes of NS knockdown (Nikpour et al., 2009). It also suggests that some cells may survive the consequence of NS depletion better than others in the events where NS and GNL3L share redundant functions, such as MDM2 regulation.
Conclusion
MDM2 is a key regulator of p53, which arguably is the most important tumor suppressor gene in the genome. Our data demonstrate that GNL3L stabilizes MDM2 by inhibiting its ubiquitylation and that depletion of GNL3L causes G2/M arrest and up- regulation of Bax, 14-3-3σ, and p21. Compared to the NS effect, the MDM2-regulatory activity of GNL3L does not so much depend on the nucleolar release mechanism as NS does and behaves as a basal regulator of MDM2. Our work shows that NS and GNL3L are two nucleolar proteins that uniquely stabilize MDM2 and suppress p53 activities, and may play a role in promoting tumor formation in human colorectal carcinomas.
Materials and Methods
Cell culture and antibodies
HCT116 and U2OS cells were cultured in McCoy’s 5A medium plus 10% FBS. Other cell lines were maintained in DMEM plus 10% FBS. Early S-phase synchronization was achieved by incubation with 2mM thymidine for 20h. Mitotic arrest was done by incubation with 0.5uM nocodazole for 20h. Primary antibodies include anti-HA (HA.11), Myc (9E10), FLAG (M2), α-tubulin (Sigma), MDM2 (SMP-14), p53 (DO-1), p-Cdc2 (Y15, Cell Signaling), p-Histone H3 (S10, Upstate), NS (Ab2438, Ab1164, and Ab138), and GNL3L (Ab134 and Ab3404).
Coimmunoprecipitation
Cells were harvested in NTEN buffer. Lysates were incubated with primary antibody for 1 hour at 4C, followed by incubation with protein G sepharose beads (Pharmacia) for an additional 4 hours at 4C. Immunoprecipitates were washed 3 times with RIPA buffer before SDS-PAGE and western detection.
siRNA knockdown of NS and GNL3L
Transient knockdown experiments were performed by either Lipofectamine-mediated transfection of shRNAmir constructs or Oligofectamine-mediated transfection of siRNA duplexes. Designs for NS and GNL3L-specific siRNAs and shRNAs were described previously (Meng et al., 2008; Zhu et al., 2009).
FLIP (Fluorescence Loss In Photobleaching)
FLIP experiments were performed in HCT116-8 cells using the same setup as described previously (Meng et al., 2007). A nucleolus of 3-um diameter was bleached with repetitive pulses at 70% power of the 488 nm Argon laser. The relative fluorescence index (RFI) in the nucleoplasm of bleached cells was normalized to the nucleoplasmic intensity of neighboring non-bleached cells after background subtraction using the following calculation: RFI=(I(t)/I(0))*(C(0)/C(t)), where I(t) and I(0) are the background-subtracted intensities in the bleached cell at time point t and before photobleaching. C(t) and C(0) are the background-subtracted intensities in the neighboring control cell at time point t and before photobleaching. FLIP data represent the average of 27–30 cells from 4–5 independent experiments.
Protein degradation and in vivo ubiquitylation assays
The cycloheximide protein degradation and in vivo ubiquitylation assays were performed as described before (Meng et al., 2008). For overexpression protein degradation experiments, Myc-tagged MDM2 was coexpressed with GNL3L in H1299 cells, which express little MDM2. For knockdown protein degradation experiments, the endogenous MDM2 and p53 proteins were measured in HCT116-8 cells.
Cell-cycle analysis
Cell-cycle profiles were analyzed by counting the PI-labeled cells with a COULTER EPICS XL flow cytometer and the XL System II software. Each cell-cycle profile was compiled from 2×104-gated events and analyzed using the Multi Cycle AV software.
Quantitative RT-PCR (qRT-PCR) analyses
Total RNAs (5ug) were reversed transcribed into 1st strand cDNAs using random hexamers and M-MLV reverse transcriptase. For qPCR, the ΔC(t) values between the target message and the reference message (Rplp0) were determined by the MyiQ single-color real-time PCR detection system and supermix SYBR green reagent. The ΔΔC(t) values were measured from three biological replicates and two technical repeats (n=6) to compare the relative expression levels of target sequences between different groups. All final results were confirmed by comparing to a second reference message, HMG-14.
Supplementary Material
Acknowledgments
We gratefully acknowledge Bert Vogelstein of the Johns Hopkins Oncology Center for providing the HCT116 cells. This work is supported by NCI-PHS grant R01 CA113750 to R.Y. Tsai.
References
- Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, Phinney DG. J Cell Biochem. 2003;89:1235–49. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- Beekman C, Nichane M, De Clercq S, Maetens M, Floss T, Wurst W, Bellefroid E, Marine JC. Mol Cell Biol. 2006;26:9291–301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H. Mol Cell Biol. 2008;28:4365–76. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Mol Cell Biol. 2004;24:7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. J Biol Chem. 2000;275:8945–51. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Fu D, Collins K. Mol Cell. 2007;28:773–85. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- Huang M, Itahana K, Zhang Y, Mitchell BS. Cancer Res. 2009;69:3004–12. doi: 10.1158/0008-5472.CAN-08-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A, Itahana K, O'Keefe K, Zhang Y. Mol Cell Biol. 2004;24:7669–80. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Ludwig RL, Levine AJ, Vousden KH. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Cancer Cell. 2004;5:465–75. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- Ma H, Pederson T. Mol Biol Cell. 2007;18:2630–5. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki N, Takechi K, Sano S, Tarui H, Sasai Y, Agata K. Dev Dyn. 2007;236:941– 50. doi: 10.1002/dvdy.21027. [DOI] [PubMed] [Google Scholar]
- Meng L, Lin T, Tsai RY. J Cell Sci. 2008;121:4037–46. doi: 10.1242/jcs.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Yasumoto H, Tsai RY. J Cell Sci. 2006;119:5124–36. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Zhu Q, Tsai RY. Mol Cell Biol. 2007;27:8670–82. doi: 10.1128/MCB.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpour P, Mowla SJ, Jafarnejad SM, Fischer U, Schulz WA. Cell Prolif. 2009;42:762–9. doi: 10.1111/j.1365-2184.2009.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura M, Naka K, Hoshii T, Muraguchi T, Shugo H, Tamase A, Uema N, Ooshio T, Arai F, Takubo K, Nagamatsu G, Hamaguchi I, Takagi M, Ishihara M, Sakurada K, Miyaji H, Suda T, Hirao A. Stem Cells. 2008;26:3237–46. doi: 10.1634/stemcells.2008-0506. [DOI] [PubMed] [Google Scholar]
- Pederson T, Tsai RY. J Cell Biol. 2009;184:771–6. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S, Gude N, Hosoda T, Muraski J, Rubio M, Emmanuel G, Fransioli J, Vitale S, Parolin C, D'Amario D, Schaefer E, Kajstura J, Leri A, Anversa P, Sussman MA. Circ Res. 2008;103:89–97. doi: 10.1161/CIRCRESAHA.107.169334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. Proc Natl Acad Sci U S A. 1999;96:6937–41. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Oncogene. 2001;20:1803–15. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Tsai RY, McKay RD. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Meng L. Int J Biochem Cell Biol. 2009;41:2122–4. doi: 10.1016/j.biocel.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Mol Cell. 2006;23:251–63. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Yasumoto H, Meng L, Lin T, Zhu Q, Tsai RY. J Cell Sci. 2007;120:2532–43. doi: 10.1242/jcs.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Mol Cell Biol. 2003;23:8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Meng L, Hsu JK, Lin T, Teishima J, Tsai RY. J Cell Biol. 2009;185:827– 39. doi: 10.1083/jcb.200812121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yasumoto H, Tsai RY. Mol Cell Biol. 2006;26:9279–90. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.