Abstract
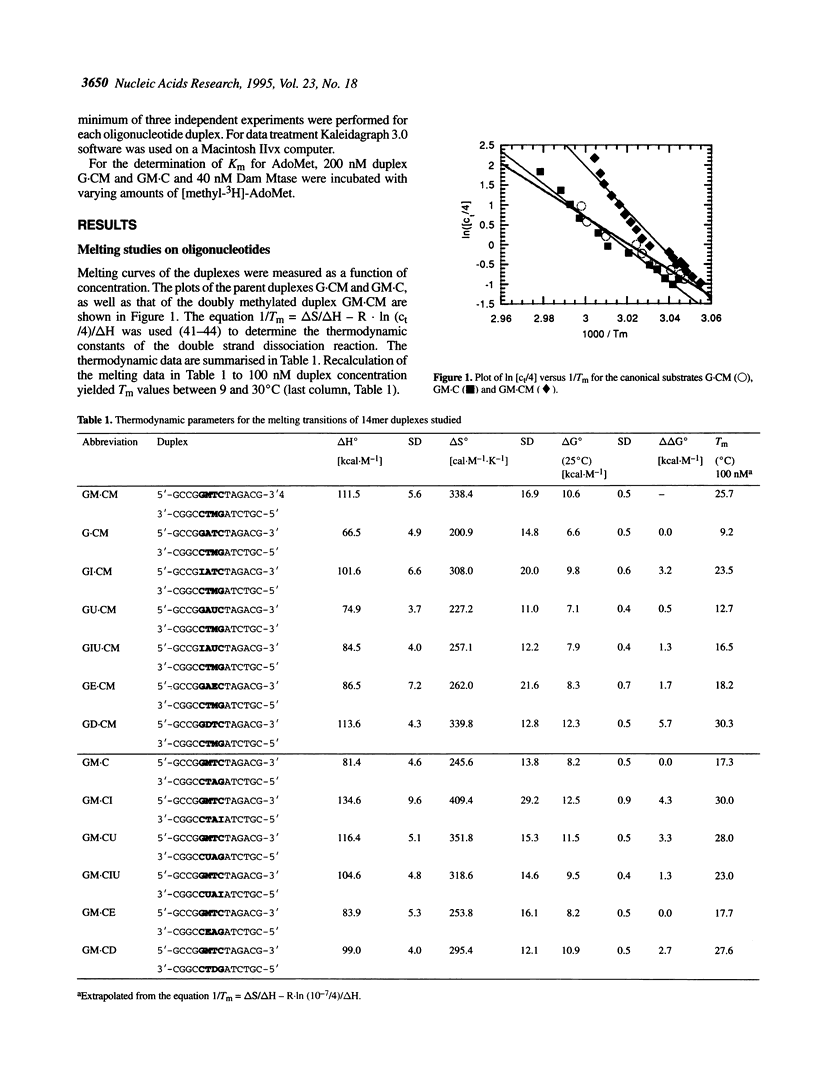
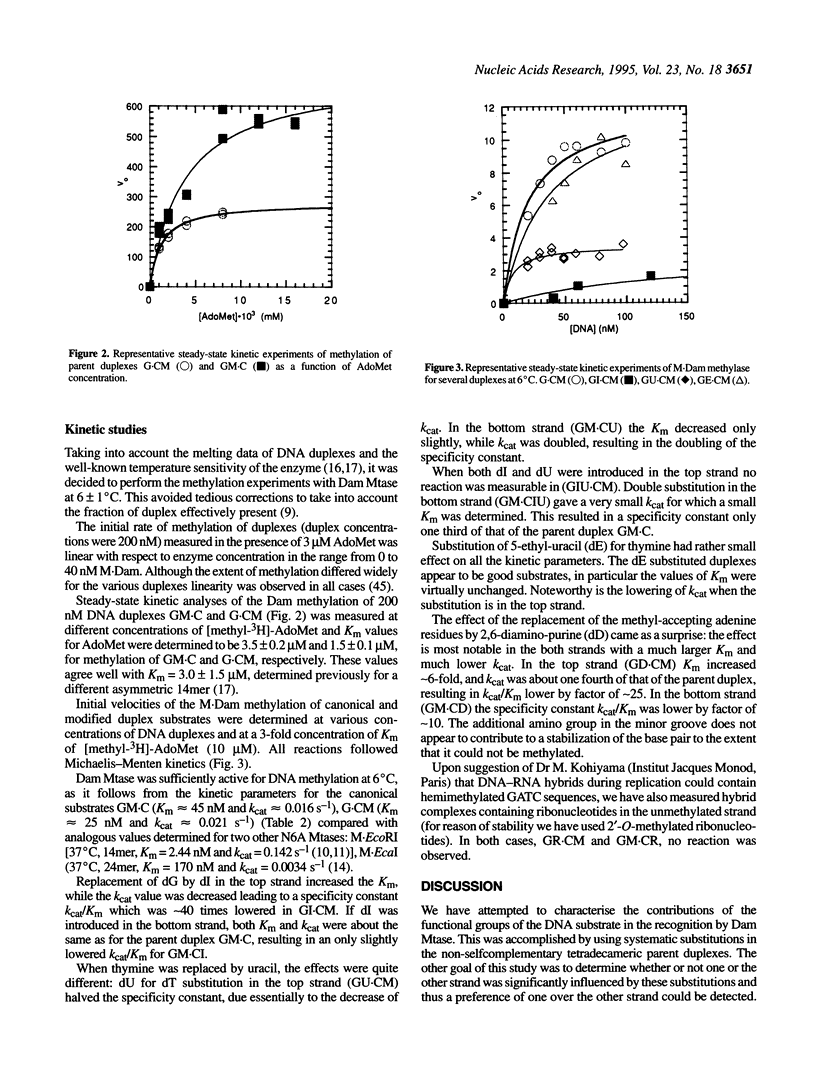
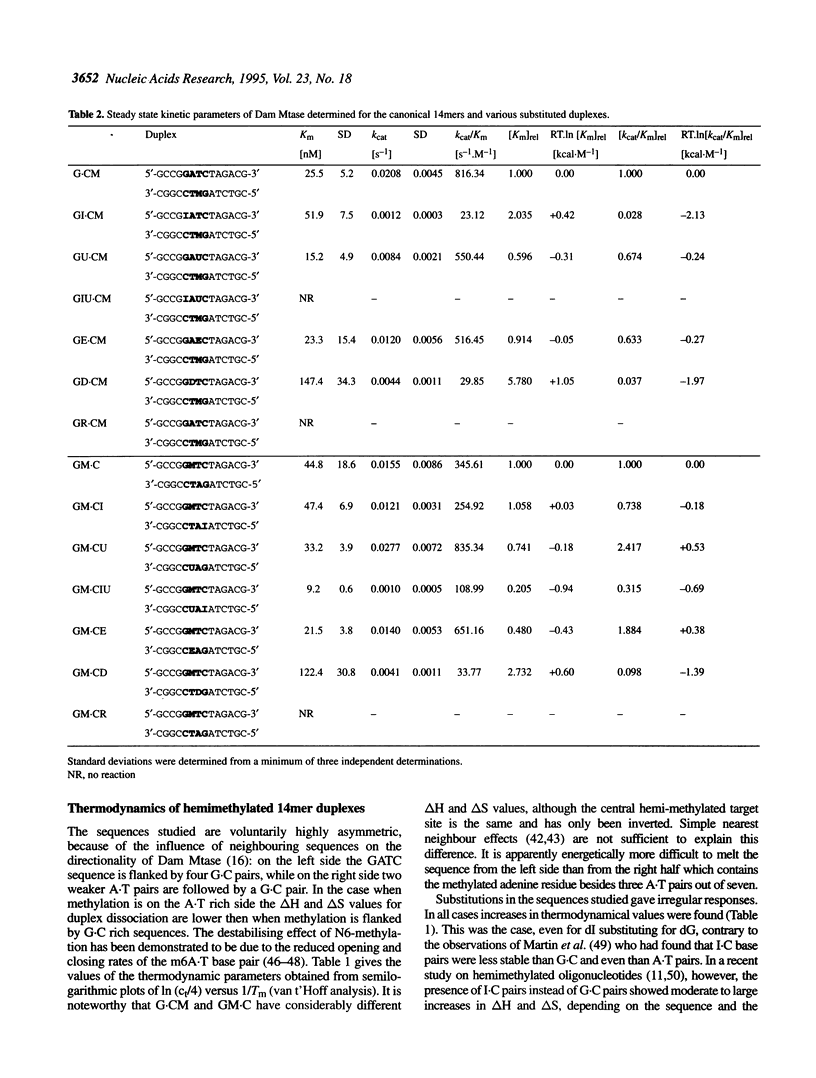
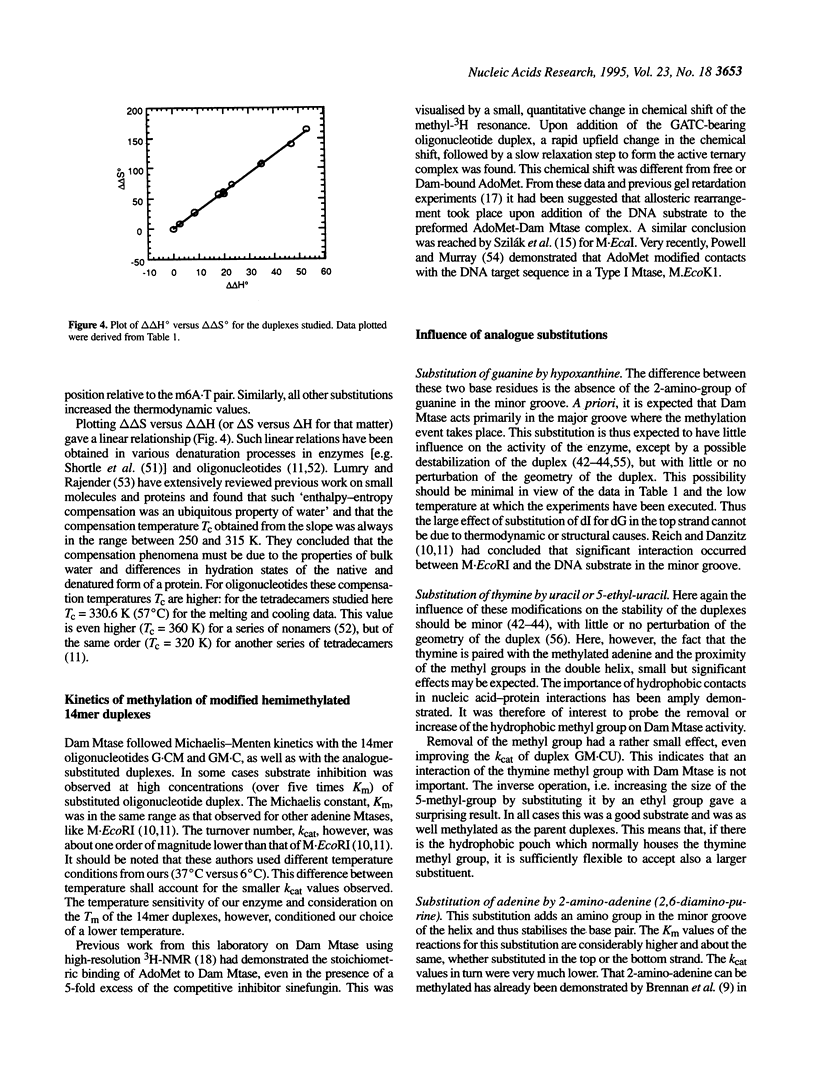
We have measured steady-state kinetics of the N6-adenine methyltransferase Dam Mtase using as substrates non-selfcomplementary tetradecamer duplexs (d[GCCGGATCTAGACG]-d[CGTCTAGATCC-GGC]) containing the hemimethylated GATC target sequence in one or the other strand and modifications in the GATC target sequence of the complementary strands. Modifications included substitution of guanine by hypoxanthine (I), thymine by uracil (U) or 5-ethyl-uracil (E) and adenine by 2,6-diamino-purine (D). Thermodynamic parameters were obtained from the concentration dependence of the melting temperature (Tm) of the duplexes. Large differences in DNA methylation of duplexes containing single dI for dG substitution of the Dam recognition site were observed compared with the canonical substrate, if the substitution involved the top strand (on the G.C rich side). Substitution in either strand by uracil (dU) or 5-ethyluracil (dE) resulted in small perturbation of the methylation patterns. When 2,6-diamino-purine (dD) replaced the adenine to be methylated, small, but significant methylation was observed. The kinetic parameters of the methylation reaction were compared with the thermodynamic free energies and significant correlation was observed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras F., Marinus M. G. The great GATC: DNA methylation in E. coli. Trends Genet. 1989 May;5(5):139–143. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- Bergerat A., Guschlbauer W., Fazakerley G. V. Allosteric and catalytic binding of S-adenosylmethionine to Escherichia coli DNA adenine methyltransferase monitored by 3H NMR. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6394–6397. doi: 10.1073/pnas.88.15.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., Guschlbauer W. The double role of methyl donor and allosteric effector of S-adenosyl-methionine for Dam methylase of E. coli. Nucleic Acids Res. 1990 Aug 11;18(15):4369–4375. doi: 10.1093/nar/18.15.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., Kriebardis A., Guschlbauer W. Preferential site-specific hemimethylation of GATC sites in pBR322 DNA by Dam methyltransferase from Escherichia coli. J Biol Chem. 1989 Mar 5;264(7):4064–4070. [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the methylation by the EcoRI modification methylase of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7279–7286. [PubMed] [Google Scholar]
- Carbonnaux C., Fazakerley G. V., Sowers L. C. An NMR structural study of deaminated base pairs in DNA. Nucleic Acids Res. 1990 Jul 25;18(14):4075–4081. doi: 10.1093/nar/18.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Fazakerley G. V., Quignard E., Teoule R., Guy A., Guschlbauer W. A two-dimensional 1H-NMR study of the dam methylase site: comparison between the hemimethylated GATC sequence, its unmethylated analogue and a hemimethylated CATG sequence. The sequence dependence of methylation upon base-pair lifetimes. Eur J Biochem. 1987 Sep 15;167(3):397–404. doi: 10.1111/j.1432-1033.1987.tb13351.x. [DOI] [PubMed] [Google Scholar]
- Fazakerley G. V., Téoule R., Guy A., Fritzsche H., Guschlbauer W. NMR studies on oligodeoxyribonucleotides containing the dam methylation site GATC. Comparison between d(GGATCC) and d(GGm6ATCC). Biochemistry. 1985 Aug 13;24(17):4540–4548. doi: 10.1021/bi00338a009. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Gromova E. S., Oretskaya T. S., Eritja R., Guschlbauer W. Kinetic studies of MvaI DNA methyltransferase interaction with modified oligonucleotide duplexes. Biochem Mol Biol Int. 1995 Jun;36(2):247–255. [PubMed] [Google Scholar]
- Guyot J. B., Grassi J., Hahn U., Guschlbauer W. The role of the preserved sequences of Dam methylase. Nucleic Acids Res. 1993 Jul 11;21(14):3183–3190. doi: 10.1093/nar/21.14.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T., Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing 5-modified uracils and their interactions with restriction endonucleases Bgl II, Sau 3AI and Mbo I (nucleosides and nucleotides 82). Nucleic Acids Res. 1988 Jun 10;16(11):4761–4776. doi: 10.1093/nar/16.11.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Ho D. K., Wu J. C., Santi D. V., Floss H. G. Stereochemical studies of the C-methylation of deoxycytidine catalyzed by HhaI methylase and the N-methylation of deoxyadenosine catalyzed by EcoRI methylase. Arch Biochem Biophys. 1991 Feb 1;284(2):264–269. doi: 10.1016/0003-9861(91)90294-s. [DOI] [PubMed] [Google Scholar]
- Hülsmann K. H., Bergerat-Coulaud A., Hahn U. E. coli Dam activity in Hepes buffer asks for a new unit definition. Nucleic Acids Res. 1990 Dec 11;18(23):7189–7189. doi: 10.1093/nar/18.23.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Roberts R. J. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res. 1995 Apr 25;23(8):1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossykh V. G., Schlagman S. L., Hattman S. Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-[N6-adenine]-methyltransferase is important for S-adenosyl-L-methionine binding. Nucleic Acids Res. 1993 Oct 11;21(20):4659–4662. doi: 10.1093/nar/21.20.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labahn J., Granzin J., Schluckebier G., Robinson D. P., Jack W. E., Schildkraut I., Saenger W. Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10957–10961. doi: 10.1073/pnas.91.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumry R., Rajender S. Enthalpy-entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9(10):1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Kozlowski S., Breslauer K. J. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983 Apr;22(4):1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Castro M. M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 1985 Dec 20;13(24):8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988 Sep 9;54(6):735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Nielsen J., Taagaard M., Marugg J. E., van Boom J. H., Dahl O. Application of 2-cyanoethyl N,N,N',N'-tetraisopropylphosphorodiamidite for in situ preparation of deoxyribonucleoside phosphoramidites and their use in polymer-supported synthesis of oligodeoxyribonucleotides. Nucleic Acids Res. 1986 Sep 25;14(18):7391–7403. doi: 10.1093/nar/14.18.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Petruska J., Goodman M. F., Boosalis M. S., Sowers L. C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Ono A., Subramaniam R., Santi D. V. On the mechanism of DNA-adenine methylase. J Biol Chem. 1988 Jun 5;263(16):7461–7464. [PubMed] [Google Scholar]
- Pohl F. M. Thermodynamics of the helix-coil transition of (dG-dC) oligomers. Eur J Biochem. 1974 Mar 1;42(2):495–504. doi: 10.1111/j.1432-1033.1974.tb03364.x. [DOI] [PubMed] [Google Scholar]
- Powell L. M., Murray N. E. S-adenosyl methionine alters the DNA contacts of the EcoKI methyltransferase. Nucleic Acids Res. 1995 Mar 25;23(6):967–974. doi: 10.1093/nar/23.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignard E., Fazakerley G. V., Teoule R., Guy A., Guschlbauer W. Consequences of methylation on the amino group of adenine. A proton two-dimensional NMR study of d(GGATATCC) and d(GGm6ATATCC). Eur J Biochem. 1985 Oct 1;152(1):99–105. doi: 10.1111/j.1432-1033.1985.tb09168.x. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Danzitz M. J., Jr EcoRI DNA methyltransferase-DNA interactions. Biochemistry. 1992 Feb 25;31(7):1937–1945. doi: 10.1021/bi00122a006. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Danzitz M. J., Jr Non-additivity of sequence-specific enzyme-DNA interactions in the EcoRI DNA methyltransferase. Nucleic Acids Res. 1991 Dec 11;19(23):6587–6594. doi: 10.1093/nar/19.23.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. O., Mashhoon N. Kinetic mechanism of the EcoRI DNA methyltransferase. Biochemistry. 1991 Mar 19;30(11):2933–2939. doi: 10.1021/bi00225a029. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Sweetnam K. R. Sequence-dependent effects on DNA stability resulting from guanosine replacements by inosine. Nucleic Acids Res. 1994 Jun 11;22(11):2089–2093. doi: 10.1093/nar/22.11.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Meeker A. K., Freire E. Stability mutants of staphylococcal nuclease: large compensating enthalpy-entropy changes for the reversible denaturation reaction. Biochemistry. 1988 Jun 28;27(13):4761–4768. doi: 10.1021/bi00413a027. [DOI] [PubMed] [Google Scholar]
- Szilák L., Dér A., Deák F., Venetianer P. Kinetic characterization of the EcaI methyltransferase. Eur J Biochem. 1993 Dec 1;218(2):727–733. doi: 10.1111/j.1432-1033.1993.tb18427.x. [DOI] [PubMed] [Google Scholar]
- Szilák L., Venetianer P., Kiss A. Purification and biochemical characterization of the EcaI DNA methyltransferase. Eur J Biochem. 1992 Oct 1;209(1):391–397. doi: 10.1111/j.1432-1033.1992.tb17301.x. [DOI] [PubMed] [Google Scholar]
- Taylor I., Watts D., Kneale G. Substrate recognition and selectivity in the type IC DNA modification methylase M.EcoR124I. Nucleic Acids Res. 1993 Oct 25;21(21):4929–4935. doi: 10.1093/nar/21.21.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Halford S. E. The activity of the EcoRV restriction endonuclease is influenced by flanking DNA sequences both inside and outside the DNA-protein complex. Biochemistry. 1992 Jan 14;31(1):90–97. doi: 10.1021/bi00116a014. [DOI] [PubMed] [Google Scholar]
- Wagner R. W., Matteucci M. D., Lewis J. G., Gutierrez A. J., Moulds C., Froehler B. C. Antisense gene inhibition by oligonucleotides containing C-5 propyne pyrimidines. Science. 1993 Jun 4;260(5113):1510–1513. doi: 10.1126/science.7684856. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wilson G. G. Organization of restriction-modification systems. Nucleic Acids Res. 1991 May 25;19(10):2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Yang A. S., Shen J. C., Zingg J. M., Mi S., Jones P. A. HhaI and HpaII DNA methyltransferases bind DNA mismatches, methylate uracil and block DNA repair. Nucleic Acids Res. 1995 Apr 25;23(8):1380–1387. doi: 10.1093/nar/23.8.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]


