Abstract
Motivation: Network diagrams are commonly used to visualize biochemical pathways by displaying the relationships between genes, proteins, mRNAs, microRNAs, metabolites, regulatory DNA elements, diseases, viruses and drugs. While there are several currently available web-based pathway viewers, there is still room for improvement. To this end, we have developed a flash-based network viewer (FNV) for the visualization of small to moderately sized biological networks and pathways.
Summary: Written in Adobe ActionScript 3.0, the viewer accepts simple Extensible Markup Language (XML) formatted input files to display pathways in vector graphics on any web-page providing flexible layout options, interactivity with the user through tool tips, hyperlinks and the ability to rearrange nodes on the screen. FNV was utilized as a component in several web-based systems, namely Genes2Networks, Lists2Networks, KEA, ChEA and PathwayGenerator. In addition, FVN can be used to embed pathways inside pdf files for the communication of pathways in soft publication materials.
Availability: FNV is available for use and download along with the supporting documentation and sample networks at http://www.maayanlab.net/FNV.
Contact: avi.maayan@mssm.edu
1 INTRODUCTION
Pathway databases such KEGG (Ogata et al., 1999), BioCarta (http://www.biocarta.com), WikiPathways (Pico et al., 2008) Science Signaling Connection Maps (Gough, 2002) and UCSD-Nature Signaling Gateway (Saunders et al., 2008) communicate over the web: cell signaling, transcriptional and metabolic pathways, as diagrams made of nodes and links. Such diagrams are visualized using different layout algorithms embedded in network viewers implemented with a variety of technologies. The majority of web-based network viewers make use of the Java web technologies. For example, PATIKAweb (Dogrusoz et al., 2006) uses Java Server Pages (JSP) to retrieve information stored in the manually curated PATIKA database, or passed through a file to generate pathway diagrams using a force-directed algorithm to arrange static images of nodes and edges. Tools such as WebInterViewer (Han et al., 2004) and VisANT (Hu et al., 2008) are useful for large protein–protein interaction networks utilizing JavaWebStart. However, JavaWebStart runs in a sandbox and does not easily communicate with the browser. Other tools, such as jSquid (Klammer et al., 2008), are powerful but since they utilize Java Applets they are slow to start and are inconsistent across browsers. Several web-based pathway viewers have been implemented without the use of Java. For example, CellDesigner (Funahashi et al., 2003) used by KEGG creates static network images with hyperlinks. It was used, for example, by BioPP (Viswanathan, 2007) with Perl and CGI displaying static images with annotations mapped to the nodes as hyperlinks. AVIS (Berger et al., 2007a), a network drawing tool that we developed, uses Asynchronous JavaScript and XML (AJAX) and underlying Perl libraries to draw static networks that are rendered using GraphViz. GraphViz (Gansner and North, 1999) is one of the most commonly used graph drawing tools for displaying pathways. Written in C, GraphViz takes text files as input to render nodes and links for generating a static network image that can be saved in different formats, i.e. scalable vector graphics (SVG), JPEG/PNG/GIF or as a postscript file. While network images can be mapped to hyperlinks, such mapping lacks interactivity such as panning and zooming, or movement of nodes and links with the mouse by the user. SVG can be made interactive but is not universally supported across browsers and is a fading technology. Although somewhat limited, GraphViz has been widely used by leading pathway databases to display their pathways online. For example, Science Signaling Connection Maps and UCSD-Nature Signaling-Gateway databases use SVG diagrams created with GraphViz. WikiPathways and BioCarta, on the other hand, use static images drawn by hand. The static images in WikiPathways are created manually with PathVisio, a tool that is a part of GenMAPP (Salomonis et al., 2007). Recently, a light-weight flash network viewer (FNV) for web applications called Cytoscape Web (Lopes et al., 2010) was launched. This viewer provides a variety of dynamic features. The use of vector graphics, provided through Flash, makes the implementation much more responsive and sharp looking. With Cytoscape, Web uploaded pathways can be altered without changing the input file, whereas a JavaScript application programming interface (API) is provided to alter uploaded networks.
2 IMPLEMENTATION
Here we present FNV. Implemented first 2 years ago, the viewer attempts to take the positives of all the existing viewers and provide users with a light-weight tool for incorporating biological pathways into their web applications. Written in Adobe ActionScript 3.0, FNV is a flash-based pathway viewer that is cross-platform and cross-browser compatible with little to no extra overhead. It contains several features that are not currently included in most existing pathway viewers, such as a content box for displaying descriptive information about nodes and edges, which includes support for hyperlinks and images, and the ability to use images as nodes.
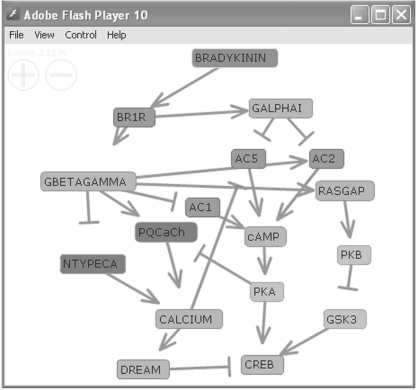
In an effort to maintain simplicity and readability, FNV uses a custom XML format to generate the networks/pathways. In addition, we provide users with the ability to easily embed the viewer within pdf files for the communication of pathways within publications. FNV is delivered as a small web format file. This allows for the complete abstraction of the ActionScript code requiring that the user only have knowledge of extensible markup language (XML). The XML file is the only input to FNV and therefore controls all of the layout and viewing options as well as all node and edge information. If the viewer is embedded in HTML, flash vars can be used to pass in parameters to FNV. Users can choose from five graph layout algorithms: force, spectral, hierarchy, spectral-hierarchy and radial. The force layout algorithm treats the nodes as objects that repel each other and the edges as springs with the objective of making edges relatively equal in length with few intersecting edges. Spectral layouts use eigenvalues of the adjacency matrix to generate coordinates for node placement. The hierarchy layout algorithm arranges the nodes so that directed edges flow from source nodes to sink nodes. The spectral-hierarchy layout option combines both hierarchical layout and spectral layout by first computing a hierarchy and then arranging nodes in each layer of the hierarchy using the spectral algorithm. Lastly, the radial algorithm positions nodes around the center of a circle depending on an inputted level. Descriptive text can include links to relevant databases and images. Interactions between nodes can be represented using three edge types: ortholine, arrow or bidirectional. Edge thicknesses, edge colors as well as node type, shape, size and color can be adjusted. Additionally, images can be used to represent nodes with or without text labels. Once rendered, the network can be rearranged dynamically by dragging nodes. In addition, zooming and panning support is implemented. The network can be saved as a pdf file or directly printed. A pathway created with FNV can be embedded inside pdf files as dynamic interactive figures in publications. FNV was used for several web-based systems: Genes2Networks (Berger et al., 2007b), a tool for finding relationships among an input list of genes; KEA (Lachmann and Ma'ayan, 2007), a tool for kinase enrichment analysis; ChEA (Lachmann et al., 2010), a tool to compute transcription factor binding enrichment; and PathwayGenerator, a tool that contains 5000 automatically generated mammalian cell signaling pathways connecting extracellular ligands and receptors to downstream effectors (Fig. 1).
Fig. 1.
Bradykinin to CREB pathway automatically created using PathwayGenerator (http://amp.pharm.mssm.edu/maayan-lab/pg) visualized with FNV.
Funding: National Institute of Health (grants 5P50GM071558-03, 1R01DK088541-01A1, KL2RR029885-0109 and RC2OD006536-01).
Conflict of Interest: none declared.
REFERENCES
- Berger S.I., et al. AVIS: AJAX viewer of interactive signaling networks. Bioinformatics. 2007a;23:2803–2805. doi: 10.1093/bioinformatics/btm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.I., et al. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007b;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogrusoz U., et al. PATIKAweb: a web interface for analyzing biological pathways through advanced querying and visualization. Bioinformatics. 2006;22:374–375. doi: 10.1093/bioinformatics/bti776. [DOI] [PubMed] [Google Scholar]
- Funahashi A., et al. CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. Biosilico. 2003;1:159–162. [Google Scholar]
- Gansner E.R., North S.C. An open graph visualization system and its applications to software engineering. Softw. Pract. Exp. 1999;S1:1–5. [Google Scholar]
- Gough N.R. Science's signal transduction knowledge environment: the connections map database. Ann. N Y Acad. Sci. 2002;971:585–587. doi: 10.1111/j.1749-6632.2002.tb04532.x. [DOI] [PubMed] [Google Scholar]
- Han K., et al. WebInterViewer: visualizing and analysing molecular interaction networks. Nucleic Acids Res. 2004;32:89–95. doi: 10.1093/nar/gkh462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., et al. VisANT: an integrative framework for networks in systems biology. Brief. Bioinform. 2008;9:317–325. doi: 10.1093/bib/bbn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klammer M., et al. jSquid: a Java applet for graphical on-line network exploration. Bioinformatics. 2008;24:1467–1468. doi: 10.1093/bioinformatics/btn213. [DOI] [PubMed] [Google Scholar]
- Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A., et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C.T., et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico A.R., et al. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders B., et al. The molecule pages database. Nucleic Acids Res. 2008;36:D700–D706. doi: 10.1093/nar/gkm907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan G.A., et al. BioPP: a tool for web-publication of biological networks. BMC Bioinformatics. 2007;8:168. doi: 10.1186/1471-2105-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]