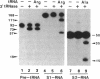
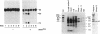Abstract
The spermidine-dependent, sequence-specific endoribonuclease (RNase 65) activities in mammalian cell extracts require both protein and 3' truncated tRNA, species of which direct their substrate sequence specificity. Computer analysis for searching possible base pairing between substrate RNAs and their corresponding 3' truncated tRNA, suggested a unified model for substrate recognition mechanism, in which a four-nucleotide (nt) sequence in the target tRNAs 1 nt upstream of their cleavage site, base pairs with the 5' terminal 4 nt sequence of their corresponding 3' truncated tRNA. This model was supported by experiments with several RNA substrates containing a substituted nucleotide in the target 4 nt sequence. In this model, the tRNA substrates and their corresponding 3' truncated tRNA form a complex resembling a 5' processed tRNA precursor containing a 3' trailer, suggesting that the protein component of RNase 65 is identical to tRNA 3' processing endoribonuclease (3' tRNase). Actually, 3' tRNase purified from pig liver cleaved the target RNAs at the expected sites only in the presence of their corresponding 3' truncated tRNA. These results show that the 3' tRNase can be converted to 4 nt specific RNA cutters using the 3' truncated tRNAs.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Castaño J. G., Tobian J. A., Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3' terminus of human pre-tRNA-Met(i) (3' pre-tRNase). J Biol Chem. 1985 Jul 25;260(15):9002–9008. [PubMed] [Google Scholar]
- Clayton D. A. A nuclear function for RNase MRP. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4615–4617. doi: 10.1073/pnas.91.11.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. Ribonuclease multiplicity, diversity, and complexity. J Biol Chem. 1993 Jun 25;268(18):13011–13014. [PubMed] [Google Scholar]
- Forster A. C., Altman S. External guide sequences for an RNA enzyme. Science. 1990 Aug 17;249(4970):783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R., Hicks M., Cruz P. 'Hairpin' catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990 Jan 25;18(2):299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Ando Y., Shiba T. Unusual priming mechanism of RNA-directed DNA synthesis in copia retrovirus-like particles of Drosophila. 1986 Oct 30-Nov 5Nature. 323(6091):824–826. doi: 10.1038/323824a0. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Sasaki N. Hyperprocessing of tRNA by the catalytic RNA of RNase P. Cleavage of a natural tRNA within the mature tRNA sequence and evidence for an altered conformation of the substrate tRNA. J Biol Chem. 1992 Jun 15;267(17):11972–11976. [PubMed] [Google Scholar]
- Liu F., Altman S. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell. 1994 Jul 1;77(7):1093–1100. doi: 10.1016/0092-8674(94)90448-0. [DOI] [PubMed] [Google Scholar]
- Nashimoto M. 3' truncated tRNAArg is essential for in vitro specific cleavage of partially synthesized mouse 18S rRNA. Nucleic Acids Res. 1993 Oct 11;21(20):4696–4702. doi: 10.1093/nar/21.20.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M. Characterization of the spermidine-dependent, sequence-specific endoribonuclease that requires transfer RNA for its activity. Nucleic Acids Res. 1992 Jul 25;20(14):3737–3742. doi: 10.1093/nar/20.14.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M., Kominami R., Nishi S., Mishima Y. A novel spermidine-dependent endoribonuclease activity caused by RNA-protein complex in mouse FM3A cell extracts. Biochem Biophys Res Commun. 1991 May 15;176(3):1163–1169. doi: 10.1016/0006-291x(91)90407-x. [DOI] [PubMed] [Google Scholar]
- Nashimoto M., Sakai M., Nishi S. Transfer RNA lacking its 3' terminus is required for spermidine-dependent ribonuclease 65 activity in mouse FM3A cell extracts. Biochem Biophys Res Commun. 1991 Aug 15;178(3):1247–1252. doi: 10.1016/0006-291x(91)91027-a. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Altman S. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease P. Science. 1994 Mar 4;263(5151):1269–1273. doi: 10.1126/science.8122108. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]