Abstract
Protein–protein interactions are key to most cellular processes. Tandem mass spectrometry (MS/MS)-based proteomics combined with co-immunoprecipitation (CO-IP) has emerged as a powerful approach for studying protein complexes. However, a majority of systematic proteomics studies on protein–protein interactions involve the use of protein overexpression and/or epitope-tagged bait proteins, which might affect binding stoichiometry and lead to higher false positives. Here, we report an application of a straightforward, label-free CO-IP-MS/MS method, without the use of protein overexpression or protein tags, to the investigation of changes in the abundance of endogenous proteins associated with a bait protein, which is in this case insulin receptor substrate-1 (IRS-1), under basal and insulin stimulated conditions. IRS-1 plays a central role in the insulin signaling cascade. Defects in the protein–protein interactions involving IRS-1 may lead to the development of insulin resistance and type 2 diabetes. HPLC-ESI-MS/MS analyses identified eleven novel endogenous insulin-stimulated IRS-1 interaction partners in L6 myotubes reproducibly, including proteins play an important role in protein dephosphorylation [protein phosphatase 1 regulatory subunit 12A, (PPP1R12A)], muscle contraction and actin cytoskeleton rearrangement, endoplasmic reticulum stress, and protein folding, as well as protein synthesis. This novel application of label-free CO-IP-MS/MS quantification to assess endogenous interaction partners of a specific protein will prove useful for understanding how various cell stimuli regulate insulin signal transduction.
Keywords: HPLC-ESI-MS/MS, Insulin receptor substrate-1, Insulin signaling complexes, Label-free, Protein-protein interactions
Introduction
The majority of biological processes are regulated by precisely controlled protein–protein interactions. Tandem mass spectrometry (MS/MS) based proteomics combined with co-immunoprecipitation (CO-IP) offers a powerful approach for the study of complex protein–protein interactions [1]. A number of strategies have been developed for protein–protein interaction quantification, using either label-free or using stable-isotopic labeling approaches, such as stable isotope labeling with amino acids in cell culture (SILAC) [2, 3] and iTRAQ [4]. Although labeling approaches offer excellent reproducibility and short instrumentation time, much interest has been attracted to the label-free approach, due to its simplicity and cost efficiency [5–9]. In addition, in the label-free approach, there is no need for chemical reactions that might be a source of experimental variability. The label-free approach allows for quantification of protein abundance based on either the peak area/intensity of a peptide [6–9] or spectral counting [5, 10–12]. Rinner et al. analyzed sequentially diluted immunoprecipitation samples from control cells and cells stably expressing human hemagglutin tagged FoxO3A using the label-free quantification by mass spectrometry approach [8]. Their results indicated that background proteins showed a constant pattern in these sequentially diluted samples while real interaction proteins showed a varying pattern. This approach might require a large amount of starting material due to the serial dilution, a limiting factor for human tissue samples. More recently, Wepf et al. have designed a novel, label-free absolute quantification strategy to assess the stoichiometry of protein complexes using an isotope-labeled peptide corresponding to the sequence of an affinity tag [6]. In addition, Sardiu et al. built a sophisticated human protein interaction network using label-free mass spectrometry [5]. Vector algebra and statistical methods were combined with normalized spectral counting (NSAF) to analyze proteins affinity-purified with 27 different epitope-tagged baits.
The majority of proteomic studies that have focused on protein–protein interactions have involved the use of protein overexpression and/or epitope-tagged bait proteins, which might affect binding stoichiometry and lead to a higher rate of false positives. A label-free approach, without the utilization of protein overexpression or protein tags, may offer advantages for studying protein complexes at the endogenous level. Recently, Malovannaya et al. reported a large-scale CO-IP-MS/MS analysis of endogenous protein complexes in nuclear extracts of HeLa S3 cells under the basal state [13]. They built a human protein network in HeLa S3 cells based on more than 1000 CO-IP-MS/MS experiments they performed. Although the results obtained were comprehensive, a large number of samples were required to generate the requisite dataset.
Insulin signaling is crucial to many biological processes in muscle, including glycogen synthesis, glucose transport, mitogenesis, protein synthesis, and degradation. The intracellular actions of insulin are mediated by interactions between key regulatory proteins and enzymes (kinases and phosphatases). Defects in the insulin signaling pathway have been implicated in the development of insulin resistance and type 2 diabetes [14, 15]. An excellent prototypical example of a protein known to be involved in insulin-induced complex formation is insulin receptor substrate-1 (IRS-1). IRS-1 has a pleckstrin homology (PH) domain and a phosphotyrosine binding (PTB) domain, through which it interacts with the insulin receptor and insulin-like growth factor 1 receptor (IGF1R). IRS-1 also has several YXXM motifs. Upon phosphorylation, IRS-1 interacts with the p85 regulatory subunit of phosphatidylinositide (PI) 3-kinase (PI 3-K), which leads to the activation of this enzyme and subsequent activation of Akt, resulting in enhanced glucose uptake, and increased glycogen synthesis and protein synthesis. Abnormal protein–protein interactions involving IRS-1 may interfere with proper insulin transduction and lead to the development of insulin resistance and type 2 diabetes.
The present study was undertaken to use a proteomic approach to discover insulin-stimulated endogenous IRS-1 interaction partners in L6 myotubes. L6 Myotubes were either left untreated or treated with 100 nm insulin for 15 min. One mg of cell lysate protein from basal or insulin-stimulated myotubes was immunoprecipitated with an antibody specific to IRS-1, respectively. The resulting immunoprecipitates were separated on the same sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel in two separate lanes, which were then cut into 10 slices each, digested with trypsin, and the resulting peptides subjected to HPLC-ESI-MS/MS analysis. Using this straightforward approach, without the use of labeling or protein overexpression/tags, a number of endogenous insulin-stimulated IRS-1 interaction partners were detected reproducibly.
Materials and Methods
Antibodies and Reagents
Anti-protein phosphatase 1 regulatory subunit 12A (PPP1R12A) was purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA; anti-IRS1 and anti-p85 antibodies were purchased from Millipore, Temecula, CA, USA; anti-rabbit IgG and anti-mouse IgG-HRP linked secondary antibody were ordered from GE Healthcare UK Ltd. and ECL from Perkin Elmer, Shelton, CT, USA. Protein A sepharose beads and all other reagents were obtained from Sigma, St Louis, Mom, USA.
Cell Culture
The parental L6 rat myoblasts (ATCC, Manassas, VA, USA) were maintained in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 5% penicillin/streptomycin in a humidified atmosphere containing 5% CO2 and 95% air at 37 °C. Cells were subcultured by trypsinization of subconfluent cultures using 0.25% trypsin with EDTA. L6 myoblasts were seeded at a density of 8×105 cells per 10 cm dish. For differentiation into myotubes, myoblasts were placed in fresh medium containing 2% fetal bovine serum and penicillin/streptomycin every 24 h for 7 d. The myotubes were serum starved for 4 h at 37 °C before insulin stimulation.
Immunoprecipitation and Western Blotting Analysis
L6 myotubes were stimulated with or without insulin (100 nM) for 15 min at 37 °C. The cells were lysed with 1 mL of triton lysis buffer to detect protein–protein interactions (50 mM HEPES [pH 7.6], 150 mM NaCl, 20 mM sodium pyrophosphate, 10 mM NaF, 20 mM β-glycerophosphate, 1% Triton, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/mL leupeptin and aprotinin). Protein concentration was estimated by the Bradford Assay (Bio-Rad) using bovine serum albumin (Sigma, St. Louis, MO, USA) as a standard. Cell lysates (1 mg) were diluted in lysis buffer and incubated with 2 μg of IRS-1 antibody (Millipore, Temecula, CA, USA). The immunoprecipitates were collected with protein A agarose beads (Sigma, St. Louis, MO, USA) overnight at 4 °C with gentle rotation and were then washed three times with phosphate buffered saline (PBS). Samples were boiled in SDS-PAGE sample buffer and resolved on 7.5% 1D-SDS-PAGE. The proteins were visualized with Coomassie blue (Sigma Chemical Co., St. Louis, MO, USA). One basal and one insulin treated sample harvested on the same day were paired and resolved on a gel side by side to minimize gel to gel variations.
For Western blotting analysis, samples were boiled in SDS-PAGE sample buffer and resolved on 7.5% SDS-PAGE, transferred onto nitrocellulose membranes (Bio-Rad), and analyzed by Western blotting with the appropriate antibodies and the immune complex was detected by chemiluminescence (Perkin Elmer, Shelton, CT, USA).
In-Gel Trypsin Digestion, Mass Spectrometry, Data Analysis, and Bioinformatics
All were performed as previously described in references [16–18] with the exception that all tryptic peptides were desalted by solid-phase extraction instead of using an on-line nanotrap. Briefly, the gel lanes resulting from each experiment were cut into 10 slices of approximately equal size. Each slice was cut into 1 mm cubes prior to digestion. The gel pieces were destained and subjected to trypsin digestion at 37 °C overnight. The resulting peptides were extracted and purified by solid-phase extraction (C18 ZipTip; Millipore, Billerica, MA, USA), followed by online HPLC-ESI-MS/MS analyses on a hybrid linear ion trap (LTQ)-Fourier Transform Ion Cyclotron Resonance (FTICR) mass spectrometer (LTQ FT; Thermo Fisher, San Jose, CA, USA) fitted with a PicoView nanospray source (New Objective, Woburn, MA, USA). The HPLC-ESI-MS/MS runs were set up as below to minimize day to day experimental variations: slices #1–5 from the basal sample, followed by a blank, then slices #1–5 from the corresponding insulin sample resolved on the same gel, followed by a blank, then slices #6–10 from the basal sample, followed by a blank, then finally slices #6–10 from the insulin treated sample.
Tandem mass spectra were extracted from Xcalibur “RAW” files and charge states were assigned using the Extract MSN script (Xcalibur 2.0 SR2; Thermo Fisher, San Jose, CA, USA). The fragment mass spectra were then searched against the IPI RAT v3.59 database (39,866 entries, http://www.ebi.ac.uk/IPI/) using Mascot (Matrix Science, London, UK; ver. 2.2). The false discovery rate was determined by selecting the option to search the decoy randomized database. The search parameters used were: 10 ppm mass tolerance for precursor ion masses and 0.5 Da for product ion masses; digestion with trypsin; a maximum of two missed tryptic cleavages; variable modifications of oxidation of methionine and phosphorylation of serine, threonine, and tyrosine. Probability assessment of peptide assignments and protein identifications were made through use of Scaffold (ver. Scaffold 2 00 06, Proteome Software Inc., Portland, OR, USA). Only peptides with ≥95% probability were considered. Proteins that contained identical peptides and could not be differentiated based on MS/MS analysis alone were grouped. Criteria for protein identification included detection of at least two unique peptides and a probability score of ≥99%. Multiple isoforms of a protein were reported only if they were differentiated by at least one unique peptide with ≥95% probability, based on Scaffold analysis.
Statistical Analysis
Statistical significance was assessed by comparing basal and insulin-stimulated protein abundance (normalized to IRS-1 protein abundance) using the sign test developed by Frank Wilcoxon [19, 20]. The sign test is a non-parametric test that can pin-point directional changes while makes few assumptions about the nature of the distributions under test. If paired samples from two continuous random variables X and Y is drawn, the sign test is used to test the hypothesis that there is “no difference” between random variables X and Y. As detailed in the Results section, we have paired normalized spectrum counts from the basal (NormSC BAS) and corresponding insulin-stimulated conditions (NormSC INS) in our experiments. For a protein of interest, suppose X = NormSC BAS and Y = NormSC INS, we let P = Pr(Y > X). Then we test the null hypothesis that there is no difference between X and Y is equivalent to test H0: P = 0.50. That is, under the null hypothesis, it says that given a random pair of measurements (Xi, Yi), Xi and Yi are equally likely to be larger than the other. To perform the sign test, n independent pairs of sample data, (X1, Y1), (X2, Y2), …, (Xn, Yn), are collected from the populations. Note that those pairs with no difference are omitted. Let M be the number of pairs for which Yi is greater than Xi. Under the assumption that H0 is true, the probability of observing the number of pairs for which Yiis greater than Xi follows a binomial distribution, that is M ~ bin(n, 0.5). The probability to observe the number of pairs with Yi greater than Xi, Pr(M), can be calculated as . If the total number of pairs of independent measurement is 5, the probability to observe 5 pairs with Yi greater than Xi, can be calculated as
Results and Discussion
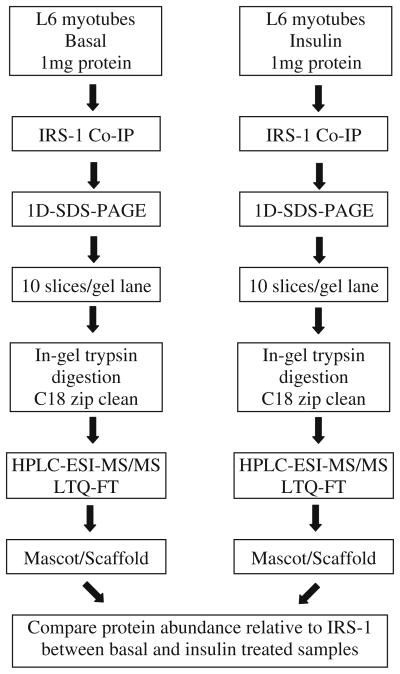
The outline of our approach is shown in Figure 1.
Figure 1.
Experimental workflow for the analysis of endogenous insulin stimulated IRS-1 interacting partners in L6 myotubes
From five independent biological comparisons (five basal and five insulin stimulated samples, 10 slices/gel lane, 100 HPLC-ESI-MS/MS runs), a total of 214 unique proteins were identified in at least one sample after reduction for redundancy. There were at least two unique peptides (95% confidence) assigned to each identified protein, all with a confidence level ≥99% based on the Scaffold analysis. The false discovery rate, as assessed by Mascot searching of a randomized database, was 10.9% at the peptide level. Since there were at least two unique peptides assigned to each identified protein, this gave a false discovery rate of 1.2% (10.9%*10.9% = 1.2%) at the protein level. A detailed list of all proteins identified in this study together with their IPI ID, molecular weight, and the number of tandem mass spectrum assigned to each protein is provided as Supporting Information (Supplemental Table 1). For each identified peptide sequence, we have included the following in Supplemental Table 2: modifications, flank residues, precursor mass, charge, and mass error observed, and the best Mascot score (Supplemental Table 2).
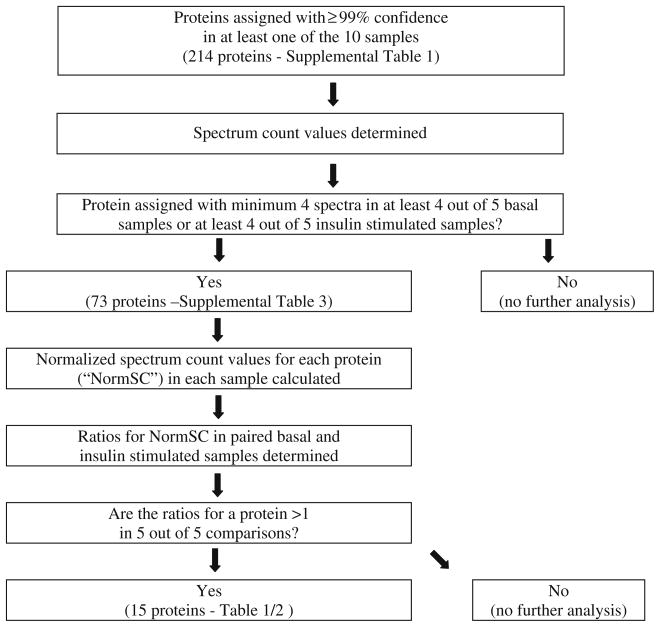
In order to minimize false positives, a series of filters were used to narrow the number of proteins that were used in comparisons between the basal and insulin stimulated conditions. This approach is diagrammed in Figure 2. First, the spectrum count, which is the number of MS/MS spectra assigned to a protein, was determined for each protein in each sample. In samples for which no spectra for a protein were observed, the spectrum count value for that protein in that sample was set to 0, and 0 s were included in all analyses of protein abundance differences. The first filter applied was to exclude all proteins from further analysis that were not detected with a minimum of four assigned spectra in at least four out of five basal samples or at least four out of five insulin stimulated samples. For example, a protein found in four out of five basal samples with a minimum of four assigned spectra in each sample would be included in the analysis regardless of how many times it was detected in the five insulin stimulated samples. There were 73 such proteins (Supplemental Table 3). The next step was to calculate the normalized spectrum values for each protein (termed “NormSC”) in each sample by dividing the spectrum count for a specific protein by the spectrum count for IRS-1 in the same sample, in order to correct for sample loading: .
Figure 2.
Data analysis workflow for endogenous insulin stimulated IRS-1 interacting partners in L6 myotubes
The normalization strategy is the same concept used in Western blotting, in which the Western blot signal for an interaction protein is normalized against that for the protein serving as the “bait.” As mentioned in the Methods section, the basal and insulin-stimulated samples that were harvested on the same day, resolved on the same gel, and analyzed by HPLC-ESI-MS/MS during the same period of time were paired to minimize day to day variations. After normalization, the ratio of the NormSC for the basal samples (NormSC BAS), and the paired insulin stimulated samples (NormSC INS), was calculated for each of the 73 proteins. There were five such ratios for each protein. A ratio greater than one indicates an increased association of a protein with IRS-1 upon insulin stimulation in that particular comparison experiment, while a ratio less than one indicates a decrease, and a value of one indicates no insulin induced change in association of the protein with IRS-1. If a protein has zero spectrum counts in the basal sample, while possessing the minimum value of four spectrum counts in the insulin stimulated sample, we did not calculate the ratio and, instead, considered the association as an insulin induced increase by default. If a protein has five increases out of the five comparisons performed, the P value of the test that this protein to be associated with IRS-1 randomly upon insulin-stimulation is 0.03125 based on the sign test (please see Methods for details). Fifteen proteins were identified as insulin-stimulated IRS-1 interaction partner (P = 0.03125) and they were listed in Table 1 and Table 2. No proteins with five decreases out of the five comparisons performed were detected. In summary, to be classified as a specific insulin stimulated interaction partner of IRS-1, a protein has to satisfy all three criteria:
Table 1.
HPLC-ESI-MS/MS analysis of endogenous PI 3-K subunits association with IRS-1 in L6 myotubes upon insulin stimulation. Data shown are mean ± SEM, n=5
| (A) Spectrum count and number of MS/MS spectra assigned to IRS-1 and PI 3-K subunits in the basal (BAS) and insulin (INS) stimulated samples | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbola | Molecular weight (AMU) | Spectrum count BAS | Spectrum count INS | Exp.1 BAS | Exp.1 INS | Exp.2 BAS | Exp.2 INS | Exp.3 BAS | Exp.3 INS | Exp.4 BAS | Exp.4 INS | Exp.5 BAS | Exp.5 INS |
| IRS-1 | 131160 | 105±10 | 100±8 | 93 | 108 | 85 | 85 | 105 | 126 | 145 | 91 | 99 | 88 |
| PIK3R1 | 83516 | 0±0 | 8±1 | 0 | 4 | 0 | 10 | 0 | 19 | 0 | 6 | 0 | 2 |
| PIK3R2 | 81251 | 11±2 | 74±15 | 12 | 88 | 16 | 83 | 12 | 100 | 2 | 71 | 11 | 29 |
| PIK3CA | 124233 | 1±1 | 20±4 | 0 | 17 | 0 | 17 | 0 | 26 | 0 | 28 | 3 | 12 |
| PIK3CB | 122595 | 0 ±0 | 6±1 | 0 | 6 | 0 | 7 | 0 | 6 | 0 | 6 | 0 | 3 |
| (B) Normalized spectrum count (NormSC) of PI 3-K subunits in the basal (BAS) and insulin (INS) stimulated samples | |||||||||||||
| Gene Symbol a | NormSC BAS | NormSC INS | Exp.1 BAS | Exp.1 INS | Exp.2 BAS | Exp.2 INS | Exp.3 BAS | Exp.3 INS | Exp.4 BAS | Exp.4 INS | Exp.5 BAS | Exp.5 INS | |
| PIK3R1* | 0.000±0.000 | 0.079±0.024 | 0.000 | 0.037 | 0.000 | 0.118 | 0.000 | 0.151 | 0.000 | 0.066 | 0.000 | 0.023 | |
| PIK3R2* | 0.111±0.028 | 0.739±0.108 | 0.129 | 0.815 | 0.188 | 0.976 | 0.114 | 0.794 | 0.014 | 0.780 | 0.111 | 0.330 | |
| PIK3CA* | 0.006±0.006 | 0.202±0.030 | 0.000 | 0.157 | 0.000 | 0.200 | 0.000 | 0.206 | 0.000 | 0.308 | 0.030 | 0.136 | |
| PIK3CB* | 0.000±0.000 | 0.057±0.008 | 0.000 | 0.056 | 0.000 | 0.082 | 0.000 | 0.048 | 0.000 | 0.066 | 0.000 | 0.034 | |
IRS-1 = insulin receptor substrate-1; PIK3R1 = PI 3-K p85 regulatory subunit ↦; PIK3R2 = PI 3-K p85 regulatory subunit β; PIK3CA = PI 3-K p110 catalytic subunit ↦; PIK3CB = PI 3-K p110 catalytic subunit β
P<0.05 by the sign test
Table 2.
HPLC-ESI-MS/MS analysis of endogenous IRS-1 interaction partners in L6 myotubes upon insulin stimulation. Data shown are mean ± SEM, n=5
| (A) Spectrum count (SC) of endogenous IRS-1 interaction partners in the basal (BAS) and insulin (INS) stimulated samples | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbol a | Molecular weight (Da) | SC BAS | SC INS | Exp.1 BAS | Exp.1 INS | Exp.2 BAS | Exp.2 INS | Exp.3 BAS | Exp.3 INS | Exp.4 BAS | Exp.4 INS | Exp.5 BAS | Exp.5 INS |
| ACTN1* | 102946 | 10±4 | 22±6 | 13 | 34 | 20 | 28 | 15 | 32 | 0 | 6 | 0 | 10 |
| ACTN4* | 104900 | 20±8 | 35±7 | 21 | 49 | 34 | 44 | 38 | 46 | 4 | 14 | 1 | 20 |
| CALD1* | 60567 | 11±4 | 21±6 | 23 | 41 | 14 | 27 | 17 | 22 | 1 | 13 | 1 | 3 |
| EEF2* | 95268 | 6±3 | 11±3 | 4 | 9 | 12 | 16 | 13 | 17 | 0 | 5 | 1 | 6 |
| HSP90AB1* | 83266 | 16±4 | 27±4 | 16 | 25 | 29 | 32 | 14 | 23 | 4 | 17 | 16 | 38 |
| HSPA5* | 72331 | 30±5 | 44±5 | 34 | 52 | 36 | 42 | 41 | 59 | 12 | 33 | 25 | 32 |
| HTRA1* | 51312 | 4±2 | 10±2 | 4 | 8 | 10 | 14 | 4 | 12 | 1 | 2 | 1 | 15 |
| MPRIP* | 257845 | 53±18 | 92±10 | 71 | 95 | 100 | 105 | 73 | 117 | 11 | 58 | 11 | 83 |
| PABPC1* | 70683 | 3±1 | 5±1 | 4 | 5 | 4 | 6 | 6 | 9 | 1 | 1 | 1 | 4 |
| PPP1R12A* | 115268 | 5±2 | 12±1 | 5 | 12 | 12 | 13 | 7 | 16 | 1 | 9 | 0 | 9 |
| TNNT3* | 30734 | 17±7 | 25±6 | 18 | 22 | 25 | 28 | 38 | 46 | 0 | 21 | 2 | 6 |
| (B) Normalized spectrum count (NormSC) of endogenous IRS-1 interaction partners in L6 myotubes from basal (BAS) and insulin (INS) stimulated samples | |||||||||||||
| Gene Symbol a | NormSC BAS | NormSC INS | Exp.1 BAS | Exp.1 INS | Exp.2 BAS | Exp.2 INS | Exp.3 BAS | Exp.3 INS | Exp.4 BAS | Exp.4 INS | Exp.5 BAS | Exp.5 INS | |
| ACTN1* | 0.104±0.046 | 0.216±0.062 | 0.14 | 0.32 | 0.24 | 0.33 | 0.14 | 0.25 | 0.00 | 0.07 | 0.00 | 0.11 | |
| ACTN4* | 0.206±0.081 | 0.344±0.088 | 0.23 | 0.45 | 0.40 | 0.52 | 0.36 | 0.37 | 0.03 | 0.15 | 0.01 | 0.23 | |
| CALD1* | 0.12±0.048 | 0.21±0.068 | 0.25 | 0.38 | 0.17 | 0.32 | 0.16 | 0.18 | 0.01 | 0.14 | 0.01 | 0.03 | |
| EEF2* | 0.062±0.029 | 0.108±0.03 | 0.04 | 0.08 | 0.14 | 0.19 | 0.12 | 0.14 | 0.00 | 0.06 | 0.01 | 0.07 | |
| HSP90AB1* | 0.166±0.05 | 0.282±0.069 | 0.17 | 0.23 | 0.34 | 0.38 | 0.13 | 0.18 | 0.03 | 0.19 | 0.16 | 0.43 | |
| HSPA5* | 0.302±0.063 | 0.432±0.083 | 0.37 | 0.48 | 0.42 | 0.49 | 0.39 | 0.47 | 0.08 | 0.36 | 0.25 | 0.36 | |
| HTRA1* | 0.044±0.02 | 0.106±0.032 | 0.04 | 0.07 | 0.12 | 0.17 | 0.04 | 0.10 | 0.01 | 0.02 | 0.01 | 0.17 | |
| MPRIP* | 0.566±0.209 | 0.926±0.189 | 0.76 | 0.88 | 1.18 | 1.24 | 0.70 | 0.93 | 0.08 | 0.64 | 0.11 | 0.94 | |
| PABPC1* | 0.034±0.01 | 0.05±0.013 | 0.04 | 0.05 | 0.05 | 0.07 | 0.06 | 0.07 | 0.01 | 0.01 | 0.01 | 0.05 | |
| PPP1R12A* | 0.054±0.025 | 0.118±0.023 | 0.05 | 0.11 | 0.14 | 0.15 | 0.07 | 0.13 | 0.01 | 0.10 | 0.00 | 0.10 | |
| TNNT3* | 0.172±0.072 | 0.24±0.064 | 0.19 | 0.20 | 0.29 | 0.33 | 0.36 | 0.37 | 0.00 | 0.23 | 0.02 | 0.07 | |
ACTN1 = ↦-actinin-1; ACTN4 = ↦-actinin-4; CALD1 = caldesmon; CENTG2 = centaurin γ 2; EEF2 = elongation factor 2; FLII = flightless I homolog; HSP90AB1 = heat shock protein HSP 90-β; HSPA5 = 78 kDa glucose-regulated protein; HTRA1 = HtrA serine peptidase 1 isoform CRA a; LRRFIP2 = leucine-rich repeat flightless-interacting protein 2; MPRIP = myosin phosphatase Rho-interacting protein; PABPC1 = polyadenylate-binding protein 1; PPP1R12A = protein phosphatase 1 regulatory subunit 12A; TNNT3 = isoform 1 of troponin T, fast skeletal muscle
Proteins have five increases out of the five comparisons
P<0.05 based on the sign test. There were eleven such proteins
Detected with at least two unique peptides assigned (each with 95% confidence based on the Scaffold analysis).
Detected with a minimum of four assigned spectra in at least four out of five basal samples or at least four out of five insulin stimulated samples,
Has five increases out of the five comparisons performed.
IRS-1 was detected from basal and insulin-stimulated cells (n = 5; 105±10 and 100±8 assigned MS/MS spectra, respectively). Out of the five comparisons, IRS-1 spectrum counts were higher in the insulin stimulated samples than those in the respective basal samples twice, lower twice, and equal once, indicating that there were no substantial changes in IRS-1 protein abundance after insulin treatment. However, we identified the p85↦ and β subunits of PI 3-K, a well known insulin-stimulated binding partner of IRS-1, in the basal samples with 0 ± 0 and 11 ± 2 assigned spectra, respectively, while in the insulin-stimulated samples these proteins were identified with 8±1 and 74±15 assigned spectra, respectively, indicating increased association of IRS-1 and p85 upon insulin stimulation (n = 5, Table 1). After correcting for sample loading by dividing the spectrum count for each of the subunits by the corresponding spectrum count of IRS-1 from each sample, the NormSC of the PI 3-K subunits showed an obvious increase after insulin treatment in all five comparisons (P = 0.03125, Table 1). Since p85 and p110 subunits of PI 3-K are well-known insulin stimulated interaction partners of IRS-1, these results served as a positive control and validated this experimental approach. It is noted that some proteins may not bind to IRS-1 directly, but indirectly through other proteins that do interact with IRS-1 directly, such as the p110 catalytic subunit of PI 3-K.
Furthermore, eleven additional proteins possessed substantially more assigned spectra and normalized spectrum counts upon insulin stimulation (n = 5, P = 0.03125, Table 2), indicating increased association with IRS-1. All of these identified proteins have five increases out of the five comparisons. No previous reports on the association of these proteins with IRS-1 in corresponding co-immunoprecipitates have been reported thus far.
As shown in Table 2, α-actinin-1 and α-actinin-4 are among the proteins that showed a reproducible increase in association with IRS-1 upon insulin stimulation; α-actinin-1 and α-actinin-4 were found to interact with GLUT4 overexpressed in L6 myotubes in response to insulin stimulation using stable isotope amino acid labeling and mass spectrometry techniques [21]. Interestingly, knockdown of α-actinin-4 but not α-actinin-1 impaired insulin-induced GLUT4 translocation [22]. Our results provide the first evidence indicating that α-actinin-4 may be regulated by insulin through IRS-1, or vice versa.
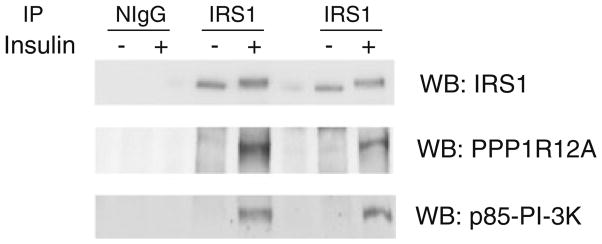
The protein PPP1R12A also underwent insulin-stimulated association with IRS-1 reproducibly. PPP1R12A, also known as the myosin phosphatase target subunit (MYPT1 or MBS), is highly expressed in non-muscle- and smooth-muscle-cells [23]. Recently, it has been reported that PPP1R12A is highly expressed in mouse EDL and soleus muscles [24]. PPP1R12A can bind to the catalytic subunit of protein phosphatase 1 (PP1c) and anchor PP1c to specific substrates such as myosin II [23, 25]. Protein phosphatase 1 (PP1) is a serine/threonine phosphatase that plays a role in a variety of cellular processes [26, 27]. Previous reports have shown that treatment of cells with okadaic acid, an inhibitor of both PP1 and the protein phosphatase 2A (PP2A), increases serine phosphorylation of IRS-1 and results in reduced insulin stimulated IRS-1 tyrosine phosphorylation [28–30]. Since our results indicate that IRS-1 may associate with the regulatory subunit of PP1, PPP1R12A, our data might provide insight as to how IRS-1 undergoes insulin induced dephosphorylation. We have performed Western blot analysis to confirm the insulin stimulated interaction of IRS-1 with PPP1R12A and p85. Serum starved L6 myotubes were either left untreated or stimulated with 100 nm insulin for 15 min and immunoprecipitated with either normal IgG (NIgG) negative control antibodies, which lack specific binding capability for IRS-1, or anti-IRS-1 antibodies, followed by immunoblotting with IRS-1, PPP1R12A, and p85 antibodies. As shown in Figure 3, PPP1R12A showed an increased abundance in the immunoprecipitates of IRS-1 after insulin stimulation by Western blot analysis (Figure 3, middle panel). As expected, p85 also showed an increased association with IRS-1 (Figure 3, lower panel), while there were no detectable bands in the NIgG immunoprecipitates in the regions of IRS-1, PPP1R12A, and p85, indicating the specificity of the antibodies.
Figure 3.
PPP1R12A and the p85 regulatory subunit of PI 3-K interact with IRS-1. L6 myotubes were serum-starved for 4 h and either left untreated or stimulated with insulin (100 nM) for 15 min at 37 °C. One mg of lysate proteins were immunoprecipitated with normal IgG or IRS-1 antibodies and Western blotted with antibodies specific to either IRS-1, PPP1R12A, or p85; each graph is representative of data obtained from three independent experiments
It is noted that Myosin phosphatase Rho-interacting protein (MPRIP) [31], a known interaction protein of PPP1R12A, is also among the proteins identified to exhibit an increased association with IRS-1 reproducibly upon insulin stimulation, although MPRIP may bind to IRS-1 indirectly through PPP1R12A.
Two other proteins involved in muscle contraction and actin cytoskeleton dynamics also showed insulin stimulated association with IRS-1 reproducibly, including Caldesmon (CALD1) and troponin T (TNNT3). Hanke and Mann used tyrosine phosphorylated and corresponding non-phosphorylated IRS-1 peptides as bait to compare the respective interacting proteins in complex whole lysates from murine C2C12 muscle cells in the basal state [32]. They identified a number of proteins involved in cytoskeletal rearrangement that bind specifically to tyrosine phosphorylated IRS-1 peptides. Although these proteins were not identified as insulin stimulated interaction partners of IRS-1 in the present study, both studies indicate the importance of the interaction between IRS-1 and cytoskeleton.
It has been reported that elongation factor 2 (EEF2) plays an important role in insulin stimulated protein synthesis [33]. In addition, polyadenylate-binding protein 1 (PABPC1) can bind to nuclear pre-mRNA poly(A) tails to play an essential role in mRNA translation [34]. The present study provides evidence for the first time that EEF2 and PABPC1 may undergo insulin stimulated association with IRS-1 and may provide new insight into the IRS-1 regulated protein synthesis.
Three proteins involved in protein folding and degradation were found to possess an insulin-induced interaction with IRS-1, 78 kDa glucose-regulated protein (HSPA5) that is also known as GRP78 or BiP, heat shock protein HSP 90-β (HSP90AB1) [35], as well as HtrA serine peptidase 1 (HTRA1) [36]. Endoplasmic reticulum (ER) stress plays a crucial role in insulin resistance [37]. HSPA5 expression is induced in response to ER stress to assist in protein folding and assembly [38], which serves as a mechanism to protect cells. Recently, GRP78(+/−) mice were shown to exhibit resistance to high fat diet-induced obesity and type 2 diabetes [39]. GRP78 also mediates the unfolded protein response (UPR) caused by ER stress, which results in a transient reduction of new protein synthesis and may serve as a negative feed-back loop for IRS-1 mediated insulin signaling, since insulin stimulates protein synthesis through IRS-1. In addition, it has been shown that UPR activates inflammatory signaling pathways, leading to increased serine phosphorylation of IRS-1 and subsequent reduced insulin action [37]. Our novel findings indicate the potential new interaction between IRS-1 and the ER and its related signaling networks.
The approach we used included immunoprecipitation of a “bait” protein, IRS-1, at the endogenous level, followed by 1D-SDS-PAGE, to separate co-interaction proteins and contaminants, in-gel trypsin digestion to generate peptide fragments, and HPLC-ESI-MS/MS analysis to identify co-immunoprecipitating proteins. Multiple biological comparisons and rigorous criteria were used to minimize false positives. The normalization strategy used is identical to that used in Western blotting, which normalizes co-interaction protein abundance to that of the “bait” protein. The strategy is straightforward and has the ability to detect endogenous protein complexes, without the use of labeling or protein overexpression/tags. This approach may be applicable to the study of other protein complexes in cells, animal models, and even in human tissue samples.
It is noted that none of the downstream kinases of PI 3-K, such as PDK1 and Akt, were identified in the present study. This might due to the fact that their interaction with IRS-1 is transient and highly dynamic. Chemical cross-linking might be used to “freeze” and capture these dynamic interactions [40, 41]. However, so far, chemical cross-linking is still not fully developed, as this technique suffers from high false positives due to nonspecific cross linking.
We used normalized spectral counts to assess insulin-stimulated IRS-1 interaction partners. An alternative is to use spectral counts directly without normalization. Using spectral counts, 16 proteins have an increased abundance with IRS-1 under insulin stimulated conditions, compared to 15 proteins using normalized spectral counts. Fourteen of these proteins were identical using either approach. LIM domain only 7b (LMO7b) and isoform 2 of ankycorbin (RAI14) were only classified as insulin-stimulated IRS-1 interaction partners when using spectral counts, while polyadenylate-binding protein 1 (PABPC1) was only classified as an insulin-stimulated IRS-1 interaction partner when using normalized spectral counts. The good agreement between using spectral counts versus normalized spectral counts may be contributed to the similar IRS-1 protein loading.
Spectral counting has emerged as an effective means to assess protein abundance under different conditions [5, 10–12], especially for detecting moderate to large changes. Sardiu et al. have shown that normalized spectral counting (NSAF), an approach based on spectral counting, can be used to built a sophisticated human protein interaction network [5]. In the present work, we have demonstrated that spectral counting is capable of detecting endogenous insulin-stimulated IRS-1 complexes. However, isotopic labeling methods are better at detecting small changes that might be missed with spectral counting [42].
The false discovery rate (FDR) associated with multiple statistical comparisons has become an area of active research [43, 44]. FDR is defined as [43, 44]:
We performed 72 sign tests using normalized spectrum counts and observed 15 proteins were significantly different between basal and insulin treated samples (P = 0.03125). Among these 15 proteins with P = 0.03125, there may be two potential false positives (72*0.03125 = 2.25). Thus, the estimated FDR for this study is .
Conclusions
The present project analyzes proteins isolated from L6 myotubes under basal and insulin-stimulated conditions utilizing state-of-the-art HPLC-ESI-MS/MS to assess endogenous interaction partners of IRS-1, a key player in the insulin signaling cascade. Eleven novel IRS-1 interaction partners were identified upon insulin stimulation, including proteins that play a role in protein serine/threonine dephosphorylation, muscle contraction and actin cytoskeleton rearrangement, ER stress and protein folding, as well as protein synthesis. These results indicate that the combination of co-immunoprecipitation of IRS-1 followed by 1D-SDS-PAGE and HPLC-ESI-MS/MS can be used to assess insulin stimulated changes in the abundance of proteins associated with IRS-1 at the endogenous level.
Supplementary Material
Acknowledgments
The authors acknowledge support for this work by NIH grants R01DK081750 (Z.Y.) and R01DK47936 (L.J.M.). The authors thank Lohith Madireddy, Alex Chao, James Dillion, and Kimberly Pham for their help with the cell culture and sample preparation.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-010-0051-2) contains supplementary material, which is available to authorized users.
References
- 1.Vermeulen M, Hubner NC, Mann M. High confidence determination of specific protein–protein interactions using quantitative mass spectrometry. Curr Opin Biotechnol. 2008;19:331–337. doi: 10.1016/j.copbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Mann M. Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Meth Mol Biol. 2007;359:37–52. doi: 10.1007/978-1-59745-255-7_3. [DOI] [PubMed] [Google Scholar]
- 3.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, silac, as a simple and accurate approach to expression proteomics. Mol Cell Proteom. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Zieske LRA. Perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 5.Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wepf A, Glatter T, Schmidt A, Aebersold R, Gstaiger M. Quantitative interaction proteomics using mass spectrometry. Nat Meth. 2009;6:203–205. doi: 10.1038/nmeth.1302. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Friedman A, Nagpal S, Perrimon N, Asara JM. Use of a label-free quantitative platform based on Ms/Ms average tic to calculate dynamics of protein complexes in insulin signaling. J Biomol Technol. 2009;20:272–277. [PMC free article] [PubMed] [Google Scholar]
- 8.Rinner O, Mueller LN, Hubalek M, Muller M, Gstaiger M, Aebersold R. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
- 9.fNittis T, Guittat L, Leduc RD, Dao B, Duxin JP, Rohrs H, Townsend RR, Stewart SA. Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol Cell Proteom. 2010;9:1144–1156. doi: 10.1074/mcp.M900490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 11.Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Cluster analysis of mass spectrometry data reveals a novel component of Saga. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 13.Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O’Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouzakri K, Koistinen HA, Zierath JR. Molecular mechanisms of skeletal muscle insulin resistance in type 2 diabetes. Curr Diab Rev. 2005;1:167–174. doi: 10.2174/1573399054022785. [DOI] [PubMed] [Google Scholar]
- 15.DeFronzo RA, Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diab Care. 2009;32(Suppl 2):S157–S163. doi: 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hojlund K, Yi Z, Hwang H, Bowen B, Lefort N, Flynn CR, Langlais P, Weintraub ST, Mandarino LJ. Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol Cell Proteom. 2008;7:257–267. doi: 10.1074/mcp.M700304-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Hojlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi Z, Bowen BP, Hwang H, Jenkinson CP, Coletta DK, Lefort N, Bajaj M, Kashyap S, Berria R, De Filippis EA, Mandarino LJ. Global relationship between the proteome and transcriptome of human skeletal muscle. J Proteome Res. 2008;7:3230–3241. doi: 10.1021/pr800064s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karas J, Savage R. Publications of Frank Wilcoxon (1892–1965) Biometrics. 1967;23:1–10. [PubMed] [Google Scholar]
- 20.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bull. 1945;1:80–83. [Google Scholar]
- 21.Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A. Insulin-dependent interactions of proteins with Glut4 revealed through stable isotope labeling by amino acids in cell culture (Silac) J Proteome Res. 2006;5:64–75. doi: 10.1021/pr0502626. [DOI] [PubMed] [Google Scholar]
- 22.Talior-Volodarsky I, Randhawa VK, Zaid H, Klip A. ↦-Actinin-4 is selectively required for insulin-induced Glut4 translocation. J Biol Chem. 2008;283:25115–25123. doi: 10.1074/jbc.M801750200. [DOI] [PubMed] [Google Scholar]
- 23.Matsumura F, Hartshorne DJ. Myosin phosphatase target subunit: many roles in cell function. Biochem Biophys Res Commun. 2008;369:149–156. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryder JW, Lau KS, Kamm KE, Stull JT. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J Biol Chem. 2007;282:20447–20454. doi: 10.1074/jbc.M702927200. [DOI] [PubMed] [Google Scholar]
- 25.Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, Hartshorne DJ, Matsumura F. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev Cell. 2008;14:787–797. doi: 10.1016/j.devcel.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- 27.Hartshorne DJ, Ito M, Erdodi F. Role of protein phosphatase type 1 in contractile functions: myosin phosphatase. J Biol Chem. 2004;279:37211–37214. doi: 10.1074/jbc.R400018200. [DOI] [PubMed] [Google Scholar]
- 28.Hartley D, Cooper GM. Role of mTor in the degradation of IRS-1: regulation of Pp2a activity. J Cell Biochem. 2002;85:304–314. doi: 10.1002/jcb.10135. [DOI] [PubMed] [Google Scholar]
- 29.Mothe I, Van Obberghen E. Phosphorylation of insulin receptor substrate-1 on multiple serine residues, 612, 632, 662, and 731. Modulates insulin action. J Biol Chem. 1996;271:11222–11227. doi: 10.1074/jbc.271.19.11222. [DOI] [PubMed] [Google Scholar]
- 30.Tanti JF, Gremeaux T, van Obberghen E, Le Marchand-Brustel Y. Serine/threonine phosphorylation of insulin receptor substrate 1 modulates insulin receptor signaling. J Biol Chem. 1994;269:6051–6057. [PubMed] [Google Scholar]
- 31.Surks HK, Richards CT, Mendelsohn ME. Myosin phosphatase-Rho interacting protein. A new member of the myosin phosphatase complex that directly binds RhoA. J Biol Chem. 2003;278:51484–51493. doi: 10.1074/jbc.M305622200. [DOI] [PubMed] [Google Scholar]
- 32.Hanke S, Mann M. The phosphotyrosine interactome of the insulin receptor family and its substrates Irs-1 and Irs-2. Mol Cell Proteomics. 2009;8:519–534. doi: 10.1074/mcp.M800407-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proud CG. Regulation of protein synthesis by insulin. Biochem Soc Trans. 2006;34:213–216. doi: 10.1042/BST20060213. [DOI] [PubMed] [Google Scholar]
- 34.Hosoda N, Lejeune F, Maquat LE. Evidence that Poly(a) binding protein C1 binds nuclear Pre-mRNA Poly(a) tails. Mol Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan CT, Paulmurugan R, Gheysens OS, Kim J, Chiosis G, Gambhir SS. Molecular imaging of the efficacy of heat shock protein 90 inhibitors in living subjects. Cancer Res. 2008;68:216–226. doi: 10.1158/0008-5472.CAN-07-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 37.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, Lee AS. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein–protein interactions. J Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 41.Toews J, Rogalski JC, Clark TJ, Kast J. Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal Chim Acta. 2008;618:168–183. doi: 10.1016/j.aca.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 42.Mallick P, Kuster B. Proteomics: a pragmatic perspective. Nat Biotechnol. 2010;28:695–709. doi: 10.1038/nbt.1658. [DOI] [PubMed] [Google Scholar]
- 43.Pounds SB. Estimation and control of multiple testing error rates for microarray studies. Brief Bioinform. 2006;7:25–36. doi: 10.1093/bib/bbk002. [DOI] [PubMed] [Google Scholar]
- 44.Storey JD, Tibshirani R. Statistical significance for genome-wide experiments. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.