Abstract
Scaffold proteins influence cellular signalling by binding to multiple signalling enzymes, receptors or ion channels. Although normally devoid of catalytic activity, they have a big impact on controlling the flow of signalling information. By assembling signalling proteins into complexes, they play the part of signal processing hubs. As we learn more about the way signalling components are linked into natural signalling circuits, researchers are becoming interested in building non-natural signalling pathways to test our knowledge and/or to intentionally reprogram cellular behaviour. In this review, we discuss the role of scaffold proteins as efficient tools for assembling intracellular signalling complexes, both natural and artificial.
Introduction
Approximately 15 years ago, Ste5 became established as one of the first well-characterized scaffold proteins [1] (Figure 1). At that time, it was a novelty to discover a protein that binds several components of a signalling cascade simultaneously. (Ste5 binds all components of a three-tiered mitogen-activated protein [MAP] kinase cascade from Saccharomyces cerevisiae.) We now know that proteins similar to Ste5 are abundant. Scaffold proteins are molecules that bind multiple signalling components and promote their communication or interaction with each other. They bind at least two signalling enzymes (e.g. kinases or phosphatases), receptors or ion channels. Classical scaffolds usually do not possess any type of enzymatic activity and they can be regarded as specificity elements that selectively facilitate signalling between their bound components. Of note, the term adaptor protein is also widely used to describe proteins that are functionally similar to scaffolds. Adaptors in the literature, however, are assigned to have amore limited role compared with scaffolds: they bind to only two other proteins and frequently direct these into specific cellular locations. Unfortunately, the scaffold definition given here does not enable straightforward identification of novel scaffold proteins. Thus, scaffold proteins so far have been mainly discovered on an individual basis, after they had been shown to bind to a better known protein kinase or ion channel, or to other effector proteins (Table 1). Historically, many active signalling components with kinase, phosphatase, GTPase, ion channel, protease or secondary messenger synthesis or degradation activity – and even some scaffolds (e.g. Ste5) – were discovered through systematic genetics screens on model organisms [2,3]. Because scaffolds assemble signalling proteins into functional modules through various protein–protein interactions (PPIs), it might be possible to search for them based on their PPI interaction profiles. Therefore, reliable PPI databases and sophisticated PPI discovery techniques will probably be instrumental to indentify scaffold candidates systematically in the future (Box 1). Some proteins that are clearly considered scaffolds because they bind to multiple enzymatic partners also contain a catalytic domain, illustrating how it is possible for a protein to have both a catalytic and scaffolding function. Because scaffolds possessing catalytic domains might operate differently from classical signalling scaffolds, this current review focuses on the classical group only.
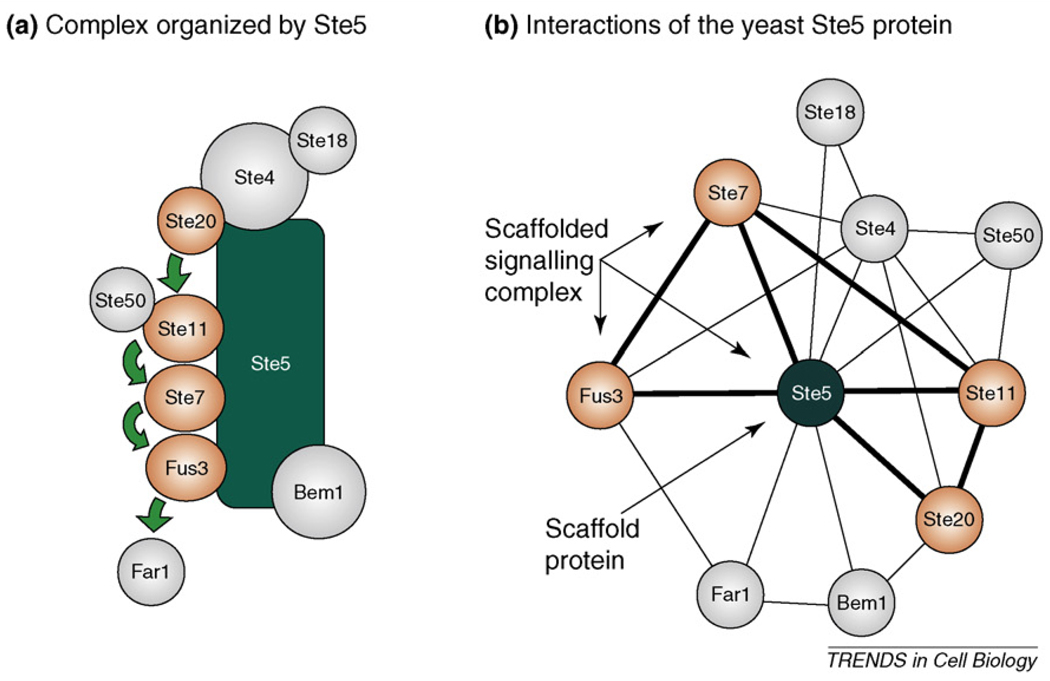
Figure 1.
Ste5, a classical signalling scaffold. (a) Our current view on the scaffolding interactions of the yeast Ste5 protein. Ste4 and Ste18 are the β and γ subunits of the heterotrimeric G-protein with a role in recruiting components of the pathway to the plasma membrane upon pheromone receptor activation. Ste20, Ste11, Ste7 and Fus3 are protein kinases, phosphorylating each other in successive steps (illustrated with green arrows). The Fus3 MAP kinase, as the output of the scaffolded module, phosphorylates substrate targets (such as the cyclin-dependent kinase inhibitor Far1). Other proteins, such as Ste50 and Bem1, have additional regulatory roles in this cascade. (b) The Ste5 signalling scaffold in its PPI network context. Proteins possessing intrinsic enzymatic activity are coloured orange, passive components are gray and the edges of the full 3-graphs that represent scaffolded signalling complexes are drawn with a thick line. These interactions were taken directly from the STRING database (http://string.embl.de), with the following settings: confidence level = 0.90, type: experimental only, maximum interactors = 100. (Some interacting partners are not presented for simplicity.).
Table 1.
Examples of classical signalling scaffolds with their known interacting partners
| Scaffold protein | Selected binding partners (type) | Refs |
|---|---|---|
| KSR1,2 | Raf-1 (protein kinase) MEK1/2 (protein kinase) ERK1/2 (protein kinase) |
[20] |
| JIP1,2,3,4 | MLKs (protein kinase) MKK3/4/7 (protein kinase) JNKs/p38s (protein kinase) |
[44] |
| β-arrestin 2 | ASK1 (protein kinase) MKK4 (protein kinase) JNK3 (protein kinase) |
[45] |
| Ste5 (S. cerevisiae) | Ste11 (protein kinase) Ste7 (protein kinase) Fus3 (protein kinase) Ste18 and Ste4 heterotrimeric G protein subunits (GTPase) |
[1,3,46] |
| Paxillin | FAK (protein kinase) ILK (protein kinase) PTP-PEST (protein phosphatase) ERK1/2 (protein kinase) |
[47] |
| Gab1,2,3 | EGFR (receptor) c-Met (receptor) PI3 kinase (lipid kinase) SHP2 (protein phosphatase) Ras-GAP (Ras GTPase activator) |
[48] |
| PSD95 | NMDAs (ion channels) Kainate receptors (ion channels) SynGAP (Ras GTPase activator) nNOS (nitric oxyde synthase) Fyn (protein kinase) |
[22,49] |
| Homer1,2,3 | mGluR1/5 (receptors) Cdc42 (GTPase) Ins(1,4,5)P3 receptor (ion channel) |
[50,51] |
| InaD (D. melanogaster) | Rhodopsin (receptor) PLC β (phospholipase) PKC (protein kinase) TRP and TRPL (ion channel) |
[11,12] |
| mAKAP | PKA (protein kinase) PDE4D3 (phosphodiestherase) RyR (ion channel) PP2A (protein phosphatase) Epac1 (Rap GDP/GTP exchanger) |
[9,10,52] |
| AKAP79 | PKA (protein kinase) PKC (protein kinase) Calcineurin (protein phosphatase) β2 adrenergic receptor (receptor) |
[53] |
| RACK1 | PKC β (protein kinase) PDE4D5 (phosphodiestherase) Src (protein kinase) Integrin β chain (receptor) IFN receptor type I (receptor) |
[54] |
Box 1. Interactome-based search and definition for classical scaffold proteins.
To assess the real importance and abundance of classical scaffold proteins, it would be important to devise methods that would be able to predict them. Naturally, classical biochemical characterization – the major method by which scaffolds have been identified in the past – will be still required to experimentally validate the role of scaffold candidates in cellular signalling. In contrast to enzymes and to other active signalling components, known classical scaffolds do not share common signature motifs. Therefore, their identification based on sequence is currently not possible. Known scaffolds, however, often contain modular PPI domains (e.g. SH3, PDZ) that can be easily identified. Moreover, related scaffolds might contain similar domain signatures (Table 2). The other common feature among known scaffolds is that they interact with at least two signalling proteins possessing some signalling related activity. This latter property can be identified from PPI databases if reliable data are available (Figure 1b). Domain architecture and sequence analysis tools are reliable; data in PPI databases, however, are often incomplete and this greatly limits identification of putative scaffolds.
Fortunately, PPI detection tools have been greatly developed: high-throughput screening methods for protein–protein interactions (HTS-PPI) were successfully used to map out protein interaction networks in model organisms [55,56]. HTS-PPI methods are based on two basic technologies: yeast two hybrid (Y2H) screens and affinity purification coupled to mass spectrometry (AP-MS) [57,58].
HTS-PPI methods might not be suitable to capture low affinity or dynamic interactions. They also produce many false positive leads. For this reason, functional interactome studies can greatly complement HTS-PPI data that mostly represent physical interactions [2,59–62]. As an example for the power of the combined approach, HTSPPI, gene expression and protein phosphorylation data were combined to identify functional motifs in the regulatory networks of S. cerevisiae [60]. Most interestingly, the kinase scaffold motif, where a protein interacts with a kinase and with its substrate, was the most abundant followed by interacting kinase substrates, kinase cascades and transcription factor regulated kinase-substrate pairs.
Despite being incomplete, high-throughput experimental interactome datasets greatly contribute to data deposited in curated PPI network databases (e.g. STRING [63], HPRD [64], BIND [65], MINT [66], BioGRID [67]), which present more reliable interactome data.
Based on interactome data, signalling scaffolds can be defined as proteins that: (i) lack intrinsic catalytic activity relevant for signalling; (ii) have at least two binding partners with catalytic activity relevant for signalling; and (iii) have binding partners that interact with each other in a direct or indirect way. Proteins satisfying the above criteria are classical signalling scaffolds.
In this review, we demonstrate how classical signalling scaffolds operate as ‘active’ signalling components even without possessing direct enzymatic activity. Moreover, the ever-increasing list of classical signalling scaffold proteins forces us to ask questions about their origin as a distinct group of signalling proteins. Because scaffolds determine connections between signalling enzymes and influence properties of signalling cascades, it is also intriguing to ask how they can be of potential use for the engineering of cells with novel therapeutic or biotechnological functions.
Scaffolding in signalling: insight into the mechanism
In the past, signalling scaffolds have been regarded as ‘passive’ components. However, this point of view is gradually changing as more scaffolds turn out to have an ‘active’ role (see later). But even if a scaffold just simply binds to two or more other proteins, this enforced proximity immediately results in several interesting properties (Figure 2a). The scaffolded complex can be regarded as a micro-environment on its own, where the concentrations of participants are vastly enriched in a small location (Box 2). Therefore, the enforcement of proximity results in an increase of specificity as well, because potential targets of the enzyme are recruited selectively by other proteins [4]. Furthermore, co-recruitment of positive and negative regulators into the same scaffolded complex will give rise to complex dynamic behaviour (see later). Moreover, it is possible to form a different signalling complex without the use of a completely different set of proteins. Because scaffolds can regulate both enzyme kinetics and target specificity, cellular networks can make an economical use of a limited set of signalling proteins without compromising specificity. Through this combinatory use of components, several pathways can share the same signalling components [5] (Figure 2b). Assembly of ‘signalosomes’ through the use of scaffolds also opens up the way for a network-level dynamic regulation of signalling modules involving feedback. Entire modules can be easily assembled on specific cellular locations because components of the complex can be delivered in a ‘package’. In turn, it is also easy to shut down signalling modules by disruption or by degradation of scaffolds (Figure 2c). This is particularly advantageous if assembly of scaffolded signalling modules is a combinatorial process: module properties are rather determined by the overall module composition than by the independent activity of the recruited components. Computational modelling of the kinetics of scaffolded systems indeed shows high sensitivity of the output amplitude to scaffold availability – something that has been confirmed by numerous experiments since the discovery of these proteins [6]. Tethering several components more or less tightly, these complexes open up the possibility for the introduction of several new regulatory mechanisms. Enzymes interacting with the scaffold might incur a change in their conformation and their activity through allosteric effects upon binding with the scaffold [7] (Figure 2d). Apart from this type of conformational fine-tuning of signalling enzymes, naturally evolved scaffolds might contain flexible joints that can bring components together in optimal orientation [8].
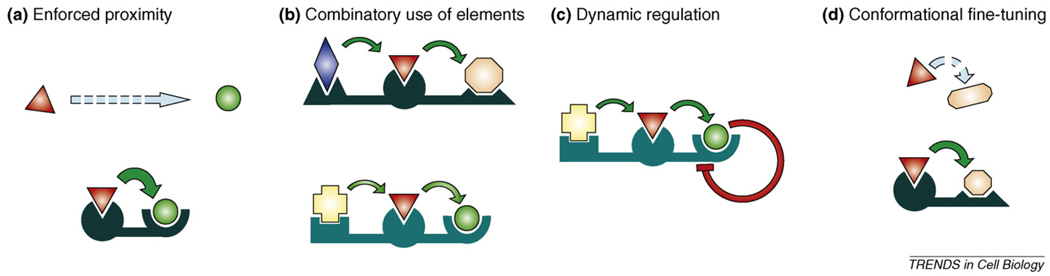
Figure 2.
Schematic representation of the four main scaffold mechanisms. Known examples of signalling scaffold proteins display four distinct mechanisms through which they can modify signalling between active components. (a) Scaffolds tether enzymes close in space and enhance effective local concentrations. (b) They can mediate assembly of signalling complexes in a combinatorial manner, meaning that a certain active component (depicted as red triangle) can participate in signalling through different pathways using distinct scaffold proteins. (c) The function of full signalling modules can be dynamically regulated if the turnover or accessibility of the scaffold is dynamically regulated, without the need to execute the same type of regulation individually on the active components. (d) Some scaffolds can also modify the conformation of enzymes binding to them, or in turn the conformation of the scaffold can also be modified.
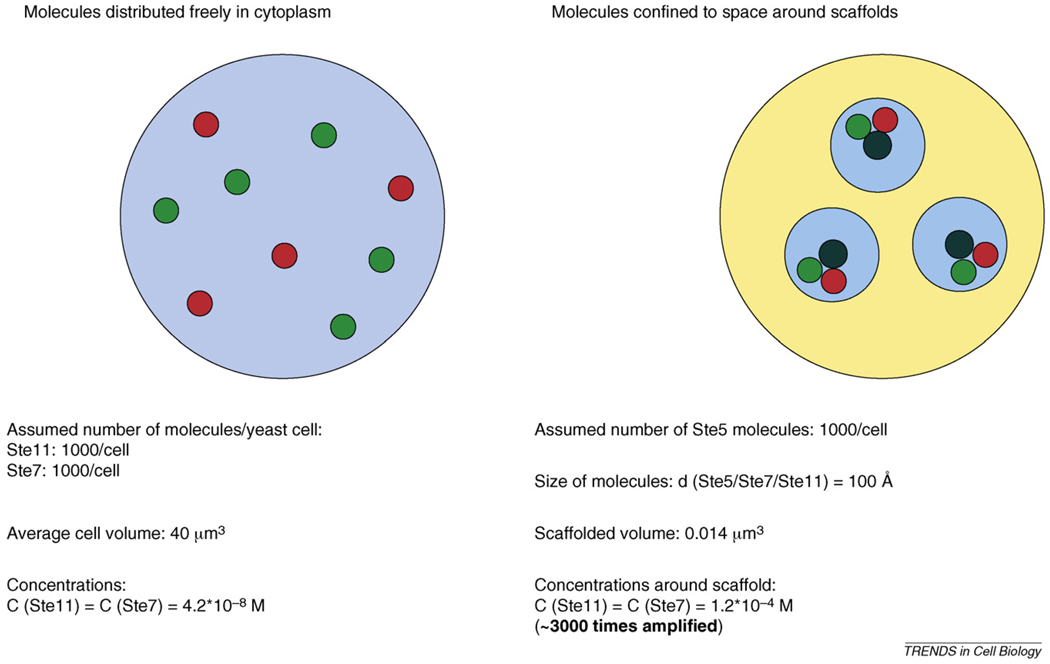
Box 2. Demonstration of the power of enforced proximity through a theoretical experiment.
Enzyme activity depends on the concentration of substrates. Therefore, an increase in the local concentration of substrates around enzymes has a great impact in cellular signalling (Figure I). A very simple calculation aiming to determine the effective concentrations for a kinase cascade pair shows that local concentrations become amplified by at least 3000-fold when both proteins are bound to a general scaffold compared with when they are freely distributed in the eukaryote cell. Ste11 and Ste7 are two kinase components of a three-tiered MAP kinase cascade from yeast, whereas Ste5 is a classical scaffold protein binding to both kinases. Values used in the calculation are realistic and they were approximated for simplicity. Number of molecules per cells for Ste11, Ste7 and Ste5 were assumed to be 1000. These numbers have been approximated by studies aimed to determine the number of signalling molecules per cell [68]. The average diameter of a yeast cell is ~4 µm, and the average volume is ~40 µm3 [69], meaning that the approximate concentration of the Ste11 and Ste7 kinases are ~0.04 µM. If there are 1000 molecules of Ste5 in the cell and we estimate the average diameter of the kinase and scaffold molecules to be 100 Ǻ (1Ǻ = 0.1 nM) [70], then the scaffolded volume into which the kinases are recruited is 0.014 µm3. Assuming that all Ste11 and Ste7 kinases bind to Ste5, the ratio of 40/0.014 = 3000 gives the fold increase in local concentrations for these two proteins when they all bind to Ste5 compared with when they are all freely distributed in the cytoplasm.
Naturally, it is not realistic to assume 100% occupancy of the scaffold by the interacting kinases. In vivo studies using fluorescence cross correlation spectroscopy (FCCS) have suggested that the actual occupancy of Ste5 scaffolds in vivo is much lower [71,72]. Nevertheless, moving only 10% of each kinase into the scaffolded volume would still dramatically increase the number of Ste11–Ste7 complexes present at steady state (~300-fold).
Scaffolds can modify signalling events in space, time and in dose–response relationships. Figure 3 demonstrates how this is accomplished by three classical scaffold proteins. Muscle-specific A kinase anchoring protein (mAKAP) can localize cyclic adenosine monophosphate (cAMP) level changes in mammalian cardiomyocytes [9,10], InaD enables fast adaptation to take place in the fly visual system [11,12] and Ste5 facilitates a certain MAP kinase signalling event in yeast [13]. These examples do not show all aspects of scaffold-facilitated processes but instead give a demonstration about what scaffolds can potentially do. Moreover, it is also important to note that unscaffolded signalling circuits might also be able to demonstrate properties similar to scaffolded ones. Circuits capable of displaying diverse signalling phenotypes commonly use positive or negative feedback (or feed-forward) loops [14]. Therefore, what makes scaffolded systems distinct is that they can efficiently tether these feedback regulatory components close in space (Figure 3a,b). Apart from being passive tethering elements for the assembly of signalling complexes, scaffolds can have a more active role: their conformation can change during repeated signalling events or they can change the conformation of enzymes binding to them and allosterically modify their function [12,13,15] (Figure 2d and Figure 3c,d).
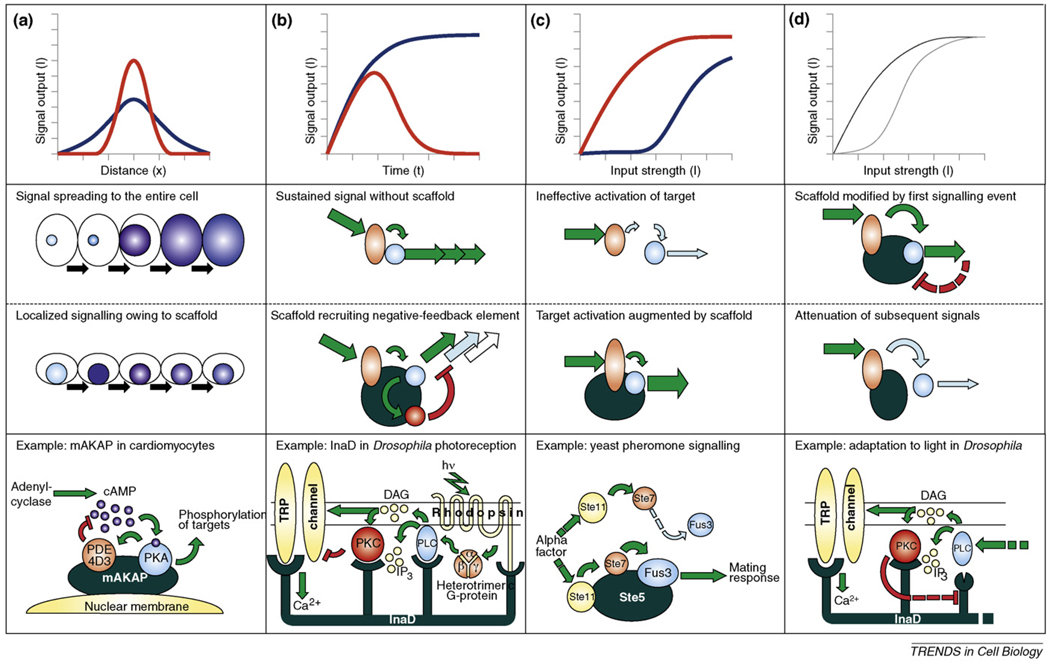
Figure 3.
Scaffold proteins regulate characteristics of intracellular signalling in space, time or input strength. Top panels (a–c) display presumed signalling profiles of certain circuits in the absence (blue) or in the presence (red) of a scaffold. In (d), signalling scenarios are compared before and after scaffold modification took place through feedback. Panels in the middle rows schematically depict the circuits that generate behaviours shown in the top panels. Bottom panels demonstrate examples. (a) Scaffolds can provide highly localized signalling. AKAP proteins confine the activity of protein kinase A (PKA) into well-defined cellular regions, and they also help inactivate the adenyl cyclase-generated cAMP signal through binding to distinct phosphodiesterase (PDE) isoforms [9]. The local assembly of all these signalling elements serves to create localized ‘pulses’ of cAMP-dependent phosphorylations that translates into perinuclear Ca2+-pulses in the case of mAKAP [10]. (b) Scaffolds can control the dynamics of signalling. In the phototransduction system of the fly Drosophila melanogaster, the seven-transmembrane-type rhodopsin photoreceptor and its downstream effectors (including the TRP ion channel) are organized into a single multimolecular complex with the help of the multi-PDZ protein InaD (Inactivation no afterpotential D). When the receptor is switched on by light, it acts as an exchanger of GDP for GTP on its associated heterotrimeric G-protein, dissociating it into α- and β–γ subunits. The binding of free β–γ subunit activates phospholipase C (PLC), which in turn hydrolyses the phosphoinositides contained in the membrane to inositol (1,4,5)-trisphosphate [Ins(1,4,5)P3] and diacylglycerol (DAG). Although Ins(1,4,5)P3 seems to have no direct role in this system, DAG activates both the transient receptor potential (TRP) channels to generate Ca2+-influx and the enzyme protein kinase C (PKC). Once turned on, the latter readily phosphorylates the TRP channel, rendering it inactive, thus terminating the signal. The recruitment of both positive effectors (phospholipase C) and feed-forward inhibitors (PKC) into a signalling complex accounts for the generation of ion flux pulses separated in time by barely a few milliseconds – creating one of the fastest known heterotrimeric G-protein-based signalling cascades [11]. (c) Scaffolds can change dose–response relationships. The mating pathway of S. cerevisiae relies on the presence of the scaffold protein Ste5, which organizes the protein kinases Ste11, Ste7 and the Fus3 MAP kinase into a single complex. Surprisingly, after binding to Ste5, Fus3 becomes a 5000-fold better substrate for its upstreamkinase, Ste7. It seems that Ste5makes Fus3 a better substrate by unlocking its activation loop for more efficient phosphorylation by Ste7 [13]. (d) Scaffolds can provide memory effects. In addition to the immediate feed-forward mechanisms in Drosophila photoreception, the presence of the scaffold enables a sophisticated mechanism for the adaptation to high-input and low-input conditions. In low-light conditions, the fifth PDZ domain of the InaD protein resides in an open conformation. During repeated stimulation of the pathway (under high-light conditions), the activation of PKC also results in the phosphorylation of the scaffold, which in turn suffers a conformational change, turning the fifth PDZ domain into a closed conformation, and releasing its previously bound partner – most probably PLC. This decreases the flux through the pathway, resulting in long-term adaptation [12].
Although classical scaffolds lack catalytic activity, they have an ‘active’ role in signalling: apart from increasing local concentrations of signalling proteins they might also change their conformation. In turn, scaffolds can be modified by their bound components. In addition, scaffolds can assemble functionally distinct sets of signalling cascades that might share components.
Scaffold (r)evolution?
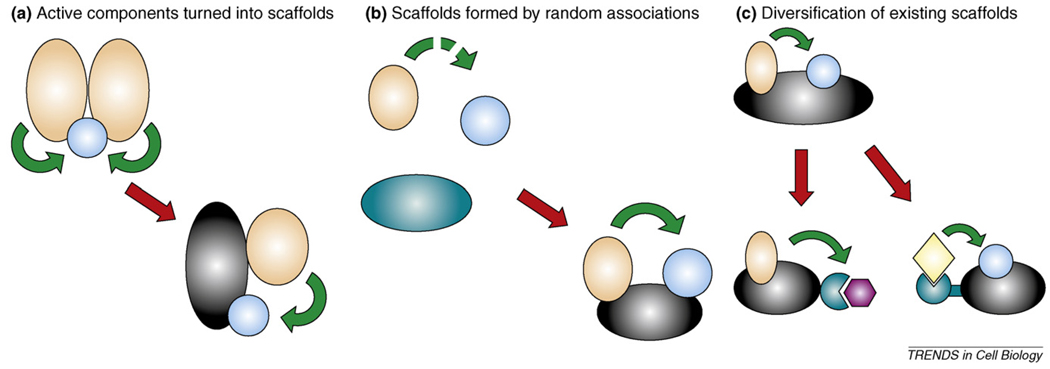
Because classical scaffolds influence signalling network behaviour in diverse ways, it is intriguing to speculate that they might have had an important role in the evolution of signalling circuits. At present, it is possible to identify some, but probably not all, mechanisms responsible for the emergence of scaffolds (Box 3). Looking at the known examples of known scaffold proteins, it seems that this group of signalling proteins is heterogeneous and it is unlikely that all scaffolds are linked through common ancestry. This is supported by the diverse, unrelated ways scaffolds can come into existence (e.g. active components turn into scaffolds or scaffolds that form by random associations). Based on their domain composition, it is also unlikely for example that the scaffold protein POSH – a multi-SH3 domain containing protein – and multi-PDZ domain containing scaffolds have ever shared common ancestry. Naturally, some scaffolds can have common origin, because diversification of scaffolds can frequently occur (see classical scaffold families with domain signatures in Table 2).
Box 3. Scaffold evolution.
Although the evolution of scaffolds is much harder to trace back in time compared with that of enzymes, there are a few generalizations that can be made about their origin (Figure I). Active components of signalling (such as naturally self-dimerizing enzymes and receptors) could be turned into scaffolds by retaining their original connections, but losing their enzymatic or other associated activities (Figure Ia). A classic example is the mammalian MAPK scaffold kinase suppressor of Ras (KSR). Because KSR is so highly homologous to one of its binding partners, the Ser/Thr kinase Raf, it is very likely that the two originally formed part of a single homomeric complex [73].
Scaffolds could also possibly arise from apparently randomly associated proteins (Figure Ib). An example of such a ‘randomly created’ scaffold is the Gephyrin protein, which has an organizational role in the inhibitory synapses of the mammalian central nervous system by clustering glycine (and probably GABA) receptors together with other signalling proteins such as GEFs. This is apparently a new invention: Gephyrin has still kept its molybdene-coenzyme-synthase activity, hinting at its origin [74]. The association of proteins with such distant functions probably happened by pure chance.
Once established, scaffolds might have evolved further by changing their structure to help establish new connections (Figure Ic). For example, InaD is a member of the multi-PDZ-domain proteins and is closely related to another multi-PDZ protein: PATJ (also known as InaD-like protein). PATJ is well-conserved in animals, being a part of the cell polarity complex that forms the tight junctions between epithelial cells. In contrast to InaD, PATJ is not involved in GPCR-mediated signalling but it binds to the atypical protein kinase C, and this interaction is important for creating and maintaining cellular polarity [75]. It seems that certain insects have invented a version of this protein (InaD) that could also associate with rhodopsin, phospholipase C and ion channels.
Table 2.
| Family | Domain signature | Number of proteins with domain signature | ||||
|---|---|---|---|---|---|---|
| D. discoideum | C. elegans | D. melanogaster | D. rerio | H. sapiens | ||
| JIP1-like | SH3-PTB | 0 | 1 | 1 | 2 | 2 |
| Paxillin-like | LIM-LIM-LIM-LIM | 7 | 4 | 6 | 15 | 11 |
| PSD95-like | PDZ-PDZ-PDZ-SH3-GuKc | 0 | 2 | 3 | 4 | 9 |
| InaD-like | PDZ-PDZ-PDZ-PDZ-PDZ | 0 | 2 | 2 | 6 | 7 |
Data were obtained by carrying out a composition search with the listed domains in the SMART database (http://smart.embl-heidelberg.de/smart). (Data were manually filtered to exclude duplicates and different isoforms transcribed from the same ORF.)
Abbreviations: D. discoideum, slime mould; C. elegans, roundworm; D. melanogaster, fruit fly; D rerio, zebrafish; H. sapiens, human.
Abbreviations: GuKc, guanylate kinase-like domain; LIM, Lin-1; Isl-1, Mec-3 domain; PDZ, PSD95, DlgA, Zo-1 domain; PTB, phosphotyrosine-binding domain; SH3: Src kinase homology 3 domain.
In contrast to scaffolds, tyrosine kinases share both origin and function. It seems that tyrosine phosphorylation was invented only once during evolution and the explosion of this new system (such as tyrosine kinases, phosphatases and Src-homology 2 [SH2] domains) somewhat coincides with the emergence of multicellular organisms [16,17]. We can say this with high confidence because all tyrosine kinases known today possess common signature motifs – some conserved sequence features essential for activity – that can be easily identified in different genomes.
Apart from the presence of some ubiquitous and frequently occurring PPI domains (see later), the sequence of putative scaffolds does not contain common signature motifs similar to the ones found in enzymes. (Common domain compositions, however, do occur among some scaffolds, but prediction of function based on these domain signatures is not possible.) Even MAPK scaffolds that organize the highly conserved set of MAPK cascade components into functional modules do not possess any sequence similarity, although the kinases that they bind to share high (50–80%) degree of sequence similarity from yeast to human. It seems that currently known MAPK protein scaffolds are unrelated (e.g. Ste5, KSR, JIP, β-arrestin) [18]. This suggests that scaffolds might have emerged several times independently as ad hoc solutions during the evolution of signalling systems.
So far, only a few scaffolds have been identified in primitive, unicellular eukaryotes. As for higher plants, the number of identified scaffolds or scaffold families is also very low [19]. S. cerevisiae does possess a classical scaffold protein, Ste5, which is however found only in other closely related yeast species and has no other homologue in higher eukaryotes. In mammals, however, there are almost a dozen of MAPK scaffolds and some of them can be traced back to Caenorhabditis elegans [20]. The same can be observed for multi-PDZ domain proteins, which have an important role in epithelial polarisation and synapse formation. These proteins are abundant in animals (especially in mammals) but they are rarely found in low eukaryotes, fungi or in plants [21]. Another example for scaffold diversification can be found among members of the PSD95-like scaffold family (nine human paralogs, compared with the two genes found in C. elegans). It seems that as the number of signalling molecules increases from lower to higher complexity organisms, the number and complexity of known scaffold proteins also increases, as expected.
In contrast to the MAPK system, it seems that some functional components of the synapse in the nervous system emerged after the appearance of scaffold proteins. In mammalian glutamatergic synapses, a rich protein mesh lies under the postsynaptic membrane, referred to as postsynaptic density (PSD). The PSD in mammals consists of a plethora of signalling enzymes, ion-channels, receptors and cell adhesion proteins physically linked by numerous scaffold proteins (e.g. PSD95, SAP97, GRIP) [22]. The first vestiges of this system developed early in metazoan history: the demosponge Amphimedon queenslandica possesses a nearly complete set of would-be synaptic scaffolds – localized to the larval flask cells (putative sensory cells), where they form a protein mesh underneath the apical membrane [23,24]. The signalling function of these complexes, however, is less clear because these organisms completely lack the family of ionotropic glutamate receptors (e.g. NMDA, AMPA and kainate types), which are crucial signalling components in PSDs in higher organisms. Nematostella vectensis (starlet sea anemone) seems to be the first organism in which all major PSD components are present – including ion channels and scaffolding components [23]. These serve as building blocks for one of the most primitive known neuronal cells and circuits.
As scaffolds enable context-dependent fine-tuning of pre-existing signalling pathways or creation of new pathways from a combination of pre-existing components, it is possible that their increasing usage might have had a role in the evolution of multi-cellular organisms. Unfortunately, comparative genomics, where protein sequences derived from sequenced genomes are compared, has a very low chance to identify scaffolding interactions; even inferring binary connections between annotated gene products is difficult [25]. Nevertheless, we are currently learning a lot from these whole-genome sequencing studies because they are being extended to more ‘exotic’ organisms that are close to major biological transitions (such as emergence of multicellularity or the nervous system). As it is not possible to infer PPIs from genome sequence data, a more complete picture about signalling network evolution could be obtained if comparative genomics is combined with ‘comparative interactomics’ [26]. Fortunately, open reading frames (ORFs) derived from exotic organisms become readily available as the genomes of these organisms become sequenced. When the emerging interactomes of S. cerevisiae [27], Drosophila [28] or mammals [29] are compared with more exotic interactomes, we could possible gain more insight not only into the compositions of networks but also, more importantly, into how the connections within them have changed.
By looking at the examples of known scaffold proteins (e.g. MAPK scaffolds), it seems that they were invented several times during the evolution of certain signalling circuits. Because scaffolds are a composition of PPI interaction elements, whereas enzyme activity requires strict stereo-chemical alignment of residues for catalysis, intuitively it seems easier to construct a new scaffold than a new enzyme.
Engineering of signalling networks
Both scaffolds and their catalytically active signalling partners show highly modular architecture. Signalling scaffolds operate as regulatory elements in signalling networks. By contrast, binding partners of scaffolds with catalytic activity can be regarded as effectors performing signalling-related modifications (e.g. changes in phosphorylation, GTP–GDP binding states or in seconder messenger levels). Similarly, binding partners of scaffolds can often be divided up into catalytic (effector) and regulatory domains [30]. For example, protein kinases contain a compact kinase domain that phosphorylates downstream targets and guanine nucleotide exchange factors (GEFs) contain a Dbl homology (DH) domain that facilitates the GDP–GTP exchange of small GTPases [31,32]. For many GEFs and kinases, other regulatory domains keep their catalytic domain in check. This is often achieved through intramolecular PPIs by using dedicated PPI domains or peptides containing consensus linear motifs [33]. These interactions are broken upon stimulation and the respective catalytic domains then become active. Separation of effector and regulatory functions through modular design of controlling elements is also widespread in the regulatory system controlling gene expression [34].
The highly modular nature of network components has indeed facilitated the design of artificial gene-expression networks. When these artificial networks are constructed in the cell they are able to perform natural-like functions such as bistable switching or oscillation [35]. These studies were initiated by a new breed of biologist who aspires to understand natural systems through emphasizing construction rather than deduction [36,37].
It is likely that modular protein architecture is instrumental in the plasticity of systems involved in cellular control compared with metabolic networks where component architecture is far less modular and more integrated [30]. On an evolutionary scale, recombination of catalytic and regulatory domains could happen through exon shuffling, and it is probable that modular architecture is more conducive for rapid emergence of novel types of regulatory mechanisms [38]. Although it is very difficult to test this argument experimentally, it is interesting to note that organism complexity seems to correlate more with the number and diversity of regulatory domains, and not with the number of integrated components (such as catalytic domains) comprising a network [5].
Because signalling components are modular and many of the PPIs involved in the major signalling pathways are well-characterized, making a synthetic connection sometimes is very simple [39]. For example, researchers have successfully redirected an epidermal-growth-factor-stimulated proliferation signal to an apoptotic caspase pathway through a chimeric adaptor protein [40]. By using a similar approach, it was straightforward to establish new input–output relationships for yeast MAPK signalling pathways. This was achieved by constructing a chimera from scaffolds specifically involved in the mating or in the osmoregulatory pathways (Ste5 and Pbs2, respectively). Yeast cells equipped with this ‘diverter’ scaffold were able to generate mating-pathway-specific responses when stimulated with high salt, demonstrating that artificial scaffolds when introduced into the cell can be very powerful and simple tools to change fundamental properties of signalling pathways [41].
Artificial signalling molecules can be readily created if the catalytic activity of kinases, phosphatases or GEFs is controlled by non-natural protein–protein or protein– ligand interactions. When these proteins are combined into a cascade, similar to how their natural counterparts are organized into higher-order structures, these cascades can display ultrasensitivity. This has been elegantly demonstrated for a synthetic GEF cascade where stimulation by an artificial input (e.g. the addition of the cAMP inducer forskolin to cells) changes cellular morphology and efficiently rewires pathways underlying the formation of filopodia or lamellopodia [42].
Apart from being able to make new connections, putting signalling enzymes under the control of non-natural regulation or linking them into artificial cascades, it is also important to demonstrate that we can control signalling pathway dynamics. This is very important because distinct pathway dynamics are crucial for determining physiological output. Because scaffolds have a role as signal-processing hubs, they are efficient targets of feedback loops that optimize signalling amplitude and timing. In the well-studied mating pathway in the budding yeast, the scaffold Ste5 allosterically activates one of its binding partners, initiating a negative feedback loop that regulates pathway output [15]. Furthermore, Ste5 has been recently used as a platform to reshape the output of the yeast mating MAPK pathway. Synthetic positive- and negative-feedback loops were constructed by dynamically regulating the recruitment of pathway modulators to an artificial binding site on Ste5. Interestingly, these engineered circuits displayed an ultrasensitive dose response, accelerated or delayed response times and tunable adaptation [43] (Figure 4).
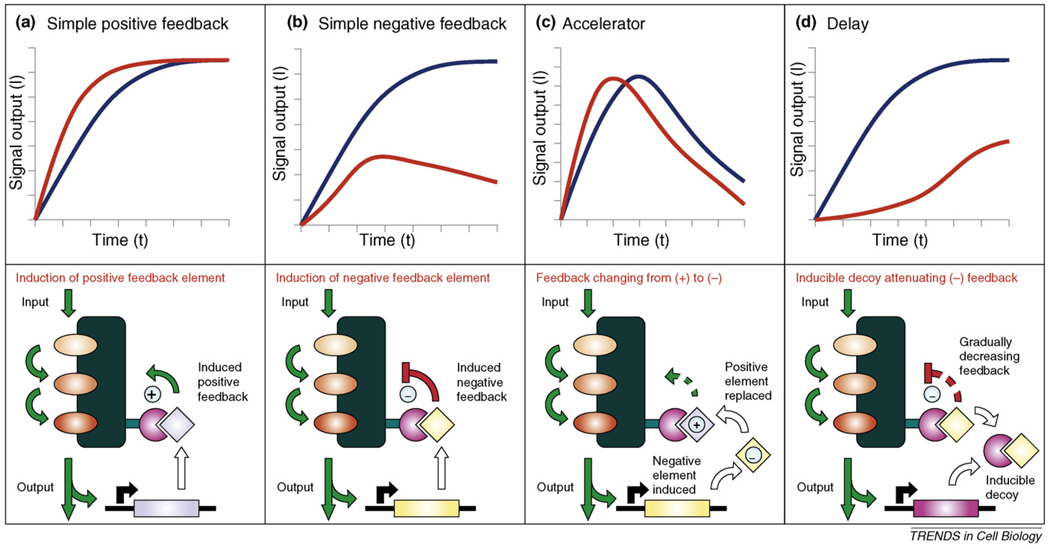
Figure 4.
Scaffold-based network architectures with diverse pathway dynamics. Top panels show experimental kinetic profiles for circuits with wild-type Ste5 (blue) and with a synthetic Ste5 scaffold (red). Lower panels schematically depict underlying circuit architectures. The Ste5 scaffold protein was engineered to include an additional binding site (e.g. by addition of a leuzine zipper, shown in magenta) to enable it to bind other proteins containing a complementary protein interaction element (e.g. another leuzine zipper). By recruiting various effectors or decoys to this scaffolded MAP-kinase module it was possible to change yeast mating pathway kinetics at will [43]. (The original components of this MAPK module: Ste11, Ste7 and Fus3 kinases are coloured in different shades of orange.) (a) Expression of a positive activator (shown in light blue) through a promoter that is responsive to pathway flux creates a simple positive feedback loop, providing more rapid responses compared with wild-type cells. (b) If pathway flux triggers the expression of a negative regulator (yellow), then a simple negative-feedback loop will result. This creates a pulse-like response upon stimulation, unlike wild-type yeast that exhibits a continuous, saturated response. (c) With a careful combination of positive- and negative-feedback elements almost any desired dynamic profile can be realized. A good example is the combination of a constitutively expressed positive regulatory element with a signal-inducible negative element. The two proteins naturally compete with each other for the same binding site, and the inhibitor element will gradually displace the enhancer element as the signalling pathway becomes activated by sustained stimulus. As expected, the system provides a pulse-like output in time, but the peak is shifted towards earlier time points when compared with a simple negative-feedback loop (containing only the inducible inhibitor). Thus, the system behaves as an accelerator. (d) A more complex way to change signalling characteristics can be achieved through the use of decoys – proteins only consisting of binding sites – that, for example, can disrupt association of a constitutively expressed negative regulatory factor with the scaffold. When compared with wild-type cells, a pronounced delay of signal propagation will become apparent. The decoy gradually diminishes the inhibition of signalling as the pathway becomes activated by a sustained stimulus.
Currently, synthetic approaches are making their mark in signal transduction research (Box 4). It is emerging that artificial protein scaffolds can be used as platforms for the design of signalling circuits with custom functions. Apart from testing our understanding on signalling circuit design principles, these studies might be useful to reprogram cellular behaviour. Many cellular processes, movement and gradient sensing for example, require rapid and spatially precise responses. This is difficult to achieve by using regulatory circuits based on gene expression networks only. Thus, protein-based networks using modified enzymes and/or scaffolds are more geared to control rapid dynamic processes.
Box 4. Synthetic biology ‘toolkit’ for designing custom signalling proteins and circuits.
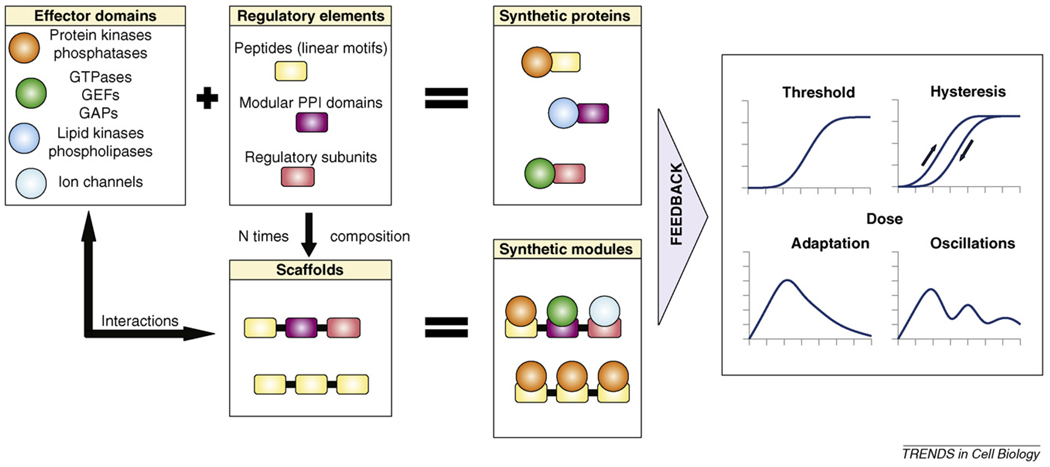
Synthetic proteins that possess novel means of regulation can be constructed by putting the activity of signalling effector domains under the control of heterologous regulatory elements (Figure I, upper panels). This is possible because the activity of many natural signalling proteins is controlled through modular allostery [76,77]. Scaffolds are composed of several regulatory elements through which they interact with effector domains. They are useful to endow original signalling proteins with new connections if their interaction element composition is carefully constructed (Figure I, lower panels). In turn, synthetic proteins and/or synthetic modules might provide opportunities for positive or negative feedback, giving rise to diverse non-linear signalling phenotypes [78]. Similar to their natural counterparts, these synthetic signalling networks might be capable of reprogramming cellular behaviour [42].
Signalling proteins can be divided up into effector domains and regulatory regions. Effector domains in signalling networks come in many forms: they are responsible for catalysis (such as carrying out protein or lipid phosphorylation or dephosphorylation, or GTP hydrolysis), or they influence secondary messenger levels in the cell (e.g. lipid kinases, phosphatases or ion-channels). Regulatory elements are also diverse: (i) linear motifs are peptides with a consensus recognition sequence towards an effector domain; (ii) modular PPI domains (such as SH2, SH3, PDZ, PTB, PH, C1, C2 domains, etc.) are compact, dedicated interaction elements; and (iii) regulatory subunits are full-length proteins (from protein phosphatases, lipid kinases or ion channels, for example) that target effector domains to their natural substrates or enable stimulus-dependent activity regulation [30,79].
Concluding remarks
We have learned a great deal about scaffolds in the last decade, but undoubtedly many important questions remain unanswered (Box 5). These could be better addressed in the near future because systematic PPI studies will probably identify many new scaffolds. So far, it has become widely accepted that scaffolds facilitate interactions between signalling enzymes through enforced proximity. This is fundamental to scaffold function. It is also becoming apparent that many known scaffolds can also do a lot more. Currently, we know of scaffolds that demonstrate all four scaffold-related mechanisms listed in this review (e.g. Ste5) and there are others for which only some of these mechanisms have been demonstrated. (This might be due to lack of experimental characterization rather than being an inherent property of the scaffold in question; for AKAPs, conformational fine-tuning and for INAD, combinatory use of elements have not been demonstrated yet, for example.) The highly heterogenous appearance of classical scaffold proteins in sequence and architecture suggests that they might have appeared independently several times during evolution. Because they can rewire connections between existing pathway components, diversify module compositions and influence signalling properties in space and time, it is tentative to speculate that scaffolds have contributed greatly to enhancing signalling complexity in an organism. In line with their pivotal role, application of modified natural scaffolds as recruitment elements for modified signalling components might be a tantalising strategy to engineer signalling circuits.
Box 5. Outstanding questions.
Do scaffold proteins possess any unique features in terms of their surrounding network topology?
Can we identify signalling scaffolds based on interactome data?
Do scaffolds that are currently regarded as passive tethers also perform dynamic regulation and conformational fine tuning?
Can any arbitrary dynamic behaviour (such as oscillation) be realized by using scaffolds? Are there any natural examples for these?
Have scaffolds had a pivotal role in the evolution of multicellularity or in the emergence of complex nervous systems?
How abundant are scaffold proteins in less studied organisms, for example in primitive eukaryotes or plants?
How did the appearance of a scaffold in a particular pathway change signalling and what costs and benefits did it confer?
Will scaffold proteins be useful to make engineered signalling circuits for further applications in biotechnology and medicine?
Figure I.
Enrichment of signalling molecules around a scaffold.
Figures I.
General mechanisms for the evolution of scaffolds.
Figures I.
A synthetic biology ‘toolkit’.
Acknowledgements
We apologize to colleagues whose work we could not cite owing to space constraints. This work was supported by a Wellcome Trust International Senior Fellowship (A.R.) and by a Marie Curie International Reintegration Grant (A.R.). The authors are grateful for other members of the Reményi laboratory for their support. We thank Mario Albrecht, István Molnár and Jesse Zalatan for critically reading the manuscript.
References
- 1.Choi KY, et al. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 2.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrrell JE, Jr, Cimprich KA. Enforced proximity in the function of a famous scaffold. Mol. Cell. 2003;11:289–291. doi: 10.1016/s1097-2765(03)00055-8. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya RP, et al. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signalling circuits. Annu. Rev. Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 6.Levchenko A, et al. Scaffold proteins might biphasically affect the levels of mitogen-activated protein kinase signalling and reduce its threshold properties. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smock RG, Lila MG. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawson T. Dynamic control of signalling by modular adaptor proteins. Curr. Opin. Cell Biol. 2007;19:112–116. doi: 10.1016/j.ceb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Dodge-Kafka KL, et al. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith FD, et al. The where’s and when’s of kinase anchoring. Trends Biochem. Sci. 2006;31:316–323. doi: 10.1016/j.tibs.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Popescu DC, et al. Scaffolding protein INAD regulates deactivation of vision by promoting phosphorylation of transient receptor potential by eye protein kinase C in Drosophila. J. Neurosci. 2006;26:8570–8577. doi: 10.1523/JNEUROSCI.1478-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montell C. Dynamic regulation of the INAD signalling scaffold becomes crystal clear. Cell. 2007;131:19–21. doi: 10.1016/j.cell.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Good M, et al. The Ste5 scaffold directs mating signalling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 2009;136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodolenko BN. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharyya RP, et al. The Ste5 scaffold allosterically modulates signalling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 16.Goldberg JM, et al. The dictyostelium kinome—analysis of the protein kinases from a simple model organism. PLoS Genet. 2006;2:e38. doi: 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pincus D, et al. Evolution of the phospho-tyrosine signalling machinery in premetazoan lineages. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cell. Signal. 2009;21:462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert HS, et al. BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J. 2009;58:109–121. doi: 10.1111/j.1365-313X.2008.03764.x. [DOI] [PubMed] [Google Scholar]
- 20.Morrison DK. KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- 21.Nourry C, et al. PDZ domain proteins: plug and play. Sci. Signal. 2003;179(re7):1–12. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- 22.Boeckers TM. The postsynaptic density. Cell Tissue Res. 2006;326:409–422. doi: 10.1007/s00441-006-0274-5. [DOI] [PubMed] [Google Scholar]
- 23.Sakarya O, et al. A post-synaptic scaffold at the origin of the animal kingdom. PLoS One. 2007;2:e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards GS, et al. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 25.Mika S, Rost B. Protein-protein interactions more conserved within species than across species. PLOS Comput. Biol. 2006;2:e79. doi: 10.1371/journal.pcbi.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R, et al. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12763–12768. doi: 10.1073/pnas.0806627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J, et al. DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figeys D. Mapping the human protein interactome. Cell Res. 2008;18:716–724. doi: 10.1038/cr.2008.72. [DOI] [PubMed] [Google Scholar]
- 30.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 31.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman GR, Cerione RA. Signalling to the Rho GTPases: networking with the DH domain. FEBS Lett. 2002;513:85–91. doi: 10.1016/s0014-5793(01)03310-5. [DOI] [PubMed] [Google Scholar]
- 33.Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu. Rev. Cell Dev. Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 34.Wagner GP, Lynch VJ. The gene regulatory logic of transcription factor evolution. Trends Ecol. Evol. 2008;23:377–385. doi: 10.1016/j.tree.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Kaern M, et al. The engineering of gene regulatory networks. Annu. Rev. Biomed. Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- 36.Drubin DA, et al. Designing biological systems. Genes Dev. 2007;21:242–254. doi: 10.1101/gad.1507207. [DOI] [PubMed] [Google Scholar]
- 37.Bhalerao KD, et al. Synthetic gene networks: the next wave in biotechnology? Trends Biotechnol. doi: 10.1016/j.tibtech.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Babushok DV, et al. Current topics in genome evolution: molecular mechanisms of new gene formation. Cell. Mol. Life Sci. 2007;64:542–554. doi: 10.1007/s00018-006-6453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pryciak PM. Designing new cellular signalling pathways. Chem. Biol. 2009;16:249–254. doi: 10.1016/j.chembiol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard PL, et al. Redirecting tyrosine kinase signalling to an apoptotic caspase pathway through chimeric adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11267–11272. doi: 10.1073/pnas.1934711100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SH, et al. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 42.Yeh BJ, et al. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature. 2007;447:596–600. doi: 10.1038/nature05851. [DOI] [PubMed] [Google Scholar]
- 43.Bashor CJ, et al. Usingengineeredscaffoldinteractions toreshape MAP kinase pathway signalling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 44.Whitmarsh AJ. The JIP family of MAPK scaffold proteins. Biochem. Soc. Trans. 2006;34:828–832. doi: 10.1042/BST0340828. [DOI] [PubMed] [Google Scholar]
- 45.McDonald PH, et al. β-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 46.Elion EA. The Ste5p scaffold. J. Cell Sci. 2001;114:3967–3978. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- 47.Deakin NO, Turner CE. Paxillin comes of age. J. Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarmay G, et al. The multiple function of Grb2 associated binder (Gab) adaptor/scaffolding protein in immune cell signalling. Immunol. Lett. 2006;104:76–82. doi: 10.1016/j.imlet.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 49.Funke L, et al. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu. Rev. Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 50.Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol. 2007;8:206. doi: 10.1186/gb-2007-8-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fagni L, et al. Homer as both a scaffold and transduction molecule. Sci. SKTE. 2002;2002:RE8. doi: 10.1126/stke.2002.137.re8. [DOI] [PubMed] [Google Scholar]
- 52.Dodge-Kafka KL, Kapiloff MS. The mAKAP signalling complex: integration of cAMP, calcium, and MAP kinase signalling pathways. Eur. J. Cell Biol. 2006;85:593–602. doi: 10.1016/j.ejcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Wang H-Y, et al. G-Protein-coupled receptor-associated A-kinase anchoring proteins: AKAP79 and AKAP250 (gravin) Eur. J. Cell Biol. 2006;85:643–650. doi: 10.1016/j.ejcb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 54.McCahill A, et al. The RACK1 scaffold protein: a dynamic cog in the cell response mechanisms. Mol. Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 55.Cusick ME, et al. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 56.Yang W, et al. Proteomic approaches to the analysis of multiprotein signalling complexes. Proteomics. 2008;8:832–851. doi: 10.1002/pmic.200700650. [DOI] [PubMed] [Google Scholar]
- 57.Suter B, et al. Two-hybrid technologies in proteomics research. Curr. Opin. Biotechnol. 2008;19:316–323. doi: 10.1016/j.copbio.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Collins MO, Choudhary JS. Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr. Opin. Biotechnol. 2008;19:324–330. doi: 10.1016/j.copbio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Zhu X, et al. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 60.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 61.Wahhout AJ, et al. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 2002;12:1952–1958. doi: 10.1016/s0960-9822(02)01279-4. [DOI] [PubMed] [Google Scholar]
- 62.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 63.Jensen LJ, et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keshava Prasad TS, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bader GD, et al. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chatr-aryamontri A, et al. MINT and IntAct contribute to the Second BioCreative challange: serving the text-mining community with high quality molecular interaction data. Genome Biol. 2008;9:S5. doi: 10.1186/gb-2008-9-s2-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 69.Jorgensen P, et al. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 70.Fischer H, et al. Average protein density is a molecular-weight-dependent function. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maeder CI, et al. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 72.Slaughter BD, et al. Mapping dynamic protein interactions in MAP kinase signalling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20320–20325. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Claperon A, Therrien M. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene. 2007;26:3143–3158. doi: 10.1038/sj.onc.1210408. [DOI] [PubMed] [Google Scholar]
- 74.Fritschy JM, et al. Gephyrin: where do we stand, where do we go? Trends Neurosci. 2008;31:257–264. doi: 10.1016/j.tins.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 75.Assemat E, et al. Polarity complex proteins. Biochim. Biophys. Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 76.Lim WA. The modular logic of signalling proteins: building allosteric switches from simple binding domains. Curr. Opin. Struct. Biol. 2002;12:61–68. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 77.Tsai CJ, et al. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol. Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tyson JJ, et al. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signalling pathways in the cell. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 79.Neduva V, et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]