Summary
Metabolism must be coordinated with development to provide the appropriate energetic needs for each stage in the life cycle. Little is know, however, about how this temporal control is achieved. Here we show that the Drosophila ortholog of the Estrogen-Related Receptor (ERR) family of nuclear receptors directs a critical metabolic transition during development. dERR mutants die as larvae with low ATP levels and elevated levels of circulating sugars. The expression of active dERR protein in mid-embryogenesis triggers a coordinate switch in gene expression that drives a metabolic program normally associated with proliferating cells, supporting the dramatic growth that occurs during larval development. This study shows that dERR plays a central role in carbohydrate metabolism, demonstrates that a proliferative metabolic program is used in normal developmental growth, and provides a molecular context to understand the close association between mammalian ERR family members and cancer.
Keywords: metabolism, glycolysis, pentose phosphate pathway, Warburg effect, transcription, gene regulation, nuclear receptor signaling
Introduction
Metabolism must be tightly coupled with developmental progression, with distinct metabolic programs supporting the nutritional and energetic requirements of each stage in the life cycle. Relatively few studies, however, have addressed the molecular mechanisms that provide this temporal coordination. One of the best characterized metabolic transitions occurs in the developing mammalian heart and is represented by a switch in substrate utilization from glucose and lactate metabolism during fetal stages to postnatal dependence on fatty acid oxidation (Lehman and Kelly, 2002). This switch is accompanied by the coordinate induction of genes involved in mitochondrial β-oxidation and oxidative metabolism and mediated by the nuclear receptors PPARα and ERRγ (Lehman and Kelly, 2002; Alaynick et al., 2007). Interestingly, this process reverts in pathological forms of cardiac hypertrophy, demonstrating that metabolic transitions can be associated with disease. In addition, the nutritional status of early developmental stages can have a profound effect on later metabolic health (Symonds et al., 2009). One of the most dramatic manifestations of this interplay between nutrition and development is the impact of childhood obesity on the incidence of type 2 diabetes and obesity in adults. Obesity also impacts the timing of sexual maturation, linking a metabolic state to a key developmental transition (Ong, 2010). In spite of this important interplay between nutrition, metabolism, and development, little is understood about how these pathways are integrated.
Nuclear receptors are a specialized family of ligand-regulated transcription factors that play central roles in controlling development, growth, and metabolism (Chawla et al., 2001). They are defined by a conserved zinc finger DNA-binding domain and a C-terminal ligand-binding domain (LBD) that can impart multiple regulatory functions. One subfamily of these receptors are the Estrogen-Related Receptors (ERRs), represented by three paralogs in mammals, ERRα, ERRβ, and ERRγ. Although some synthetic estrogen analogs can suppress the constitutive transcriptional activity of these receptors, they have no known naturally-occurring ligands (Busch et al., 2004; Willy et al., 2004; Chao et al., 2006). Genetic studies in mice have demonstrated roles for ERR family members in mitochondrial biogenesis, oxidative phosphorylation, and lipid metabolism (for review, see Tremblay and Giguere, 2007). Consistent with these functions, ERRα mutant mice are lean and resistant to diet-induced obesity (Luo et al., 2003). In addition, ERRα and ERRγ are essential for proper cardiac metabolism. ERRα is required for energy production in response to cardiac stress (Huss et al., 2007), while ERRγ directs a metabolic switch that allows the postnatal heart to metabolize fatty acids (Alaynick et al., 2007). Recent studies, however, suggest that ERRs play a broader role in metabolic homeostasis (Ao et al., 2008; Charest-Marcotte et al., 2010; Eichner et al., 2010). Moreover, all three mammalian ERRs are associated with cancer progression. ERRα is necessary for the normal growth of estrogen receptor-negative breast cancer tumor grafts (Stein et al., 2008), and elevated ERRα expression is associated with aggressive forms of breast cancer (Ariazi et al., 2002). In contrast, ERRβ inhibits the progression of prostate cancer and increased expression of ERRγ is correlated with a favorable clinical prognosis for breast cancer (Ariazi et al., 2002; Yu et al., 2008). These observations suggest that ERR family members coordinate cell growth and proliferation with metabolism. The molecular basis for this relationship, however, remains unclear.
Here we present a functional study of the Drosophila ERR ortholog, dERR. The presence of only a single ERR gene in flies eliminates the potential genetic redundancy between multiple members of the ERR subfamily, allowing us to determine its key ancestral functions. We show that dERR is an essential regulator of carbohydrate metabolism during larval stages. dERR mutants die during the second larval instar with abnormally high levels of circulating sugar and diminished concentrations of ATP and triacylglycerides (TAG). These metabolic defects result from decreased expression of genes involved in glycolysis, the pentose phosphate pathway, and other aspects of carbohydrate metabolism. These genes are coordinately induced midway through embryogenesis, in apparent direct response to the expression of activated dERR protein. Interestingly, this dERR-regulated metabolic state is ideally suited to promote larval growth by converting dietary carbohydrates into biomass, and is strikingly reminiscent of the Warburg effect. Our studies reveal that an ERR family member coordinates metabolism with growth, indicate that a proliferative metabolic program is used in the context of normal development, and suggests that mammalian ERRs are associated with cancer through their ability to promote the Warburg effect.
Results
dERR is an essential metabolic regulator
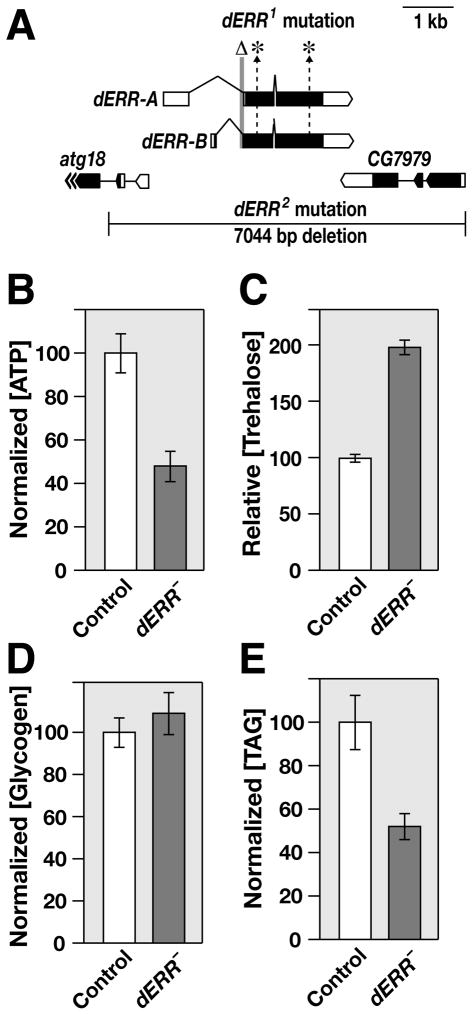
In an effort to examine dERR function, we generated two loss-of-function alleles at this locus (Figure 1A). Animals that carry a transheterozygous combination of these mutations, dERR1/dERR2, progress normally through embryogenesis and first instar development, but die abruptly during the later half of the second larval instar, with no apparent morphological defects (Figure S1A). dERR mutants, however, are severely metabolically compromised, with a two-fold decrease in ATP levels relative to control second instar larvae (Figure 1B). Despite these decreased levels of ATP, dERR mutants have elevated concentrations of the circulating sugar trehalose and normal glycogen concentrations, suggesting that they are unable to access their carbohydrate stores for energy production (Figures 1C and 1D). The high trehalose levels can be rescued by expressing wild-type dERR under the control of a ubiquitous GAL4 driver, indicating that the phenotype is due to a specific loss of dERR function (Figure S1B). TAG levels are also decreased in dERR mutants, suggesting that these animals have either increased fat catabolism or decreased TAG synthesis (Figure 1E). Similar metabolic defects are seen when comparing dERR1/dERR2 and dERR1/Df(3L)Exel6112 mutants with dERR1/+ heterozygous controls, demonstrating that they do not arise from changes in genetic background and that dERR1 represents a null allele for the locus (Figure S1C–E).
Figure 1. dERR mutants exhibit metabolic defects.
(A) A schematic representation of the dERR locus is depicted along with the flanking genes atg18 and CG7979. The dERR1 lesions are shown, including an 84 bp deletion that removes the exon 2 splice acceptor (Δ) and point mutations in exons 2 and 3 (*). The dERR2 deletion removes the entire dERR coding region and portions of the neighboring genes. The arrows at the 3′ end of atg18 indicate that it extends beyond what is depicted. (B–E) w1118 control and dERR1/dERR2 mutants (dERR–) were collected as mid-second instar larvae and whole animal homogenates were analyzed for concentrations of (B) ATP, (C) trehalose, (D) glycogen, or (E) TAG. Amounts of ATP, glycogen, and TAG were normalized to soluble protein levels. Mutant animals contain lower levels of (A) ATP (p < 1 × 10−4) and (E) TAG (p < .001), but have higher concentrations of (C) trehalose (p < 1 × 10−18) and normal levels of (D) glycogen (p = 0.44). n>20 independently collected samples per value with 25 animals per sample. Error bars are ± S.E.
Genes involved in carbohydrate metabolism are down-regulated in dERR mutants
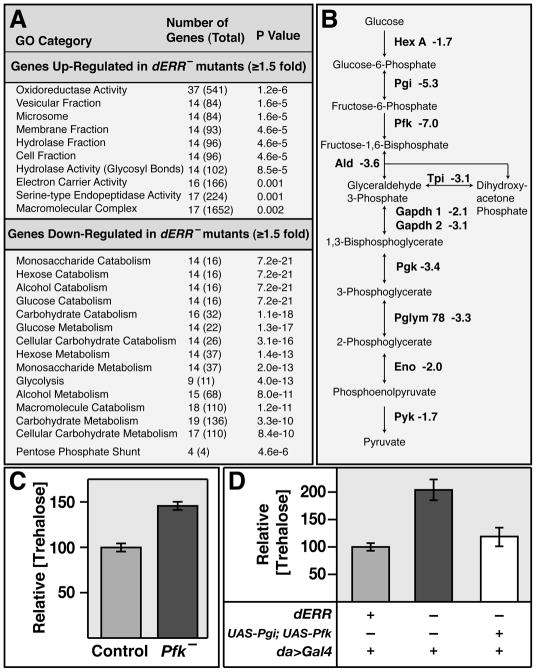
Microarray analysis was used to examine the molecular basis for the metabolic defects in dERR mutants. A total of 906 genes are misregulated ≥1.5 fold in dERR mutant second instar larvae relative to controls. GOstat analysis of this data set revealed that there are few statistically significant gene ontology classes among the 572 up-regulated genes. In contrast, the top 26 GO categories for the 334 down-regulated genes all represent different aspects of carbohydrate metabolism (Figure 2A and data not shown). Notably, these include almost the entire glycolytic pathway, with genes that encode enzymes at every step in glycolysis being significantly down-regulated in dERR mutants (Figure 2B). Northern blot hybridization was used to validate these changes in gene expression, demonstrating highly reduced transcript levels (Figure S2A). Interestingly, the Drosophila ortholog of Phosphofructokinase (Pfk), which encodes the rate-limiting enzyme in glycolysis, is the fifth most down-regulated gene in dERR mutants and the most highly affected gene in the glycolytic pathway (Table S1). These observations suggest that the elevated levels of trehalose in dERR mutants are due, at least in part, to decreased glycolytic flux. Consistent with this hypothesis, a mutation that eliminates Pfk expression results in a 48% increase in trehalose levels, demonstrating that a block in glycolysis can partially phenocopy the elevated trehalose levels observed in dERR mutants (Figure 2C and Figure S2). In addition, although expression of Pfk alone does not rescue the high trehalose levels in dERR mutants, expression of both Pgi and Pfk under the control of a ubiquitous GAL4 driver is sufficient to fully rescue this phenotype (Figure 2D). Moreover, ubiquitous expression of Pgi and Pfk in a dERR mutant background allows 8% of the animals to complete larval development and pupariate (compared with 1% of dERR mutant controls; p<0.01), suggesting that the lethality of dERR mutants is due, at least in part, to the reduced expression of these two enzymes. The reduced levels of TAG and ATP, however, are not rescued in these animals, suggesting that these phenotypes arise from defects in other pathways (Figure S2E and data not shown)
Figure 2. Genes involved in carbohydrate metabolism are down-regulated in dERR mutants.
(A) Gene ontology (GO) analysis of the 572 up-regulated and 334 down-regulated genes in dERR1/dERR2 mutant animals relative to w1118 controls. The top GO categories for each gene set are listed in order of significance along with the number of genes affected in that category, the total number of genes in that category (in parentheses), and the statistical significance of the match. (B) A diagram of glycolysis is depicted that displays the glycolytic genes that are down-regulated in dERR mutants followed by their fold-change in expression from the microarray. (C) Trehalose was measured in extracts from Pfk06339/+ (Control) and Pfk06339/Df(2R)BSC303 (Pfk−) mid-second instar larvae, revealing elevated levels in Pfk mutants. (D) Trehalose concentrations were determined for dERR2/+, da-GAL4 controls (grey box), dERR1/dERR2; da-GAL4 mutants (black box), and UAS-Pgi; dERR1/dERR2, UAS-Pfk, da-GAL4 animals (white box). Trehalose levels are rescued when both transgenes are expressed using the ubiquitous da-GAL4 driver.
The higher levels of trehalose seen in dERR mutants relative to Pfk mutants suggests that dERR plays important roles in other aspects of carbohydrate metabolism. At least one of these functions appears to be the Pentose Phosphate Pathway (PPP), where seven of the nine genes that encode PPP enzymes are down-regulated in the dERR mutant (Figure 2A and Tables S1,S2). The PPP is essential for growth and energy storage, using glucose-6-phosphate to synthesize ribose-5-phosphate for nucleotide production, and generating NADPH for fatty acid synthesis and other biosynthetic reactions. ImpL3, which encodes lactate dehydrogenase (Ldh), is also significantly affected in dERR mutants, representing the seventh most highly down-regulated gene (Table S1, S2). This enzyme converts pyruvate to lactate, regenerating NAD+ for use by the glycolytic pathway.
Metabolomic analysis of dERR mutants
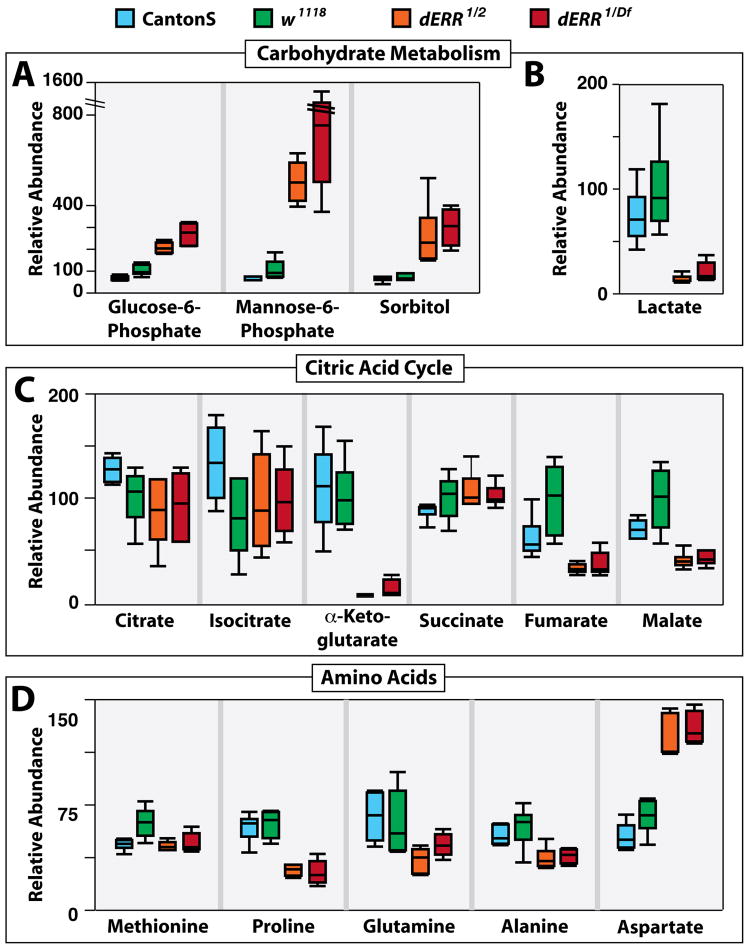
In order to characterize how these changes in gene expression might impact global metabolism, small molecule GC/MS analysis was used to compare the metabolic profiles of w1118 and CantonS controls with dERR1/dERR2 and dERR1/Df(3L)Exel6112 mutant second instar larvae. This metabolomic approach revealed that dERR mutants possess increased concentrations of glucose-6-phosphate, sorbitol, mannose-6-phosphate, and three unidentified carbohydrates, consistent with their inability to metabolize sugars (Figure 3A, Table S3). In contrast, oleic acid, stearic acid, and palmitic acid levels remained unchanged in mutant larvae, suggesting that fat metabolism is normal in dERR mutants and that the observed decrease in TAG levels are secondary to the carbohydrate defects (Table S3).
Figure 3. dERR mutants display changes in the levels of specific metabolites.
GC/MS was used to compare the relative levels of small metabolites in CantonS (blue) and w1118 (green) controls with dERR1/dERR2 (orange) and dERR1/Df(3L)Exel6112 (red) mutant second instar larvae. dERR mutants exhibit (A) elevated levels of glucose-6-phosphate, mannose-6-phosphate, and sorbital, along with (B) diminished concentrations of lactate. (C) The relative amounts of citrate, isocitrate, and succinate are similar among the four strains, while α-ketoglutarate, fumarate, and malate levels are decreased in mutant larvae. (D) Methionine levels are normal in mutant animals, while proline concentrations are significantly lower. Glutamine and alanine levels appear to be slightly decreased in mutant strains, and aspartate is the only amino acid that is elevated in dERR mutants. All data are graphically represented as a box plot, with the box representing the lower and upper quartiles, the horizontal line representing the median, and the bars representing the minimum and maximum data points. n = 6 samples collected from independent populations with 25 larvae per sample (See Table S3 for p values). Similar results were observed in two additional independent experiments (Table S3).
A number of metabolites downstream from glycolysis are also significantly reduced in mutant larvae. dERR mutants exhibit a 80–95% decrease in lactate, which confirms that there is decreased flux through the glycolytic pathway and suggests that wild-type larvae produce a relatively large amount of lactate (Figure 3B and Table S3). Some TCA intermediates are also reduced in dERR mutants, with no reproducible changes in citrate, isocitrate, or succinate levels, but a more than 90% reduction in α-ketoglutarate levels and a more than 60% reduction in fumarate and malate levels (Figure 3C and Table S3). These effects suggest that the TCA cycle is cataplerotically depleted in dERR mutants, with a major effect on α-ketoglutarate, which contributes to amino acid and purine biosynthesis.
Most amino acids, such as methionine, are relatively unaffected in dERR mutants (Figure 3D, Table S3). Interestingly, proline is the only amino acid that is reproducibly and significantly depleted in these animals. Glutamine and alanine levels are also somewhat depleted, but not consistently, or to the same extent as the effects on proline (Figure 3D, Table S3). Aspartate is the only amino acid that is consistently elevated in our analysis (Figure 3D). Taken together with the depleted malate levels and reduced glycolytic flux in the dERR mutants, this increase in aspartate concentration may be indicative of a decrease in the activity of the malate-aspartate shuttle due to low substrate availability.
dERR temporally coordinates the expression of glycolytic enzymes
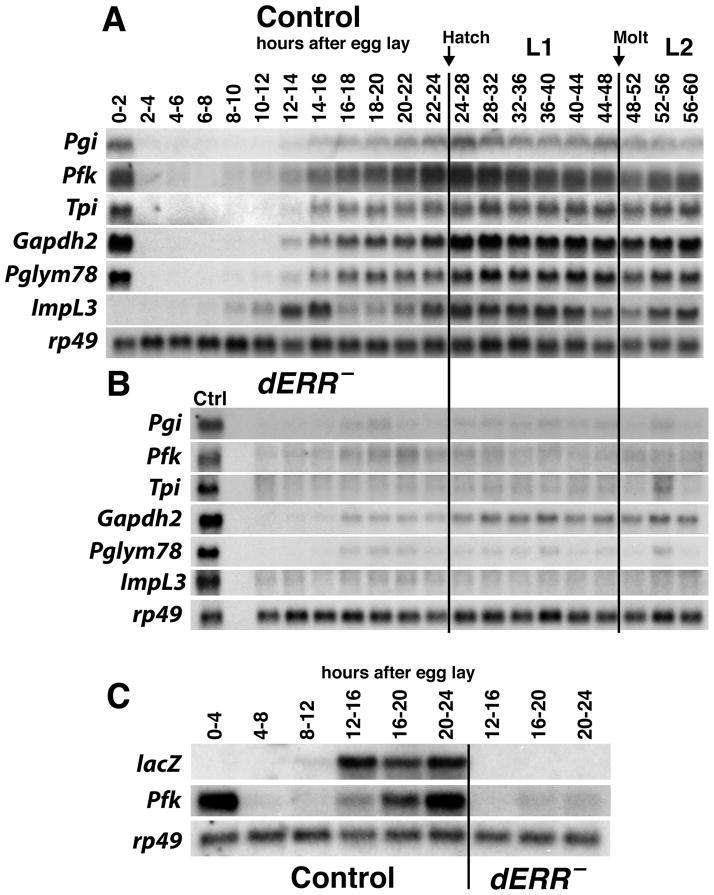
Previous studies have shown that genes involved in glycolysis are up-regulated at the end of embryogenesis, and that the encoded enzymes become active just prior to the onset of larval development (Wright and Shaw, 1970; Madhavan et al., 1972; Sun et al., 1988; Shaw-Lee et al., 1991; Roselli-Rehfuss et al., 1992; Shaw-Lee et al., 1992; Currie and Sullivan, 1994a; Currie and Sullivan, 1994b; Abu-Shumays and Fristrom, 1997). The observation that these genes are dependent on dERR function raises the possibility that this receptor coordinates their expression at the end of embryogenesis, directing a metabolic switch that promotes carbohydrate metabolism. To test this hypothesis, we examined the temporal expression patterns of six dERR target genes during embryogenesis and early larval development in control and dERR mutant animals (Figure 4A and B). Five of these genes encode glycolytic enzymes, Pgi (phosphoglucose isomerase), Pfk, Tpi (triose phosphate isomerase), Gapdh2 (glyceraldehyde 3-phosphate dehydrogenase), Pglym78 (phosphoglycerate mutase), and ImpL3. Interestingly, these genes are coordinately induced at 10–14 hours after egg laying and reach maximal levels of expression just prior to larval hatching (Figure 4A). In addition, this metabolic switch is severely disrupted in dERR mutants (Figure 4B).
Figure 4. dERR is required for the coordinate induction of glycolytic gene expression during mid-embryonic development.
(A–B) Total RNA from staged (A) w1118 control and (B) dERR1/dERR2 mutant embryos, first instar larvae (L1), and second instar larvae (L2) was analyzed by northern blot hybridization to detect the expression of transcripts encoding glycolytic enzymes. (A) Glycolytic genes are coordinately induced during mid-embryogenesis in control animals, but (B) not in dERR mutants. A sample of RNA from 24–28 hr w1118 first instar larvae (Ctrl) was included on the blot of dERR mutant RNA to facilitate comparisons between control and dERR mutant animals. (C) The temporal expression pattern of a lacZ reporter construct that carries a multimerized dERR binding site from the Pfk locus was analyzed by northern blot hybridization in staged w1118 control or dERR1 mutant larvae to detect the expression of lacZ or Pfk mRNA. The reporter is expressed in synchrony with zygotic Pfk expression in control animals and is dependent on dERR function. Hybridization to detect rp49 mRNA is included as a control for loading and transfer.
Nearly every gene that encodes a glycolytic enzyme lies near a predicted dERR binding site, suggesting that are directly regulated by the receptor (Table S4). This includes Pfk, which has a canonical dERR binding site in the fourth intron that is conserved across Drosophila species (Figure S2B and S3A, Table S4). This sequence is bound by dERR in an electrophoretic mobility shift assay, and the interaction can be efficiently competed with an excess of unlabeled wild-type binding site, but not an oligonucleotide that carries a 2 bp mutation in the core binding sequence (Figure S3B). dERR is also bound to the Pfk site in vivo, as demonstrated by chromatin immunoprecipitation, suggesting that it represents a functional regulatory interaction (Figure S3C). To test this possibility, we examined the activity of a multimerized version of the Pfk dERR binding site placed upstream from a lacZ reporter gene. The temporal pattern of lacZ mRNA accumulation in transgenic embryos parallels that of the endogenous zygotic Pfk transcript, and this expression is almost completely abolished in a dERR mutant background (Figure 4C). Taken together, these results demonstrate that dERR can directly regulate glycolytic gene expression and that dERR binding is necessary and sufficient to coordinately induce these genes during mid-embryogenesis.
dERR expression and activation is dynamically regulated during mid-embryogenesis
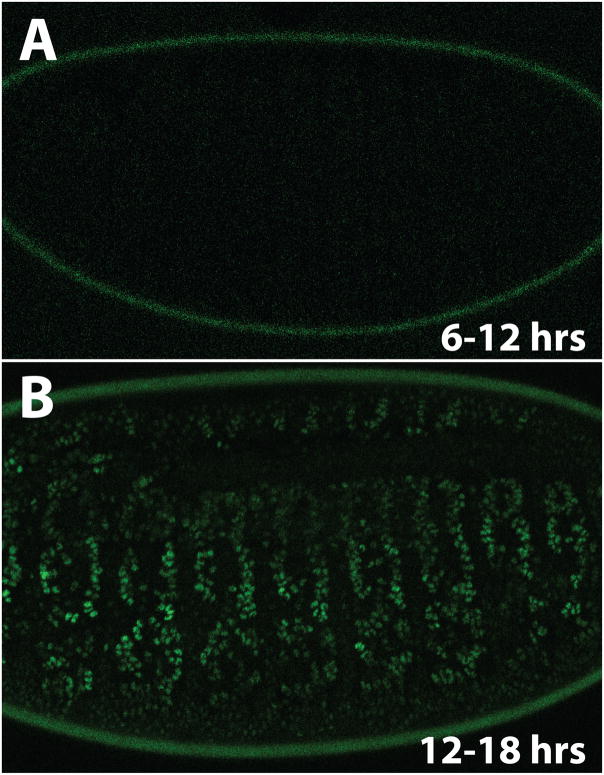
The coordinate induction of dERR target genes in 10–14 hour embryos raises the question of how this timing is achieved. dERR transcripts are present throughout embryonic development, indicating that either dERR protein levels and/or dERR activity is temporally regulated (Sullivan and Thummel, 2003). To detect dERR protein, we established a transformant line that carries a 3.3 kb genomic fragment spanning the dERR locus, with GFP inserted after the translation start site (dERR-GFP). This dERR-GFP reporter construct accurately reflects the activity of the native locus, as it is expressed throughout embryogenesis in the same manner as endogenous dERR, rescues the lethality of dERR1 mutants, and restores the normal temporal expression pattern of Pgi, Pfk, and Pglym78 in mutant embryos (Figure S4A). In spite of constant dERR-GFP mRNA expression, however, no GFP fluorescence is evident during the first 12 hours after egg laying (AEL) in dERR-GFP, dERR1 embryos (Figure 5A), and dERR-GFP protein is not detectable on a western blot during this time (Figure S4B). Rather, GFP fluorescence is detectable and dERR-GFP protein begins to accumulate at 12–16 hours AEL (Figure 5 and Figure S4B), in synchrony with the coordinate induction of dERR target genes (Figure 4A). Expression appears to be most abundant in the muscle and epidermis (Figure 5B). This timing is consistent with the appearance of activated dERR LBD, as determined using the hs-GAL4-dERR; UAS-GFP ligand sensor (Palanker et al., 2006). We confirmed this result with a second hs-GAL4-dERR; UAS-lacZ reporter strain, which displayed minimal GAL4-dERR LBD activity during the first 12 hrs of embryogenesis and became highly active in the muscle and epidermis by 12 hours AEL (Figure S5). We conclude that dERR is post-transcriptionally regulated and that the accumulation of activated dERR protein is the primary factor that drives the timing of dERR regulatory functions during embryogenesis.
Figure 5. dERR protein accumulation is temporally regulated during embryonic development.
Staged dERR1 mutant embryos that carry a dERR-GFP rescue transgene were visualized by confocal microscopy to detect GFP fluorescence. (A) No GFP was detected between 6–12 hrs after egg laying (AEL), while (B) dERR-GFP accumulates in the nuclei of several cell types, including the muscle and epidermis, between 12–18 hrs AEL.
dERR regulates carbohydrate metabolism in peripherial tissues
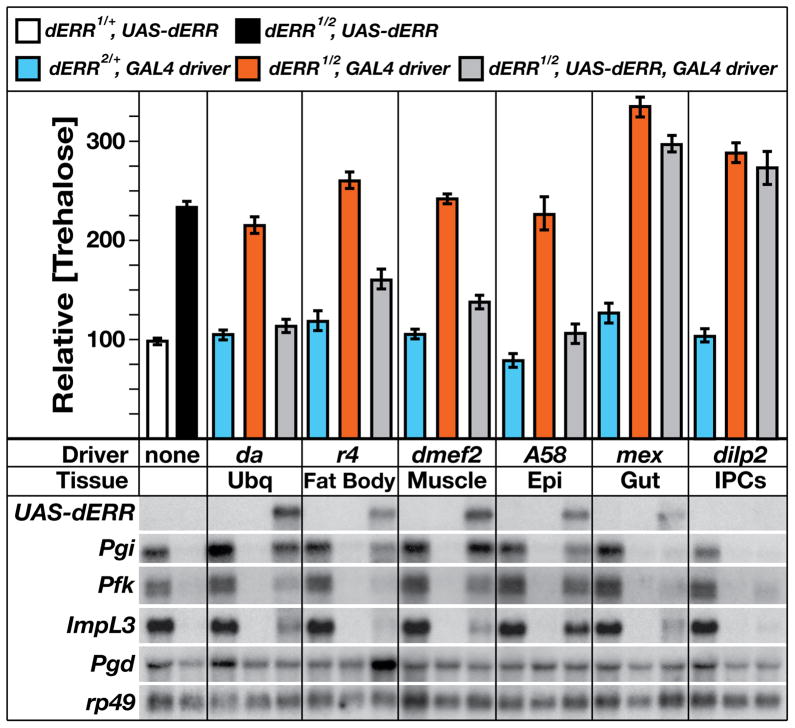
The dERR expression and activation patterns suggest that it regulates metabolism in peripheral tissues that use glucose for growth, ATP generation, and energy storage. In order to test this possibility, a series of tissue-specific GAL4 drivers were used to express a wild-type UAS-dERR rescue construct in a dERR mutant background. As described above, ubiquitous expression of dERR using the da-GAL4 driver restores normal trehalose levels in mutant animals (Figure 6). Similarly, specific UAS-dERR expression in the fat body (CG-GAL4 or r4-GAL4), muscle (dmef2-GAL4), or epidermis (A58-GAL4), significantly rescues trehalose levels in the mutant (Figure 6 and data not shown). In contrast, expression of dERR in the midgut (mex-GAL4), insulin-producing cells (IPCs) (dilp2-GAL4), prothoracic gland (phm-GAL4), corpora cardiaca (akh-GAL4), or Malphigian tubules (C42-GAL4), has no effect on the high trehalose levels (Figure 6 and data not shown).
Figure 6. dERR functions in peripheral tissues to control carbohydrate metabolism.
Mid-second instar larvae of five genotypes were tested for trehalose levels or the expression of specific dERR target genes: dERR1/+; UAS-dERR controls (white boxes), dERR1/dERR2; UAS-dERR mutants (black boxes), dERR2/+; GAL4 controls (blue boxes), dERR1/dERR2; GAL4 mutants (red boxes), or rescued dERR1/dERR2; UAS-dERR, GAL4 animals (grey boxes). Six GAL4 transgenes were used to drive UAS-dERR expression: da-GAL4 (Ubq, ubiquitous expression), r4-GAL4 (fat body), dmef2-GAL4 (muscle), A58-GAL4 (Epi, epidermis), mex-GAL4 (midgut), and dilp2-GAL4 (IPCs, Insulin-Producing Cells). Total RNA was analyzed by northern blot hybridization to detect UAS-dERR expression, three genes involved in glycolysis (Pgi, Pfk, and ImpL3), and a gene in the pentose phosphate pathway (Pgd). Hybridization to detect rp49 mRNA was used as a control for loading and transfer. The apparent reduced level of mex>dERR relative to A58>dERR is an artifact of the blot hybridization. These levels of expression are comparable. In contrast, the low level of dilp2>dERR expression likely reflects the small number of cells that express the dilp2 driver.
Consistent with dERR regulating carbohydrate metabolism by promoting transcription of glycolytic genes, expression of the UAS-dERR rescue construct by the da-GAL4 driver partially restores expression of Pgi, Pfk, and ImpL3 in a dERR mutant background (Figure 6). Similarly, Pgi, Pfk, and ImpL3 mRNA levels are restored in mutant animals when wild-type dERR is specifically expressed in the muscle or epidermis, but not in the midgut or IPCs (Figure 6). dERR, however, appears to promote a distinct metabolic program in the fat body, with expression of wild-type dERR in this tissue having only minor effects on Pgi expression. Instead, fat body-specific expression of wild-type dERR leads to abnormally high levels of Pgd mRNA, suggesting that a role for dERR in the fat body is to shuttle carbohydrates through the PPP (Figure 6). Finally, dERR may also exert a unique function in the midgut in regulating lactate metabolism. Although midgut-specific expression of wild-type dERR in mutant animals did not rescue the high trehalose phenotype or restore expression of Pgi or Pfk, it did restore partial expression of ImpL3. Interestingly, however, the ability of tissue-specific UAS-dERR expression to rescue the high trehalose levels or metabolic transcriptional defects in dERR mutants does not correlate with its effects on viability. Expression of UAS-dERR in the fat body of dERR mutants does not restore larval viability, and only 20% of mutant animals that express dERR in muscle are able to complete larval development (Figure S6). In contrast, 57% of mutants survive to form pupae when UAS-dERR is expressed using the ubiquitous da-GAL4 driver (Figure S6). These results suggest that of the lethality of dERR mutants arises from a loss of gene function in multiple tissues.
Discussion
Drosophila larval development is characterized by a ~200-fold increase in body mass. This period of dramatic growth requires the efficient conversion of dietary nutrients into cellular building blocks, such as amino acids, fatty acids, and nucleotides. The metabolic basis that supports juvenile growth, however, has not been defined. The studies described here support the model that Drosophila larval growth depends on a form of aerobic glycolysis that is similar to the Warburg effect, and that this metabolic program is established as a mid-embryonic transcriptional switch in response to activated dERR protein.
dERR regulates a metabolic program associated with cell proliferation
Our microarray study of dERR mutant larvae revealed a profound and widespread effect on genes that control carbohydrate metabolism, including highly reduced expression of almost all genes in glycolysis and the PPP (Figure 2A, 2B, and Table S1). This is consistent with the ~2-fold increase in trehalose seen in dERR mutants, along with highly reduced levels of ATP (Figure 1B and 1C). Mutation of the rate-limiting step in glycolysis, Pfk, results in elevated trehalose levels, similar to the dERR mutant, and ectopic expression of Pgi and Pfk in dERR mutants is sufficient to rescue this phenotype (Figure 2C and 2D). These studies demonstrate an essential role for dERR in carbohydrate catabolism during larval stages.
Metabolic profiling confirms these observations, showing significant accumulation of a number of sugars, including glucose-6-phosphate, the first intermediate in the glycolytic pathway (Figure 3 and Table S3). This study, however, also reveals changes in the levels of a number of other key metabolites, providing a broader understanding of dERR function. Lactate is almost completely absent in dERR mutants, consistent with lactate dehydrogenase being one of the most highly down-regulated genes in these animals. Levels of α-ketoglutarate, malate, and other late-stage TCA cycle intermediates are also significantly reduced in dERR mutant larvae, along with depletion of several amino acids, of which the most significant and reproducible is proline.
Interestingly, when taken together with the widespread effects of dERR on glycolysis and the PPP, these changes in metabolite levels are consistent with a form of aerobic glycolysis that is normally associated with cell proliferation (Wang et al., 1976; Vander Heiden et al., 2009). In the context of cancer, this metabolic signature is referred to as the Warburg effect (Warburg et al., 1928). The increase in carbohydrate metabolism is not designed to produce ATP, but rather promotes the synthesis of amino acids, lipids, and nucleotides, thereby supporting cellular proliferation. Both highly proliferative cells and Drosophila larvae shunt large quantities of glucose through the PPP, allowing them to generate ribose-5-phosphate for nucleotide synthesis and NADPH for fatty acid synthesis and other biosynthetic reactions (Geer et al., 1979; Eisenreich et al., 2004; Vander Heiden et al., 2009). Inadequate acetyl-CoA production from glycolysis in dERR mutants and reduced NADPH generation via the PPP likely results in decreased fatty acid synthesis, accounting for the reduced TAG levels in these animals (Figure 1E). In support of this hypothesis, TAG levels are reduced by 32% in animals that carry a loss-of-function mutation in glucose-6-phosphate dehydrogenase, which encodes a rate-limiting step in the PPP (data not shown, p<0.05). Moreover, mutations in PPP enzymes significantly decrease the rate of carbohydrate-dependent fatty acid synthesis in larvae (Geer et al., 1979). Similarly, elevated Ldh activity, which is a hallmark of cancer metabolism, is present in normal Drosophila larvae, which display elevated expression of the Drosophila ortholog of Ldh and high levels of Ldh enzyme activity (Warburg, 1956; Rechsteiner, 1970; Abu-Shumays and Fristrom, 1997). This enzyme converts pyruvate into lactate, preventing pyruvate from entering the mitochondria and generating NAD+ to promote maximal glycolytic flux. As a result of this diversion of pyruvate away from energy production, the TCA cycle in proliferating cells becomes dependent on amino acids derived from glutamic acid. Large amounts of glutamine are consumed by these cells to anaplerotically maintain the concentration of TCA intermediates. In an analogous manner, Drosophila larval metabolism appears to rely heavily on proline, which is significantly reduced in dERR mutants. Many insects use proline to generate energy and anaplerotically replenish the TCA cycle in the same way that proliferating yeast and cancer cells rely on glutamine (Sacktor, 1967; Arrese and Soulages, 2010). Taken together, these observations support the model that dERR establishes a metabolic state that is related to cellular proliferation, and that this function is essential for larval viability and growth.
Our studies also provide initial insights into the tissue-specific metabolic programs regulated by dERR during larval stages. When dERR is expressed in only the muscle or epidermis, it promotes transcription of the core glycolytic pathway (Figure 6). In contrast, specific expression of dERR in the fat body up-regulates Pgd, which encodes an essential enzyme in the PPP that is induced by sugar consumption (Cochrane et al., 1983). These results are consistent with the metabolic requirements of these different tissues. The dramatic expansion of the epidermis during larval growth, along with its production of cuticle, requires efficient glucose catabolism, as does the muscle to provide larval movement. In contrast, the fat body is one of the principle sites where sugar is processed by the PPP (Cochrane et al., 1983), and efficient lipid storage requires PPP activity (Geer et al., 1979). These observations indicate that dERR promotes appropriate tissue-specific metabolic programs during larval development.
dERR triggers a mid-embryonic metabolic switch that supports larval growth
A role for dERR in directing a metabolic state normally associated with cell proliferation provides a new context to understand how Drosophila larvae undergo their remarkable 200-fold increase in mass during the four days of larval development. It has long been known that many glycolytic enzymes are induced at the onset of larval development, and that the PPP and Ldh are highly active at this stage (Wright and Shaw, 1970; Madhavan et al., 1972; Cochrane et al., 1983; Sun et al., 1988; Shaw-Lee et al., 1991; Roselli-Rehfuss et al., 1992; Shaw-Lee et al., 1992; Currie and Sullivan, 1994a; Currie and Sullivan, 1994b; Abu-Shumays and Fristrom, 1997). No evidence, however, has tied these pathways together. Our results suggest that dERR directs a coordinated metabolic program that supports the unusual growth that occurs during this stage. Although yeast use a similar mechanism to support their proliferation, this is the first description of the use of this metabolic state to promote normal developmental growth (Diaz-Ruiz et al., 2009). Our studies of the dERR-regulated transcriptional program also reveal that this metabolic state is established by a coordinate switch in gene expression during mid-embryogenesis (Figure 4A and 4B). Accumulation of active dERR protein in 12–18 hr embryos directly induces Pfk transcription and, likely, other key dERR target genes (Figures 5, S3 and S4). This transcriptional switch, in turn, establishes the metabolic requirements for the next stage in development. The timing of the accumulation of active dERR protein could be regulated at a number of levels including post-translational modifications, ligand binding, and/or cofactor recruitment. Further studies are needed to define the mechanisms that regulate this response. It is interesting to note that the use of aerobic glycolysis to support developmental growth may not be restricted to Drosophila larval stages. Many animals undergo exponential growth during embryonic and fetal stages of development. It will be interesting to determine if similar metabolic states are associated with early growth in other organisms.
Implications for mammalian ERR function
Studies of mammalian ERR family members have largely focused on their roles in mitochondrial biogenesis and oxidative phosphorylation (for review, see Tremblay and Giguere, 2007). Our microarray study of RNA isolated from dERR mutant larvae, however, identified very few genes associated with β-oxidation, the TCA cycle, or the electron transport chain (Tables S1 and S2). Although genes involved in oxidoreductase activity are up-regulated in dERR mutants (Figure 2A), most encode cytochrome P450s with unknown functions. Similarly, a number of genes encoding mitochondrial ribosomal proteins are expressed at reduced levels in dERR mutants, although these effects are relatively minor (≤ 1.5-fold). dERR mutant larvae have normal mitochondrial genome number and mitochondrial morphology, demonstrating no detectable effect on mitochondrial biogenesis (data not shown). These observations are consistent with a primary function for dERR in biomass production during larval stages, rather than energy generation and oxidative phosphorylation. Rather, we speculate that dERR may play a more central role in mitochondrial function during adult stages, when the fly is highly dependent on oxidative metabolism to support its increased mobility.
Conversely, several recent studies have expanded our understanding of the metabolic functions of mammalian ERR family members to include those controlled by dERR. ERRγ regulates glycolytic gene expression in the heart, ERRα is bound to the extended promoters of genes involved in glycolysis and the TCA cycle, and ERRγ regulates several genes in these pathways (Alaynick et al., 2007; Charest-Marcotte et al., 2010; Eichner et al., 2010). Similarly, ERRα is required for the up-regulation of genes encoding glycolytic enzymes when cells are raised under hypoxic conditions (Ao et al., 2008). Our studies of dERR define carbohydrate metabolism as a key ancestral function for this nuclear receptor subclass, aspects of which have been conserved through evolution to mammals.
Our work also provides a new context to understand roles for mammalian ERR family members in cancer progression. Many studies have demonstrated a close association between ERR receptors and cancer (Ariazi et al., 2002; Stein et al., 2008; Yu et al., 2008). The molecular basis for this association, however, remains unclear, likely due to the functional redundancy and crosstalk between mammalian ERR paralogs. A recent paper has provided an initial step in this direction, showing that the miR-378* microRNA, which promotes the Warburg effect in BT-474 cancer cells, down-regulates ERRγexpression in this context (Eichner et al., 2010).Our studies of the single Drosophila ERR family member raise the important possibility that mammalian ERRs control the dramatic cellular proliferation associated with cancer through their ability to promote the Warburg effect.
Experimental Procedures
Fly stocks
Flies were maintained on standard Bloomington Stock Center medium with malt at 25°C. All larvae were raised on yeast paste at 25°C. The dERR mutant alleles were generated by ends-in homologous recombination using standard methods (Maggert, 2008). See Supplemental Experimental Procedures for a detailed description. To generate the UAS-dERR transgenic line, the dERR-A isoform was reverse transcribed and amplified from total RNA using the oligonucleotides 5′-gcaactgaataaccgatggtcg-3′ and 5′-cctaagactatatttgcacctttgc-3′. The resulting cDNA was inserted into the pUAST vector and confirmed by DNA sequencing. The Pfk-lacZ reporter construct was generated by inserting oligonucleotides containing tandem repeats of the Pfk dERR-binding site (5′-cctgaaggtcaccttg-3′) between the EcoRI/KpnI sites and SacII/BamHI sites in the pH-Pelican vector. The dERR-GFP genomic rescue construct was generated by inserting the GFP coding region within the second exon of dERR, immediately after the start codon in dERR-A (see Supplemental Experimental Procedures for a detailed description). The UAS-Pgi and UAS-Pfk constructs were generated by amplifying the corresponding cDNAs (DGRC) using the oligonucleotides pairs (Pgi) 5′-gcggccgcatggccggcccacttcctcc-3′ and 5′-gcggccgcttattacttccaattggctttgatg-3′ or (Pfk) 5′-gcggccgcatgcattcaataaaatttcgagtatttacc-3′ and 5′-gcggccgcttattaggcgacggcgtcagtgtcac-3′, and inserting the resulting PCR products into pUAST. All constructs were used for standard P-element-mediated germline transformation and maintained in a w1118 background. See Supplemental Experimental Procedures for a description of the GAL4 drivers used for tissue-specific rescue. Df(3L)Exel6112, which is a molecularly-defined deficiency that removes the entire dERR locus, was obtained from the Bloomington Stock Center.
Metabolic Analysis
Samples used for metabolic analyses were collected from independent matings on at least three different days and consisted of extracts prepared from twenty-five staged mid-second instar larvae. Metabolite levels from each sample were combined to determine the average concentration and standard error, except for the GC/MS analysis, in which the three experiments were analyzed separately. Measurements of ATP, glycogen, and TAG were performed as described and normalized to total soluble protein (Palanker et al., 2009; Wang et al., 2010). For trehalose assays, 25 larvae were homogenized in trehalase buffer (5 mM Tris pH 6.6, 137 mM NaCl, 2.7 mM KCl) and immediately incubated at 70°C for 5 minutes. Twenty μl of homogenate was incubated with either 20 μl of trehalase buffer or 20 μl of trehalase buffer + 0.06 μl of porcine trehalase (Sigma, T8778-1UN) for ~12 hours at 37°C, after which the samples were centrifuged at maximum speed for 3 minutes. Thirty μl of sample was transferred to a 96 well plate and incubated with 100 μl of glucose reagent (Sigma). The plate was incubated at 37°C for 30 min, after which the reaction was stopped by adding 100 μl of 12 N H2SO4 and the color intensity was measured using a BioTek Synergy HT microplate reader at 540 nm. Glucose and glucose plus trehalose amounts were determined using a standard curve, and the amount of trehalose alone was determined by subtracting the amount of glucose from the amount of glucose plus trehalose. GC/MS analysis was performed at the University of Utah Metabolomics Core Facility, as described (Wang et al., 2010).
DNA binding
EMSAs were preformed essentially as described (Horner et al., 1995). The oligonucleotide containing the wild-type dERR binding site in Pfk consisted of the sequence 5′-GGCGTCTCTGGCCTGAAGGTCACCTTGAAA-3′ and the oligonucleotide in which the dERR binding site was mutated contained the sequence 5′-GGCGTCTCTGGCCTGAAGTACACCTTGAAA-3′, which carries a 2 bp change in the AGGTCA core binding site. Chromatin immunoprecipitation of dERR-GFP was performed as described (Sieber and Thummel, 2009), using a mouse anti-GFP antibody (MBL) to immunoprecipitate dERR-GFP.
Developmental Northern Blots
Staged embryos and larvae were collected as described (Sullivan and Thummel, 2003). Approximately 250 dERR1/dERR2 embryos were hand-sorted for each timepoint. RNA was extracted using TriPure isolation reagent (Sigma) and northern blot analysis was conducted essentially as described (Karim and Thummel, 1991).
Microarray Analysis
Microarray analysis was performed in triplicate on independent RNA samples isolated from w1118 or dERR1/2 mid-second instar larvae. Total RNA was extracted using TriPure (Roche) followed by purification with RNAeasy columns (Qiagen). Probe labeling, hybridization to Affymetrix GeneChip® Drosophila Genome 2.0 Arrays, and scanning were performed by the University of Maryland Biotechnology Institute Microarray Core facility. Raw data were normalized using RMA (Bolstad et al., 2003; Irizarry et al., 2003) and analyzed using SAM, imposing a 1.5-fold cutoff and False Discovery Rate of ≤1.643% (Tusher et al., 2001). Microarray data can be accessed at the GEO website (http://www.ncbi.nlm.nih.gov/geo/) (accession number GSE23336).
Statistical Analyses
Statistical significance was calculated using an unpaired two-tailed Student’s t-test with unequal variance. All quantitative data are reported as the mean ± SEM.
Supplementary Material
Acknowledgments
We thank James Cox at the Health Sciences Center Metabolomics Core Facility at the University of Utah for assistance with the GC/MS metabolomic assays, and the Bloomington Stock Center for providing fly stocks. We thank A. Sullivan for generating the UAS-dERR transgenic strain, H. Xie and K. Golic for help with the gene targeting, A.F. Ruaud, M Sieber, and L. Palanker for helpful discussions and technical assistance, and M. Horner and D. Seay for comments on the manuscript. J.M.T. was supported by NIH National Research Service Award F32DK083864 from the NIDDK. This research was supported by NIH grants 1R01DK075607 and 1RC1DK086426.
Footnotes
Supplemental data include Supplemental Experimental Procedures, Supplemental Figures, and Supplemental Tables, and can be found with this article online at http://
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Shumays RL, Fristrom JW. IMP-L3, A 20-hydroxyecdysone-responsive gene encodes Drosophila lactate dehydrogenase: structural characterization and developmental studies. Dev Genet. 1997;20:11–22. doi: 10.1002/(SICI)1520-6408(1997)20:1<11::AID-DVG2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, Jonker JW, Giles W, Naviaux RK, Giguere V, Evans RM. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci U S A. 2008;105:7821–6. doi: 10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–8. [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–25. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Busch BB, Stevens WC, Jr, Martin R, Ordentlich P, Zhou S, Sapp DW, Horlick RA, Mohan R. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J Med Chem. 2004;47:5593–6. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- Chao EY, Collins JL, Gaillard S, Miller AB, Wang L, Orband-Miller LA, Nolte RT, McDonnell DP, Willson TM, Zuercher WJ. Structure-guided synthesis of tamoxifen analogs with improved selectivity for the orphan ERRgamma. Bioorg Med Chem Lett. 2006;16:821–4. doi: 10.1016/j.bmcl.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, Mootha VK, Giguere V. The homeobox protein Prox1 is a negative modulator of ERR{alpha}/PGC-1{alpha} bioenergetic functions. Genes Dev. 2010;24:537–42. doi: 10.1101/gad.1871610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–70. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Cochrane BJ, Lucchesi JC, Laurie-Ahlberg CC. Regulation of enzyme activities in Drosophila: genetic variation affecting induction of glucose 6-phosphate and 6-phosphogluconate dehydrogenase in larvae. Genetics. 1983;105:601–13. doi: 10.1093/genetics/105.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie PD, Sullivan DT. Structure and expression of the gene encoding phosphofructokinase (PFK) in Drosophila melanogaster. J Biol Chem. 1994a;269:24679–87. [PubMed] [Google Scholar]
- Currie PD, Sullivan DT. Structure, expression and duplication of genes which encode phosphoglyceromutase of Drosophila melanogaster. Genetics. 1994b;138:352–63. [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ruiz R, Uribe-Carvajal S, Devin A, Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim Biophys Acta. 2009;1796:252–65. doi: 10.1016/j.bbcan.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, Giguere V. miR-378( *) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 2010;12:352–61. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Eisenreich W, Ettenhuber C, Laupitz R, Theus C, Bacher A. Isotopolog perturbation techniques for metabolic networks: metabolic recycling of nutritional glucose in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:6764–9. doi: 10.1073/pnas.0400916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer BW, Lindel DL, Lindel DM. Relationship of the oxidative pentose shunt pathway to lipid synthesis in Drosophila melanogaster. Biochem Genet. 1979;17:881–95. doi: 10.1007/BF00504310. [DOI] [PubMed] [Google Scholar]
- Horner MA, Chen T, Thummel CS. Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev Biol. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguere V, Murphy E, Kelly DP. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Thummel CS. Ecdysone coordinates the timing and amounts of E74A and E74B transcription in Drosophila. Genes Dev. 1991;5:1067–79. doi: 10.1101/gad.5.6.1067. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–45. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol Cell Biol. 2003;23:7947–56. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan K, Fox DJ, Ursprung H. Developmental genetics of hexokinase isozymes in Drosophila melanogaster. J Insect Physiol. 1972;18:1523–30. doi: 10.1016/0022-1910(72)90231-4. [DOI] [PubMed] [Google Scholar]
- Maggert KA, Gong WJ, Golic KG. Methods for homologous recombination in Drosophila. Totowa, N.J: Humana Press; 2008. [DOI] [PubMed] [Google Scholar]
- Ong KK. Early determinants of obesity. Endocr Dev. 2010;19:53–61. doi: 10.1159/000316897. [DOI] [PubMed] [Google Scholar]
- Palanker L, Necakov AS, Sampson HM, Ni R, Hu C, Thummel CS, Krause HM. Dynamic regulation of Drosophila nuclear receptor activity in vivo. Development. 2006;133:3549–62. doi: 10.1242/dev.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–39. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner MC. Drosophila lactate dehydrogenase and alpha-glycerolphosphate dehydrogenase: distribution and change in activity during development. J Insect Physiol. 1970;16:1179–92. doi: 10.1016/0022-1910(70)90208-8. [DOI] [PubMed] [Google Scholar]
- Roselli-Rehfuss L, Ye F, Lissemore JL, Sullivan DT. Structure and expression of the phosphoglycerate kinase (Pgk) gene of Drosophila melanogaster. Mol Gen Genet. 1992;235:213–20. doi: 10.1007/BF00279363. [DOI] [PubMed] [Google Scholar]
- Sacktor BaC, CC Metabolism of proline in insect flight muscle and its significance in stimulating the oxidation of pyruvate. Archives of Biochemistry and Biophysics. 1967;120:583–588. [Google Scholar]
- Shaw-Lee R, Lissemore JL, Sullivan DT, Tolan DR. Alternative splicing of fructose 1,6-bisphosphate aldolase transcripts in Drosophila melanogaster predicts three isozymes. J Biol Chem. 1992;267:3959–67. [PubMed] [Google Scholar]
- Shaw-Lee RL, Lissemore JL, Sullivan DT. Structure and expression of the triose phosphate isomerase (Tpi) gene of Drosophila melanogaster. Mol Gen Genet. 1991;230:225–9. doi: 10.1007/BF00290672. [DOI] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–90. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RA, Chang CY, Kazmin DA, Way J, Schroeder T, Wergin M, Dewhirst MW, McDonnell DP. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68:8805–12. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AA, Thummel CS. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol. 2003;17:2125–37. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- Sun XH, Tso JY, Lis J, Wu R. Differential regulation of the two glyceraldehyde-3-phosphate dehydrogenase genes during Drosophila development. Mol Cell Biol. 1988;8:5200–5. doi: 10.1128/mcb.8.12.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds ME, Sebert SP, Hyatt MA, Budge H. Nutritional programming of the metabolic syndrome. Nat Rev Endocrinol. 2009;5:604–10. doi: 10.1038/nrendo.2009.195. [DOI] [PubMed] [Google Scholar]
- Tremblay AM, Giguere V. The NR3B subgroup: an ovERRview. Nucl Recept Signal. 2007;5:e009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lam G, Thummel CS. Med24 and Mdh2 are required for Drosophila larval salivary gland cell death. Dev Dyn. 2010;239:954–64. doi: 10.1002/dvdy.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261:702–5. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O, Posener K, Negelein E. Ueber den stoffwechsel der tumoren. Biochem Z. 1928;152:319–344. [Google Scholar]
- Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC, Jr, Martin R, Mohan R, Zhou S, Ordentlich P, Wei P, et al. Regulation of PPARgamma coactivator 1alpha (PGC–1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc Natl Acad Sci U S A. 2004;101:8912–7. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Shaw CR. Time of expression of genes controlling specific enzymes in Drosophila embryos. Biochem Genet. 1970;4:385–94. doi: 10.1007/BF00485755. [DOI] [PubMed] [Google Scholar]
- Yu S, Wong YC, Wang XH, Ling MT, Ng CF, Chen S, Chan FL. Orphan nuclear receptor estrogen-related receptor-beta suppresses in vitro and in vivo growth of prostate cancer cells via p21(WAF1/CIP1) induction and as a potential therapeutic target in prostate cancer. Oncogene. 2008;27:3313–28. doi: 10.1038/sj.onc.1210986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.