Abstract
Treatment-induced and spontaneous clearance of hepatitis C virus (HCV) infection are affected by various host factors. Polymorphisms in the region of the gene IL28B are associated with HCV clearance, implicating the gene product, interferon (IFN)-γ3, in the immune response to HCV. Although it is not clear how the IL28B haplotype affects HCV clearance, IFNγ3 upregulates interferon-stimulated genes (ISGs), similar to interferon-α and β, but via a different receptor. There is also evidence that IFNγ3 affects the adaptive immune response. The IL28B genotype can be considered, along with other factors, in predicting patient responses to therapy with pegylated interferon-α and ribavirin. We review the genetic studies that uncovered the association between IL28B and HCV clearance, the biology of IFNγ3, the clinical implications of the genetic association, and areas of future research.
Keywords: IL28B, Hepatitis C Virus, Interferon Lambda, Interferon Sensitivity, HCV Treatment
Introduction
Spontaneous clearance of hepatitis C virus (HCV) occurs in ~30% of patients with acute infections; the remaining patients develop chronic infections and have predispositions to cirrhosis and hepatocellular carcinoma.1–3 Among chronically infected persons, HCV can be cleared by interferon (IFN)-α based treatment in some cases. Researchers have searched for factors responsible for natural and treatment-induced HCV clearance for more than 10 years. In the past year, genetic studies have identified several single nucleotide polymorphisms (SNPs) in and near IL28B (which encodes IFNγ3) that are associated with clearance. We review the role of IL28B in HCV infection, clinical implications, and directions for future research.
Overview of the IFN-γ family
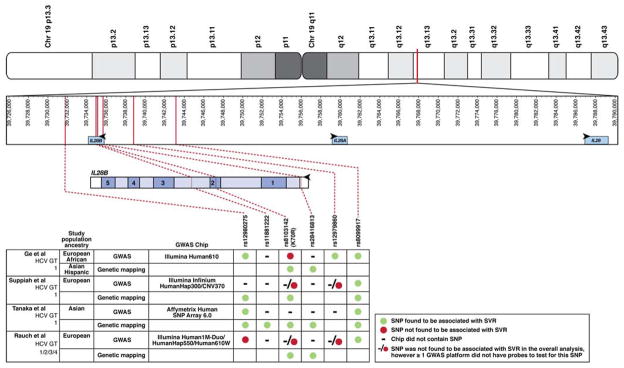
IFNγ3 belongs to the IFNγ family, along with IFNγ1 and IFNγ2, which are encoded by IL29 and IL28A, respectively. IFNγs are categorized as type 3 IFNs and are potent, endogenous antiviral cytokines. Although they are structurally most homologous to members of the IL10 family, IFNγs are more functionally similar to type 1 IFNs4; they signal via Jak–STAT intracellular pathways and upregulate transcription of IFN-stimulated genes (ISGs) that are required to control viral infection. IFNγ-like sequences have been found in most mammalian species, but many are pseudogenes.5 Phylogenetic studies of type 1 and type 3 IFNs and the IL10 family of cytokines indicate the existence of a common, ancestral IFN gene that shared the multi-exon genomic structure of the IFNγ family members; it was putatively identified in ancient fish, evolved in a series of gene duplication and retro-transposon events, and gave rise to the IFNs observed in mammals.5 In humans, the IFNγ genes cluster on chromosome 19 (Fig. 1).5
Fig. 1. SNPs in IFNγ Gene Cluster Associated with HCV Control.
The IFNγ gene cluster is shown in the top panel indicating its position on Chromosome 19. In the second panel the positions of the relevant SNPs corresponding to the text and published data are indicated in relation to the IFNγ gene cluster.10, 12–14 IL28B is upstream and in reverse orientation compared with IL28A. The third panel depicts the genomic structure of IL28B, including its 5 exons, intervening introns, and flanking putative regulatory regions. Vertical lines denote the position of individual SNPs that are associated with HCV treatment response; these are connected by hashed lines to the adjoining table. The only SNP that is in a coding region encodes K70R; it causes an A > G polymorphism on the negative, coding strand and a T > C polymorphism on the positive, non-coding strand. The bottom panel shows which SNPs were associated with response to treatment in each of the indicated studies, including those that were found by genetic mapping studies. As indicated in the legend, symbols are used to indicate if a given SNP was associated with SVR in each of 4 GWAS and genetic mapping studies. ● SNP was associated with SVR; ● SNP was not associated with SVR; −/● SNP was not found to be associated with SVR in the overall analysis, however ≥1 GWAS platform did not have probes to test for this SNP.
IFNγs inhibit HCV replication in vitro6–8 and trials of IFNγ1 in patients with chronic HCV infections have shown promising results;9 86% of treatment naïve-patients who received 4 weeks of the combination of PEG-IFNγ and RBV had a >2 log10 IU/mL decrease in HCV RNA. Therefore, associations made between IL28B variants and HCV clearance in large-scale genetic studies provide an exciting mechanistic link between innate immunity and viral clearance.
IL28B and Control of HCV Infection with Therapy
Many of the first studies that linked IL28B and HCV clearance came from studies of large cohorts of patients with chronic HCV infection who were treated with pegylated IFN-α (PEG-IFNα) and ribavirin (RBV). These cohorts were investigated in genome-wide association studies (GWAS), which allow an unbiased sampling of variations in genes across the entire genome without a hypothesis. The first GWAS was performed using an Illumina Human610quad BeadChip on the IDEAL study, in which patients infected with HCV genotype 1 were randomly assigned to groups that were treated with PEG-IFNα-2a or -2b; the study included only subjects who were treated with a minimum number of total doses.10 Although more than 500,000 SNPs were considered, the investigators found that the strongest predictor of sustained virologic response (SVR) was a SNP (rs12979860) located on the long-arm of chromosome 19, within the IFNγ gene cluster (Fig 2). IL28B is upstream and in the reverse orientation of IL28A; rs12979860 is upstream of both of these genes, closer to IL28B (Fig. 1). At this position, the C allele is the most frequently observed in Caucasians (but not in African-Americans); it is associated with SVR—persons with the CC genotype have SVR rates > 2-fold higher than those with the minor T allele (Table 1). In addition to this SNP, the 6 SNPs most strongly associated with SVR were also found at the IFNγ gene cluster, although the effects of these were no longer observed after adjusting for the presence of rs12979860. After adjusting for the association of rs12979860, the next SNP most strongly associated with SVR found in the GWAS was rs8099917, a non-coding SNP found ~7.5 kb upstream of the IL28B start codon. These findings were validated in a study that genotyped the rs12979860 SNP in patients from the Duke Hepatology Clinical Research Database and Respository, a registry of persons with HCV infections who were followed longitudinally.11 The subjects had HCV genotype 1 (80.5%) or genotype 2 or 3 (19.5%) infections and had received complete courses of PEG-IFNα and RBV therapy. Caucasians, who made up most of the study group, with the CC genotype were > 5 times more likely to achieve an SVR than subjects with the CT or TT genotypes (P=9.0×10−6). Differences in treatment outcomes among African-Americans were not significantly associated with the IL28B genotype, although the authors acknowledged the limited power of the study, given the smaller numbers of African-Americans included (n=106). The association of IL28B genotype with SVR was independent of treatment history. In contrast to treatment response, relapse was not associated with IL28B genotype.
Fig. 2. Manhattan Plot of Genome-Wide Significant Associations of SNPs with Response to Treatment for HCV Infection.
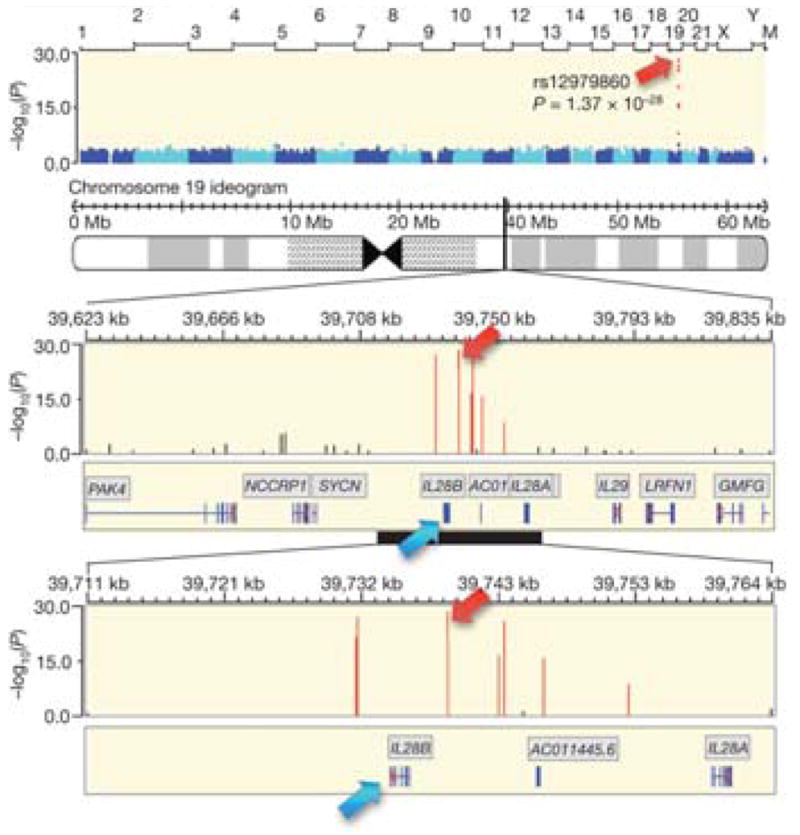
Using an Illumina Human610-quad BeadChip, Ge et al. performed a GWAS using DNA from 1671 HCV-infected persons who were treated with PEG-IFNα-2a or -2b and ribavirin for 48 weeks to find genetic determinants of treatment response. Chromosome number and relative position are indicated on the x-axis above the plot, and −log10(P value) is in the y-axis. The top 6 SNPs were found in or near the IFNγ gene cluster. (Adapted by permission from Macmillan Publishers Ltd: Nature 461, 399–401 (16 August 2009)).
Table 1.
Comparison of IL28B genotype and outcome of HCV treatment with pegylated interferon-α and ribavirina
| SNP Study | Overall SVRb(%) |
SVR in persons with indicated genotype (%) |
% of SVR explained by the favorable genotype | |
|---|---|---|---|---|
| Homozygous favorable allele | Heterozygous or homozygous unfavorable allele | |||
| rs12979860 | ||||
| Ge et al. (USA)c,d | 56 | 82 | 40 | 56 |
| rs8099917 | ||||
| Suppiah et al. (Australia)d | 46 | 56 | 36 | 63 |
| Tanaka et al. (Japan)d,e | NA | NA | NA | NA |
| Rauch et al. (Switzerland)d | 64 | 74 | 50 | 68 |
SVR rates for the indicated SNPs, selected as the most predictive polymorphisms in the respective studies, were calculated using published outcome data.10,12–14 As a clinical predictor, IL28B is similar to other determinants of HCV treatment outcome, such as race, HCV RNA level, and HCV genotype.
The proportion of persons with SVR included in the study.
While the full cohort included persons of European, African, and Hispanic descent, SVR rates are given here for Caucasians. For African Americans, the SVR rates were 53% and 18% in the protective and risk genotypes, respectively.
SVR rates are presented for HCV genotype 1 infections except for Rauch et al., which included persons infected with HCV genotypes 1/2/3/4.
Tanaka et al. was a case-control study, and therefore these percentages are not relevant.
Three other groups used similar approaches to study the genetic basis for SVR; these found a group of SNPs near IL28B that were in strong linkage disequilibrium, indicating that they were inherited as a block, rather than independently. Depending on the technology used and the racial composition of the study population, the specific findings of each study varied (detailed below; Fig. 1, Table 1). Collectively, the results showed that there is a genomic region comprising IL28B and its potential regulatory sequences that is strongly associated with interferon response (Fig. 3). Suppiah et al. studied Australians of European descent with HCV genotype 1 infections who received PEG-IFNα and RBV.12 In the first phase, a GWAS was performed on all participants and results were compared between patients who achieved an SVR while receiving treatment and those that did not respond. Several SNPs were associated with clearance, but the strongest was rs8099917. Compared with the T allele, heterozygosity for the minor G allele was associated with a 1.64-fold increase in risk for not responding to therapy and homozygosity was associated with a 2.39-fold increase in risk.
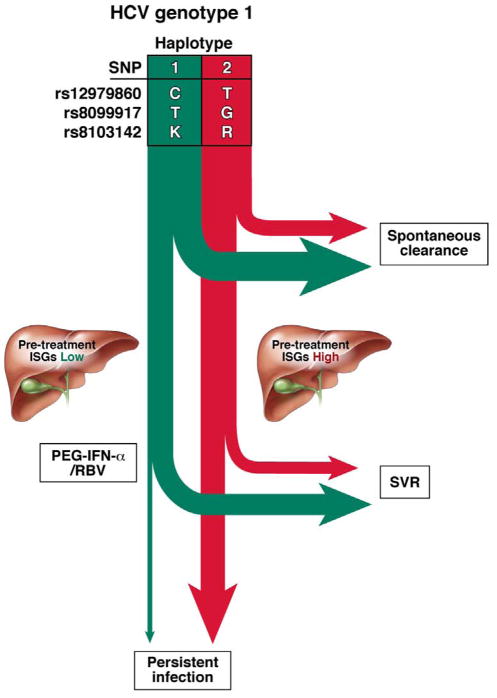
Fig. 3. Predicted Natural and Treatment History of HCV Infection based on Haplotype.
Based on both GWAS and genetic mapping data several SNPs with alleles that are in strong linkage disequilibrium have been identified in or near the IL28B gene locus. These SNPs collectively delineate haplotypes associated with HCV clearance (green) or persistence (red). After acute HCV infection, a greater proportion of persons with specific haplotypes spontaneously clear HCV, compared to patients with other haplotypes. Of the persons who develop persistent HCV infections, intrahepatic expression of ISGs is lower in the group with the HCV clearance haplotype, compared with the persistence haplotype. Patients with the haplotype associated with HCV clearance are more likely to achieve SVRs when they receive treatment with PEG-IFN-α/RBV than patients with other haplotypes.
Tanaka et al. performed a GWAS using an Affymetrix SNP Array of DNA from HCV genotype 1-infected Japanese patients who achieved an SVR to therapy with PEG-IFNα-2a or -2b and RBV, comparing data with that of non-responders.13 The SNPs rs8099917 and rs12980275 segregated with treatment response (Fig. 1). Rauch et al. performed a GWAS of the Swiss Hepatitis C Cohort, a population of European Caucasian subjects infected with HCV genotypes 1 (48%), 2 (10%), 3 (29%), and 4 (9%) who received PEG-IFNα and RBV.14 The most significant markers were clustered at the IFNγ gene loci. The SNP rs8099917 was most highly associated with response to treatment, also reported by Suppiah et al. and Tanaka et al. (Fig. 1). Only 73.9% of persons homozygous for the T allele achieved an SVR, but patients with the minor G allele were 5-fold less likely to respond to therapy (P=3.11×10−8). It is important to note that these investigators used different GWAS platforms than Ge et al. (Fig. 1). The rs12979860 was associated with SVR in subsequent studies, but this association was not fully tested; Tanaka et al. did not include rs12979860 in their GWAS platform and Suppiah et al. and Rauch et al. used multiple platforms with only limited representation of this SNP. Rauch did report that where data were available for rs12979860 it was linked to rs8099917.
IL28B Mapping and HCV Control
It appears that an IL28B haplotype can be a strong determinant of a patient’s response to treatment for HCV infection, and can be represented by a single SNP (or a small number of SNPs) (Fig. 3). Several groups performed more finely tuned genetic studies to clarify which SNP(s) had the greatest associations with treatment response. Ge et al. sequenced the IL28B gene in a subset of patients to find genetic markers that were in high linkage disequilibrium with the SNP rs12979860, identified in their GWAS.10 Two additional markers were found: a non-synonymous SNP (rs8103142) within the IL28B gene that encodes a Lysine→Arginine substitution at position 70 (K70R) and a G→C substitution (rs28416813) 37 base pairs upstream of the translation initiation site (Fig. 1). These 3 SNPs were tightly linked, so it was a challenge to associate any one, individually, with treatment response. Suppiah et al. genotyped 20 additional SNPs within the IL28B gene in an expanded cohort that included Caucasians from Europe and Australia and compared frequencies between persons who did and did not achieve an SVR.12 A G allele at rs12980275 had the strongest association with non-response (Fig. 2). In this study, a 6-allele haplotype was identified that also included K70R (T > C on the coding strand); the haplotype with an arginine (R) was associated with non-response.
Similarly, Tanaka et al. sequenced 16 SNPs in a validation cohort, based on HapMap data that characterized chromosome 19.13 Although they associated the haplotype which included K70R with response to therapy, it did not have a significantly greater association with SVR than any of the individual SNPs; thus any single SNP is sufficient to mark the association with an SVR. In a logistic regression model, rs8099917 was most predictive of an SVR. Rauch et al. sequenced the IL28B gene in subjects with the TT genotype at rs8099917 (associated with response) who achieved the expected SVR and of those who did not achieve an SVR (the unexpected outcome).14 They also sequenced DNA from subjects with the risk GG genotype with the expected outcome of non-response to therapy and in those who achieved an SVR despite carrying this genotype. Twenty-one additional SNPs were identified that fell into 1 of 2 haplotype families. The first family was associated with HCV clearance and the second was associated with persistence. As with Suppiah et al., an R at amino acid position 70 was found in the risk haplotype, along with another SNP (rs28416813) identified by Ge et al..10, 12, 14 These genetic mapping studies identified the specific alleles associated with response (or lack of response) to anti-HCV therapy (Fig. 3).
IL28B and Spontaneous Clearance
The same IL28B haplotypes associated with treatment response are also associated with spontaneous clearance of HCV. Thomas et al. genotyped the rs12979860 SNP in > 1000 persons, from 6 cohorts, with well-characterized spontaneous clearance of HCV or viral persistence and found that the CC genotype was strongly associated with HCV clearance (odds ratio 0.33, P<10−12).15 The investigators also demonstrated that the clearance was mediated by linkage of this genotype with other identified markers, because their inclusion in a multivariate model did not reduce the association between the CC genotype and viral clearance. Ge et al., had observed the clearance effect of the C allele at a higher frequency (73%) in a population with unknown HCV status, compared with patients with chronic infections that went on to receive treatment (63%; P=2.48×10−6), suggesting that this allele occurred more frequently among patients with spontaneous clearance of HCV.10 Further evidence that IL28B is involved in spontaneous clearance was provided by Rauch et al., using a GWAS.14 They associated the T allele of rs8099917 with spontaneous clearance; no SNPs outside of the IL28B/A gene loci were associated with clearance. In studies of a cohort of Spanish patients with acute HCV infection and a cohort of German patients with acute HCV infections (from the anti-D common source outbreak), researchers reported that the CC genotype at rs12979860 was observed more frequently in persons with spontaneous resolution of HCV16, 17 These results indicate the involvement of the same IL28B SNPs in both spontaneous and treatment-induced control of HCV infection (Fig. 3).
IL28B and Racial Differences in HCV Control
Because SNPs in IL28B have varied distributions among ethnic groups (Table 2; www.hapmap.org), it is intriguing to consider whether the differences at this locus account for the association between Caucasian ethnicity and increased spontaneous and treatment-induced clearances. Of the 2 alleles most strongly associated with HCV clearance (rs12979860 C and rs8099917 T), only the former is more common in persons of European, compared with African, descent (Table 2; Fig. 4; hapmap.org); it might underlie the racial differences observed. Thomas et al. genotyped the rs12979860 SNP in > 2000 people from 51 different ethnic populations worldwide and demonstrated that East Asian populations have the highest frequencies of the alleles associated with clearance, sub-Saharan Africans have the lowest frequencies, and Europeans have intermediate frequencies (Fig. 4).15 In their study population, Ge et al. also found the lowest frequencies of the allele associated with clearance among African-Americans (allele frequency=~0.42), the highest frequencies in East Asians (~0.95), and intermediate frequencies in European-Americans (~0.73) and Hispanics (~0.7); these findings were validated in a subsequent study.10, 11 This distribution of alleles could account for the high rates of treatment-induced SVR observed in East Asians.18, 19 When this SNP was studied in multivariate models of spontaneous and treatment-induced clearance, however, it accounted for only ~50%–60% of the ethnic differences observed in HCV control (Ge et al. Nature supplement; Thomas, personal communication).10 It is notable that only 53% of African-Americans with this genotype achieved an SVR, compared with 82% of Caucasians. Other genetic factors beyond IL28B genotype mediate spontaneous and treatment-associated clearance of HCV, although within a given race, IL28B genotype does predict outcome.
Table 2.
Allele Frequencya of Individual SNPs in Populations of Different Ancestry.
| Racec | rs12980275 A/G |
rs11881222 A/G |
rs12979860b C/T |
rs8099917 T/G |
|---|---|---|---|---|
| European | 0.66/0.34 | 0.69/0.31 | NA | 0.83/0.17 |
| African | 0.47/0.53 | 0.67/0.33 | 0.40/0.60 | 0.93/0.07 |
| Asian | 0.92/0.08 | 0.93/0.07 | 0.92/0.08 | 0.92/0.08 |
| Hispanic | 0.57/0.43 | NA | 0.56/0.44 | 0.69/0.31 |
Data are from the International HapMap Project (www.hapmap.org).
Allele frequencies are defined in each population as Protective/Risk. As illustrated by rs12979860, a protective allele may be the major variant in one population and the minor variant in another. rs8103142 and rs28416813 were not included in this table because of lack of available data.
Populations were sampled worldwide and were then summed to constitute each racial designation, as described here and defined by the International HapMap Project. European: Utah residents with Northern and Western European ancestry; Tuscan in Italy. African: African ancestry in Southwest USA; Luhya in Webuye, Kenya; Maasai in Kinyawa, Kenya; Yoruba in Ibadan, Nigeria. Asian: Han Chinese in Beijing, China; Chinese in Metropolitan Denver, Colorado; Japanese in Tokyo, Japan. Hispanic: Mexican ancestry in Los Angeles, California.
Allele frequencies for rs12979860 were more completely tested and are presented in Fig. 4.
NA-allele frequencies not available
Fig. 4. Allele frequencies of the SNP rs12979860 among Different Ethnic Populations.
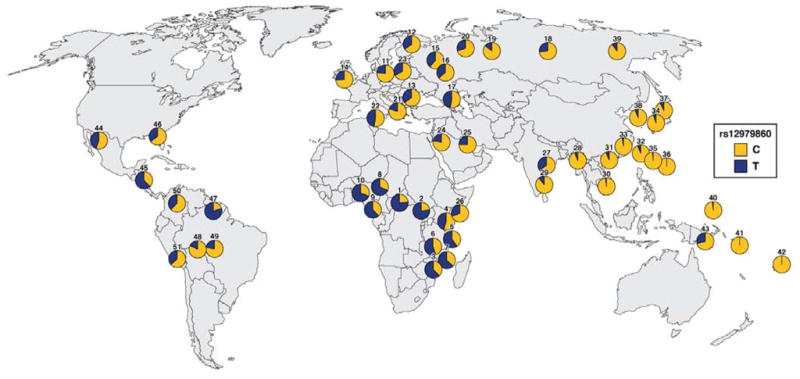
Thomas et al. genotyped the rs12979860 SNP in 2371 person from 51 distinct populations. The frequency map shows the proportional prevalence of the C (associated with HCV clearance) and T (associated with persistence) alleles. People in East and Southeast Asia have the lowest frequency of the alleles associated with HCV persistence; people in Europe have intermediate incidence, and the highest frequencies are found in sub-Saharan Africa. (Adapted by permission from Macmillan Publishers Ltd: Nature 461, 798–801 (16 September 2009)).
IL28B in HCV Genotype 2 and 3 Infections
Most of the initial studies of IL28B and HCV control were centered on persons with HCV genotype 1 infections; studies of IL28B in patients infected with HCV of other genotypes have produced conflicting data and included small numbers. The largest study included 268 persons infected with HCV genotype 2/3 who were randomly assigned to groups that were given PEG-IFNα and RBV for a standard (24 weeks) or variable duration (12 weeks if they had a rapid virologic response or 24 weeks if they did not).20 Surprisingly, the genotype of IL28B was not associated with SVR in persons who received standard duration therapy or who achieved a rapid virologic response and received variable therapy (12 weeks). There was a strong association between IL28B and treatment response only among persons who did not achieve a rapid virologic response and received variable therapy (24 weeks). Intriguingly, in this subset there appeared to be an effect of gene dose—IL28B heterozygotes had an intermediate rate of SVR, between that of patients with homozygosity for alleles that were and were not associated with clearance.
Several other studies have produced mixed results. Montes-Cano et al. associated the IL28B haplotype with an SVR in a cohort of Spanish patients with non-genotype 1 infections.16 McCarthy et al. found that the effect of the IL28B haplotype on SVR was similar between persons with HCV genotypes 1 and 2/3.11 The inclusion of IL28B genotype in a multivariate model, however, reduced only slightly the influence of HCV genotype on SVR. In contrast, Rauch et al. noted a trend in the effect of the IL28B haplotype in persons infected with HCV genotypes 2/3, but this was not statistically significant,14 whereas Rallon et al. did not associate the IL28B haplotype with treatment response in persons infected with HCV genotype 3.21 The effect of the IL28B haplotype status on treatment response might therefore be attenuated for genotypes 2 and 3, but further research is required to clarify this relationship.
IL28B in Patients Co-Infected with HIV and HCV
About one third of HIV-infected persons are also infected with HCV. HIV complicates HCV infection by increasing rates of HCV persistence.2 Despite the immunologic changes associated with HIV infection, it does not affect the association between SNPs near the IFNγ gene cluster and clearance of HCV. Thomas et al. reported that stratification based on HIV status did not modulate the effect of the SNP rs12979860 on HCV clearance.15 Similarly, in a subset of patients in the Swiss Hepatitis C Cohort with that were infected with HIV, Rauch et al. found that the SNP rs8099917 was still associated with HCV clearance; effects of this SNP did not differ markedly between HCV-infected patients with or without HIV infection.14 Rallon et al. found that among patients co-infected with HIV and HCV who completed PEG-IFNα and RBV treatment, the CC genotype at locus rs12979860 was associated with a SVR, and that the association was strongest in among patients infected with HCV genotypes 1 or 4.21 Given the low rates of spontaneous clearance and treatment-induced SVR among persons co-infected with HIV and HCV, it is striking that IL28B is still associated with HCV control in this population.
IL28B and Other Infectious Diseases
Thio et al. investigated whether IL28B SNPs are important in the control of other chronic viral infections where IFNα is important in the immune response (Thio et al, JID in press).22 A cohort of 226 individuals with persistent HBV infection was compared to 384 who had recovered from HBV infection; recovery was not associated with the rs12979860 SNP. They also studied a cohort of 2548 individuals with, or who were at high-risk for, HIV infection and found that the rs12979860 SNP was not associated with infection with HIV or disease progression. In GWAS with the Illumina HumanHap550 BeadChip, Kamatani et al. studied patients with chronic HBV infection and found no association of chronic infection with IL28B genotype. 23 Similarly, Fellay et al. studied HIV-infected persons using the Illumina BeadChip and found no association of IL28B with either HIV-1 viral RNA set-point or with disease progression.24 Polymorphisms in IL28B have therefore not been associated with clearance of other viral infections.
The Effect of IL28B on HCV RNA Levels, Viral Kinetics, and Interferon Responsiveness
Several groups studied the correlation of IL28B genotype, baseline levels of HCV RNA, and treatment response. Ge et al. found that although the C allele of rs12979860 was associated with SVR, it was also, paradoxically, associated with higher viral RNA levels, compared with the T allele (CC patients had 6.35 log10 IU/mL, TC patients had 6.33 log10 IU/mL, and TT patients had 6.16 log10 IU/mL; P=1.21×10−10).10 Higher levels of HCV RNA levels were also observed in patients that were off therapy who had the C.11 Although the differences are modest (< 0.5 log10), the higher levels of HCV RNA among patients with the CC genotype might facilitate innate immune detection and control of the virus during treatment.
In addition to baseline level of HCV RNA, the kinetics of the treatment response appears to be influenced by IL28B genotype. Thompson et al. compared viral kinetic data between persons with CC, CT, and TT genotypes at the rs12979860 SNP 2 weeks after they began therapy with PEG-IFNα and RBV.25 Irrespective of race, persons with the CC genotype had the largest reductions in levels of HCV RNA (Caucasians with CC genotype had the greatest decrease in HCV RNA). This translated to greater rapid virologic response and early virologic response rates among persons with the CC genotype. Much of the effect of IL28B genotype is evident in the first 48 hours following treatment, indicating that IL28B genotype somehow primes the host response to HCV, decreasing the threshold for virologic control with treatment. Alternatively, IL28B genotype could simply be a marker for greater baseline levels of the interferon response, consistent with the findings in patients with spontaneous clearance of the HCV.
Intriguingly, Honda et al found that persons with the TT genotype at the SNP rs8099917 (associated with SVR) had pre-treatment hepatic expression levels of ISGs that were lower than those of persons with the TG or GG genotypes.26 The fact that persons with genotypes associated with SVR have reduced expression of ISGs might account for the higher levels of HCV RNA observed before treatment. Sensitivity to exogenous interferon is inversely associated with levels of ISGs; IL28B genotypes may affect expression levels of ISGs, accounting for the association between IL28B genotype and response to therapy.27, 28 Despite the association of IL28B with clearance, there are persons who carry alleles that are not associated with clearance who clear the virus, as well as patients with alleles associated with clearance whose infection persists. Further studies should be performed with these patients to investigate IL28B genetics, ISG expression levels, and other genetic factors involved in the response to anti-HCV therapy.
IFNγ Biology and Role in HCV Infection
IL28B can determine the outcome of HCV infection, but the mechanisms that mediate the association between the different SNPs and HCV control are unclear—especially in light of the lack of association of IL28B polymorphisms with HBV or HIV infection outcomes. Investigating biology of IFNγ in HCV and other viral infections could provide mechanistic insight.
The main cellular sources of IFNγs are thought to be plasmacytoid dendritic cells (pDCs), although macrophages and conventional DCs probably also produce IFNγs.29 It is not known which cells produce IFNγs in the liver, but candidates include Kupffer cells, DCs, and liver sinusoidal endothelial cells. Hepatocytes also have active innate immune responses and probably release IFNγs upon viral infection.6, 7 Although IFNγs have many effects on a number of viruses, their site of action is constrained by expression of their cognate receptor. IFNγs signal through a heterodimer that comprises the IL28 receptor α chain (IL28Rα) and the IL10 receptor β chain (IL10Rβ). In contrast to the distribution of interferon-α receptor (IFNAR) and even IL10Rβ, which are found on a wide variety of cell types, IL28Rα is found primarily on epithelial cells. This has implications on which cells IFNγs can act on; in 1 study, livers from mice were found to express only low levels of IL28Rα.30
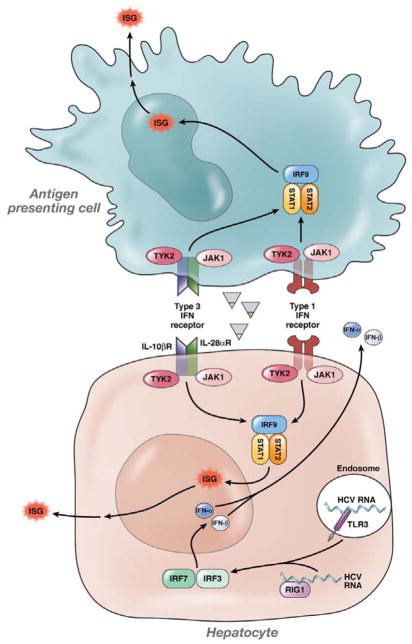
Downstream signaling after IFNγ receptor ligation, however, is similar to type 1 IFN signaling and occurs via the covalently bound tyrosine kinases Tyk2 and Jak1 (Fig. 5).29 These binding partners phosphorylate each other and also phosphorylate STAT 1 and 2 proteins. A consequence of phosphorylation of STATs 1 and 2 is the formation of the IFN-stimulated gene factor 3 complex (ISGF3) along with activated IRF9, which leads to the upregulation of canonical ISGs. ISG upregulation causes many of the innate cellular defenses against viral infection. Overall, IFNγ signaling is thought to be pro-inflammatory and unlike responses to IL10, despite sharing the IL10 receptor subunit.
Fig. 5. IFNγ, IFNα, and IFNβ Signaling in Response to HCV.
HCV RNA is sensed by pattern recognition receptors such as RIG-I and TLR3, which signal using IRF3 and 7 and activate the transcription factor NF-κB to induce expression of IFNγ (plasmacytoid Dendritic Cells), IFNα, and IFNβ (pDCs and hepatocytes). When these factors bind their receptors, IFNγR (a heterodimer of IL28Rα and IL10Rβ), IFNAR1 and IFNAr2, respectively, they induce Jak–STAT signaling, which results in the formation of ISGF3, the transcription factor that upregulates hundreds of ISGs. Exogenously administered IFNs also bind and signal through their cognate receptors; in persistent HCV infection that does not respond to therapy, a steady-state environment exists that is characterized by increased expression of ISGs. (Figure adapted from Thio and Thomas et al., Gastroenterology, 2010).45
In addition to phosphorylation of STATs 1 and 2, STATs 3, 4, and 5 can also be activated via the Il10Rβ chain, which can have immunomodulatory effects.29 IFNγ1 and 2 inhibit IL13 production by T cells following stimulation with concanavalin A, indicating that IFNγs promote the T-helper cell (Th)1 response.31, 32 IFNγ3 increases the Th1 response to an HIV DNA vaccine and simultaneously inhibits regulatory T-cell responses.33 More investigations into the effect of IFNγs on adaptive immunity could reveal that HCV control by IFNγ is the result of a multi-level response.
It is not clear how the SNPs in IL28B affect the IFN signaling pathways. The rs12979860 is 3 kb upstream of IL28B, whereas rs8099917 is nearly 8 kb upstream (Fig. 1). Although it is possible that these SNPs modulate IL28B transcription, it is more likely that they are in linkage disequilibrium with 1 or more SNPs in the IL28B coding or promoter regions. Alternatively, the SNPs could modify transcription factor binding sites. IL29 has multiple interferon-regulatory factor (IRF) and NF-κB binding sites (e.g. −214 to −172 and −98 to −89 upstream of the transcription initiation site), although those that have been reported are not polymorphic.34 The SNP that encodes K70R is important to study because it is tightly linked with SNPs associated with SVR and natural clearance. K70R is not predicted to affect binding to the receptor or signaling, but it could be involved in interactions with other signaling factors that affect viral control.4 The SNPs in IL28B might lead to expression of forms of IFNγ3 that do not function or have weak function, or even hyper-functional variants that reduce the antiviral response by negative feedback. Research is required to determine how IL28B and its variants affect HCV persistence and response to therapy.
Since IFNγs inhibit viral replication, it is logical to consider that expression of different amounts of endogenous IFNγ3 could determine whether a patient controls the virus or remains infected. Two studies compared mRNA levels of IFNγ in whole blood or PBMCs from persons with the T allele at position rs8099917 (associated with clearance) with that of persons with the G allele (associated with viral persistence).12, 13 It has been difficult to quantify IFNγ2 and 3 mRNA levels by PCR because of their sequence homology; levels of IFNγ2 and 3 measured by quantitative PCR are combined when reported. Each group found the highest expression of IFNγ2 and 3 in persons with the TT genotype, compared to TG and GG genotypes, associating higher amounts of endogenous IFNγs with HCV clearance. Using information from the SNPExpress database, Ge et al., in contrast, did not observe differences in IFNγ3 expression among persons not infected with HCV who were homozygous for an allele in linkage disequilibrium with the rs12979860 SNP (see supplemental data for Ge et al).10 Similarly, Honda et al. found no association between hepatic expression of IFNγ2 and 3 and rs8099917 genotype.26 Studies of pDCs and cells that produce IFNγ in the liver should provide insight to the relationship between the IL28B genotype and expression.
Studies of animals with viral infections indicate that the organ-specific distribution of IL28Rα determines response to IFNγ. While some studies observed that IFNγ protects mice against respiratory but not hepatic viruses35, 36 others found protection against hepatic viral infection.37 One study made the surprising observation that IFNγ did not induce expression of ISGs in livers of mice.30 Studies in mice should be interpreted cautiously, however; in human cells and tissues, IFNγ expression was shown to affect hepatotropic viruses. Robek et al. found that IFNγ1 and 2 inhibited HBV replication.6 Moreover, recombinant IFNγ3 had a more potent antiviral effect than IFNγ1 or 2 against encephalomyocarditis virus in hepatocyte cell lines.38 There has not been an extensive, published study of whether primary human hepatic cells respond to IFNγ, although in one report HCV-infected liver tissue had higher amounts of Il28Rα than uninfected liver tissue.39 Interestingly, treatment of macrophages with IFNγ1 inhibited HIV-1 infection, possibly through production of competitive ligands for HIV co-receptors—it is not clear how this finding might relate to HCV clearance in patients.40
Several in vitro studies support a direct role for IFNγ for the control of HCV replication through the innate immune pathway. Robek et al. demonstrated that sub-genomic and full-length HCV replicons were inhibited by recombinant IFNγ1 and 2, which upregulated a representative ISG.6 In a cell culture system, Marcello et al. 7 showed that IFNγ1 inhibited HCV replication with similar kinetics to that of IFNα, but that IFNγ1-induced upregulation of ISGs was stronger and lasted longer. Combinations of IFNγ1 and IFNα had the greatest inhibitory effect on HCV replication, compared with individual agents.8 IFNγ and IFNα might therefore have synergistic effects in controlling HCV infection. Type 1 interferon potentiated IFNγ release in an animal model of viral infection.41 It is possible that the putative SNPs alter the interaction of IFNγ with IFNα.
Clinical Implications
The IL28B genotype provides important, independent information about a patient’s likelihood of achieving an SVR—a commercial test became available in the US in July 2010. Results from this test could be used in combination with algorithms based on HCV genotype and viral load to predict patients’ responses to treatment; IL28B genotype could be a factor that patients and their doctors use to decide whether to initiate therapy or wait until direct anti-viral agents become available. Unfortunately, IL28B genotype does not have a positive predictive value of 100% for SVR, so it cannot be used as the only predictor of response (Table 1); HCV treatment should not be withheld based solely on IL28B genotype. In addition, the positive predictive value is influenced by the prevalence of SVR, which is difficult to extrapolate from published studies where patients with ambiguous outcomes were removed from the analyses and the proportions of patients that achieve SVRs varied. To put some numbers in perspective, the SVR rate for African-Americans with the rs12979860 CC genotype (associated with clearance) was 53%10; this is similar to that of Caucasians with genotype 1 HCV, irrespective of IL28B genotype. Among Caucasians in the same study, the SVR rate for those with the rs12979860 CC genotype was ~82%. Interestingly, when the likelihood of SVR approaches 80%, IL28B genotype can also affect the decision for liver disease staging before treatment. Just as patients with genotype 2 or 3 HCV infection can elect to undergo treatment without consideration of fibrosis stage, persons who have IL28B genotypes associated with viral control might decide not to undergo biopsy evaluation. In either case, non-invasive staging is still recommended to determine whether patients should be screened for hepatocellular carcinoma.42
IL28B genotype can affect how long a clinician should monitor someone with an acute HCV infection before treatment. Those with a haplotype associated with HCV clearance might be monitored longer, since they are more likely to spontaneously clear the virus; those with haplotypes associated with persistence might be better off receiving therapy during the acute period and be monitored for a shorter period beforehand.43 The association between kinetics of HCV response to IFN treatment and IL28B genotype might be used to identify patients that require shorter durations of therapy—further studies are required to determine if this is the case. The association between sensitivity to IFN therapy and IL28B genotype could also affect how clinicians use direct anti-viral agents. For example, IFN lead-in dosing might be the best option for patients with IL28B haplotypes that are not associated with HCV clearance—they are more likely to develop resistance to direct anti-viral agents because their response to IFNα therapy is slower.
As therapeutic agents, IFNγs might have longer and more potent effects than type 1 IFNs, with fewer adverse events, because distribution of IFNγ receptors is more restricted. Phase 1 trials of IFNγ1 in treatment-naïve patients and those with chronic HCV who relapsed after therapy have demonstrated significant reductions in HCV viremia after 4 weeks.9 It will be interesting to see if polymorphisms in IL28B predict a response to IFNγs in these trials; later-stage trials are underway.
Future Directions
Basic science studies help us understand how specific genetic features relate to immunologic function and HCV clearance. It is important to determine exactly which SNP or specific genetic feature of IL28B affects clearance—this could require sequencing of IL28A and IL29 in different ethnic groups of patients with natural and treatment-induced clearance to understand linkage in this region. Comparing the human IL28B sequence to that of chimpanzees and other primates might also provide important information about linkage. Although the same gene cluster and SNPs are associated with clearance vs. persistence and an SVR vs. no response to therapy, these gene variants might affect responses via different mechanisms. The structure of IFNγ3 has been determined, but the active-site amino acids have only been inferred from alanine scans; the role of SNPs in the IL28B coding region might be more precisely defined by resolving the structure of IFNγ3 bound to its heterodimeric receptor. Investigations into the genetics of HCV control are hampered by inadequate model systems of HCV infection; animals with phenotypes that more closely resemble patients with HCV infection would improve our understanding of the role of IFNγ3 in HCV infection. Several HCV cell culture systems exist and studies are underway to examine the role of IL28B in HCV replication using site-directed mutagenesis, to compare major and minor alleles for several SNPs. Similarly, knockout mice with humanized livers can be used to study the effects of IFNγ3 on the immune response and HCV control. Since IFNγs can have redundant effects, responses to IFNγ might need to be fully suppressed by interfering with IL28Rα signaling. Using combined approaches, ISG responses to HCV can be determined using cell culture systems, animal models, small interfering RNAs, and antibodies that inhibit IFNγ signaling.
In situ studies of liver tissues from patients with chronic HCV infection are necessary to delineate which cells release or respond to IFNγs, especially given findings from mouse studies that hepatic expression of IL28Rα is limited. Studies should be performed in human hepatic tissue to compare expression of IFNγ with its receptor. IL28B genotype is assumed to predict the early stages of HCV control, but other immunologic factors, such as pre-treatment levels of ISG, might also predict response. It will be important to determine whether IL28B SNPs also predict response to small molecule therapeutics, their utility in patients with acute HCV infections, and optimal treatment duration. There is evidence that IL28B is associated with an SVR in subjects with chronic HCV who were treated with the protease inhibitor telaprevir in addition to standard therapy.44 The paradox of the association of IL28B genotypes that promote HCV clearance and higher baseline levels of HCV RNA should be further evaluated; usually patients with poor response to treatment have high pre-treatment levels of HCV RNA. IL28B genotype might not predict clearance of all HCV genotypes; the interaction between host and viral genotypes should be further explored. Interest in IL28B genotype has extended to other chronic viral infections—researchers are investigating whether genotypes associated with HCV clearance have other effects in the immune response to pathogens. The identification of IL28B heralds the era of genomic medicine in HCV and opens the door to understanding HCV clearance. The finding has spurred intense bidirectional investigation into the clinic and the bench with the hopes of enhanced therapies against HCV. The coming months and years promise rapid fulfillment of some of these goals, and healthy excitement in the field.
Acknowledgments
Grant Support – This work is supported by NIH grant DA13324 and 1K08AI081544
Abbreviations
- HCV
hepatitis C virus
- IFN
interferons
- IFNα
interferon alfa
- IFNβ
interferon beta
- IFNγ
interferon lambda
- ISG
interferon-stimulated genes
- RBV
ribavirin
- IL29
interleukin-29
- IL28A
interleukin-28A
- IL28B
interleukin-29B
- SNP
single-nucleotide polymorphisms
- HIV
human immunodeficiency virus
- HBV
hepatitis B virus
- IL10
interleukin 10
- PEG-IFNα
pegylated interferon-alfa
- GWAS
genome-wide association studies
- SVR
sustained virologic response
- IDEAL
The Individualized Dosing Efficacy vs. Flat Dosing to Assess Optimal Pegylated Interferon Therapy study
- IL28Rα
interleukin-28 receptor alpha chain
- IL10Rβ
IL10 receptor beta chain
- IFNAR
interferon α receptor
- STAT
Signal Transducers and Activators of Transcription protein
- IRF-3
interferon-regulatory factor 3
- NF-κB
nuclear factor kappa B
- PBMC
peripheral blood mononuclear cells
- pDCs
plasmacytoid dendritic cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilson LE, Torbenson M, Astemborski J, Faruki H, Spoler C, Rai R, Mehta S, Kirk GD, Nelson K, Afdhal N, Thomas DL. Progression of liver fibrosis among injection drug users with chronic hepatitis C. Hepatology. 2006;43:788–795. doi: 10.1002/hep.21091. [DOI] [PubMed] [Google Scholar]
- 2.Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, Laeyendecker O, Boitnott J, Wilson LE, Vlahov D. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 3.Thomas DL, Seeff LB. Natural history of hepatitis C. Clin Liver Dis. 2005;9:383–98. vi. doi: 10.1016/j.cld.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family. J Biol Chem. 2009;284:20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox BA, Sheppard PO, O’Hara PJ. The role of genomic data in the discovery, annotation and evolutionary interpretation of the interferon-lambda family. PLoS ONE. 2009;4:e4933. doi: 10.1371/journal.pone.0004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 8.Pagliaccetti NE, Eduardo R, Kleinstein SH, Mu XJ, Bandi P, Robek MD. Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem. 2008;283:30079–30089. doi: 10.1074/jbc.M804296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, Gordon SC, Marotta P, Vierling JM, Carlos Lopez-Talavera J, Byrnes-Blake K, Fontana D, Freeman J, Gray T, Hausman D, Hunder NN, Lawitz E. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010 doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 10.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009 doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, Tillmann HL, Muir AJ, McHutchison JG. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–2314. doi: 10.1053/j.gastro.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 14.Rauch A, Kutalik Z, Descombes P, Cai T, di IJ, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J, Colombo S, Cerny A, Dufour JF, Furrer H, Gunthard HF, Heim M, Hirschel B, Malinverni R, Moradpour D, Mullhaupt B, Witteck A, Beckmann JS, Berg T, Bergmann S, Negro F, Telenti A, Bochud PY. Genetic variation in IL28B Is Associated with Chronic Hepatitis C and Treatment Failure - A Genome-Wide Association Study. Gastroenterology. 2010 doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 15.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montes-Cano MA, Garcia-Lozano JR, Abad-Molina C, Romero-Gomez M, Barroso N, Aguilar-Reina J, Nunez-Roldan A, Gonzalez-Escribano MF. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 2010;52:33–37. doi: 10.1002/hep.23624. [DOI] [PubMed] [Google Scholar]
- 17.Tillmann HL, Thompson AJ, Patel K, Wiese M, Tenckhoff H, Nischalke HD, Lokhnygina Y, Kullig U, Gobel U, Capka E, Wiegand J, Schiefke I, Guthoff W, Grungreiff K, Konig I, Spengler U, McCarthy J, Shianna KV, Goldstein DB, McHutchison JG, Timm J, Nattermann J. A Polymorphism Near IL28B Is Associated With Spontaneous Clearance of Acute Hepatitis C Virus and Jaundice. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Yan KK, Guirgis M, Dinh T, George J, Dev A, Lee A, Zekry A. Treatment responses in Asians and Caucasians with chronic hepatitis C infection. World J Gastroenterol. 2008;14:3416–3420. doi: 10.3748/wjg.14.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CH, Liu CJ, Lin CL, Liang CC, Hsu SJ, Yang SS, Hsu CS, Tseng TC, Wang CC, Lai MY, Chen JH, Chen PJ, Chen DS, Kao JH. Pegylated interferon-alpha-2a plus ribavirin for treatment-naive Asian patients with hepatitis C virus genotype 1 infection: a multicenter, randomized controlled trial. Clin Infect Dis. 2008;47:1260–1269. doi: 10.1086/592579. [DOI] [PubMed] [Google Scholar]
- 20.Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D, Carretta V, Minerva N, Goldstein DB, McHutchison JG. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–7. 827. doi: 10.1053/j.gastro.2010.05.079. [DOI] [PubMed] [Google Scholar]
- 21.Rallon NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, Shianna KV, Vispo E, Thompson A, McHutchison J, Soriano V. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23–F29. doi: 10.1097/QAD.0b013e3283391d6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin MP, Qi Y, Goedert JJ, Hussain SK, Kirk GD, Hoots WK, Buchbinder S, Carrington M, Thio CL. IL28B polymorphism does not determine hepatitis B virus or HIV outcomes. Journal Infect Dis. 2010 doi: 10.1086/657146. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, Puseenam A, Sura T, Daigo Y, Chayama K, Chantratita W, Nakamura Y, Matsuda K. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 24.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R, Poordad F, Lawitz EJ, McCone J, Shiffman ML, Galler GW, Lee WM, Reindollar R, King JW, Kwo PY, Ghalib RH, Freilich B, Nyberg LM, Zeuzem S, Poynard T, Vock DM, Pieper KS, Patel K, Tillmann HL, Noviello S, Koury K, Pedicone LD, Brass CA, Albrecht JK, Goldstein DB, McHutchison JG. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Yamashita T, Nakamura M, Shirasaki T, Horimoto K, Tanaka Y, Tokunaga K, Mizokami M, Kaneko S. Hepatic Interferon-Stimulated Genes Expression Is Associated With Genetic Variation in Interleukin 28B and the Outcome of Interferon Therapy for Chronic Hepatitis C. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 28.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ank N, West H, Paludan SR. IFN-lambda: novel antiviral cytokines. J Interferon Cytokine Res. 2006;26:373–379. doi: 10.1089/jir.2006.26.373. [DOI] [PubMed] [Google Scholar]
- 30.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, Gallagher G. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254–261. doi: 10.1038/sj.gene.6364382. [DOI] [PubMed] [Google Scholar]
- 32.Srinivas S, Dai J, Eskdale J, Gallagher GE, Megjugorac NJ, Gallagher G. Interferon-lambda1 (interleukin-29) preferentially down-regulates interleukin-13 over other T helper type 2 cytokine responses in vitro. Immunology. 2008;125:492–502. doi: 10.1111/j.1365-2567.2008.02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N, Weiner DB. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009;113:5868–5877. doi: 10.1182/blood-2008-11-190520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 35.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 39.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, Yao L, Liu H, Barahmand-pour F, Sivakumar P, Chan C, Birks C, Foster D, Clegg CH, Wietzke-Braun P, Mihm S, Klucher KM. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 40.Hou W, Wang X, Ye L, Zhou L, Yang ZQ, Riedel E, Ho WZ. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol. 2009;83:3834–3842. doi: 10.1128/JVI.01773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, Klucher K, Paludan SR. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- 42.Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, Management, and Treatment of Hepatitis C. Hepatology. 2009 doi: 10.1002/hep.20119. In press. [DOI] [PubMed] [Google Scholar]
- 43.Grebely J, Petoumenos K, Hellard M, Matthews GV, Suppiah V, Applegate T, Yeung B, Marks P, Rawlinson W, Lloyd AR, Booth D, Kaldor JM, George J, Dore GJ. Potential role for Interleukin-28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology. 2010 doi: 10.1002/hep.23850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Chayama K, Nakamura Y, Kumada H. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421–429. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
- 45.Thio CL, Thomas DL. Interleukin-28b: a key piece of the hepatitis C virus recovery puzzle. Gastroenterology. 2010;138:1240–1243. doi: 10.1053/j.gastro.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]