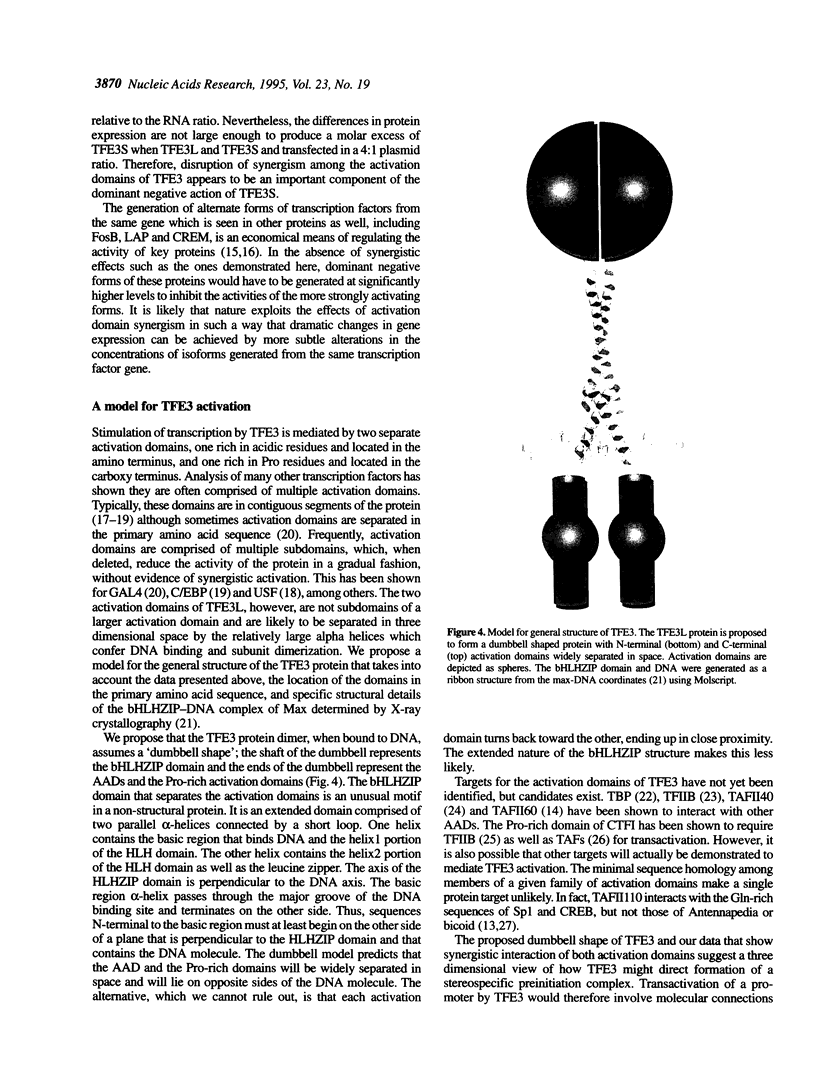
Abstract
TFE3 is a basic-helix-loop-helix-zipper (bHLHZIP) domain-containing protein that binds mu E3 sites in regulatory elements in the immunoglobulin heavy chain gene. The protein is a transcriptional activator that is expressed in vivo as two alternately spliced isoforms with different activating properties: TFE3L contains an N-terminal acidic activation domain; TFE3S lacks this activation domain and is a dominant negative inhibitor of TFE3L. We show that TFE3L and TFE3S contain a second, C-terminal activation domain rich in proline residues. This pro-rich activation domain has activity in a Gal4 fusion assay comparable to the N-terminal acidic activation domain present in TFE3L. The TFE3 pro-rich activation domain contains regions of strong homology with the related proteins microphthalmia and TFEB, suggesting that these regions are important for function. Using two different assays, we show that the N- and C-terminal activation domains of TFE3 act synergistically. This synergism explains in part the ability of TFE3S to act as a dominant negative. Our domain analysis of TFE3 is incorporated into a general structural model for the TFE3 protein that predicts that the activation domains of TFE3 will be widely separated in space.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artandi S. E., Cooper C., Shrivastava A., Calame K. The basic helix-loop-helix-zipper domain of TFE3 mediates enhancer-promoter interaction. Mol Cell Biol. 1994 Dec;14(12):7704–7716. doi: 10.1128/mcb.14.12.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Carr C. S., Sharp P. A. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990 Aug;10(8):4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Descombes P., Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991 Nov 1;67(3):569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994 Jan 1;13(1):180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Carr C. S., Parent L. A., Sharp P. A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991 Dec;5(12A):2342–2352. doi: 10.1101/gad.5.12a.2342. [DOI] [PubMed] [Google Scholar]
- Foulkes N. S., Sassone-Corsi P. More is better: activators and repressors from the same gene. Cell. 1992 Feb 7;68(3):411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- Friedman A. D., McKnight S. L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990 Aug;4(8):1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- Gill G., Pascal E., Tseng Z. H., Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Hoey T., Thut C. J., Admon A., Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993 Nov 5;75(3):519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Hodgkinson C. A., Moore K. J., Nakayama A., Steingrímsson E., Copeland N. G., Jenkins N. A., Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993 Jul 30;74(2):395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R. O., Gill G., Chen J. L., Dynlacht B. D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993 Jan 29;72(2):247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Kim T. K., Roeder R. G. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum B. J., Pognonec P., Roeder R. G. Definition of the transcriptional activation domain of recombinant 43-kilodalton USF. Mol Cell Biol. 1992 Nov;12(11):5094–5101. doi: 10.1128/mcb.12.11.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm J. D., Rould M. A., Aurora R., Herr W., Pabo C. O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994 Apr 8;77(1):21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Roman C., Cohn L., Calame K. A dominant negative form of transcription activator mTFE3 created by differential splicing. Science. 1991 Oct 4;254(5028):94–97. doi: 10.1126/science.1840705. [DOI] [PubMed] [Google Scholar]
- Roman C., Matera A. G., Cooper C., Artandi S., Blain S., Ward D. C., Calame K. mTFE3, an X-linked transcriptional activator containing basic helix-loop-helix and zipper domains, utilizes the zipper to stabilize both DNA binding and multimerization. Mol Cell Biol. 1992 Feb;12(2):817–827. doi: 10.1128/mcb.12.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989 Sep 25;17(18):7539–7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Clouston W. M., Herr W. The Oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicates of themselves to activate transcription. Mol Cell Biol. 1994 Sep;14(9):6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Thut C. J., Chen J. L., Klemm R., Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995 Jan 6;267(5194):100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]




