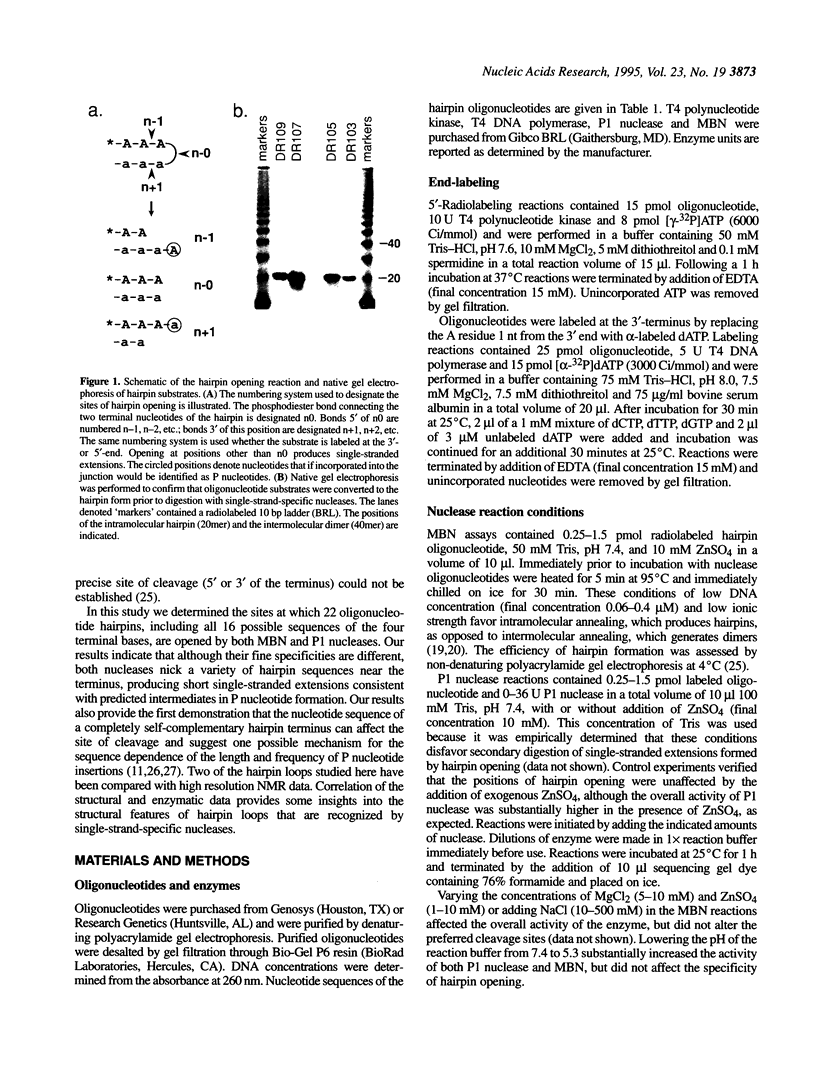
Abstract
DNA molecules with covalently sealed (hairpin) ends are probable intermediates in V(D)J recombination. According to current models hairpin ends are opened to produce short single-stranded extensions that are thought to be precursors of a particular type of extra nucleotides, termed P nucleotides, which are frequently present at recombination junctions. Nothing is known about the activities responsible for hairpin opening. We have used two single-strand-specific nucleases to explore the effects of loop sequence on the hairpin opening reaction. Here we show that a variety of hairpin ends are opened by P1 nuclease and mung bean nuclease (MBN) to leave short, 1-2 nt single-stranded extensions. Analysis of 22 different hairpin sequences demonstrates that the terminal 4 nt of the hairpin loop strongly influence the sites of cleavage. Correlation of the nuclease digestion patterns with structural (NMR) data for some of the hairpin loops studied here provides new insights into the structural features recognized by these enzymes.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumann U., Frank R., Blöcker H. Conformational analysis of hairpin oligodeoxyribonucleotides by a single-strand-specific nuclease. Eur J Biochem. 1986 Dec 1;161(2):409–413. doi: 10.1111/j.1432-1033.1986.tb10460.x. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Blunt T., Finnie N. J., Taccioli G. E., Smith G. C., Demengeot J., Gottlieb T. M., Mizuta R., Varghese A. J., Alt F. W., Jeggo P. A. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995 Mar 10;80(5):813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- Bosma G. C., Custer R. P., Bosma M. J. A severe combined immunodeficiency mutation in the mouse. Nature. 1983 Feb 10;301(5900):527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- Boubnov N. V., Wills Z. P., Weaver D. T. V(D)J recombination coding junction formation without DNA homology: processing of coding termini. Mol Cell Biol. 1993 Nov;13(11):6957–6968. doi: 10.1128/mcb.13.11.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box H. C., Budzinski E. E., Evans M. S., French J. B., Maccubbin A. E. The differential lysis of phosphoester bonds by nuclease P1. Biochim Biophys Acta. 1993 Feb 13;1161(2-3):291–294. doi: 10.1016/0167-4838(93)90227-i. [DOI] [PubMed] [Google Scholar]
- Davison A., Leach D. R. Two-base DNA hairpin-loop structures in vivo. Nucleic Acids Res. 1994 Oct 25;22(21):4361–4363. doi: 10.1093/nar/22.21.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Erie D. A., Suri A. K., Breslauer K. J., Jones R. A., Olson W. K. Theoretical predictions of DNA hairpin loop conformations: correlations with thermodynamic and spectroscopic data. Biochemistry. 1993 Jan 19;32(2):436–454. doi: 10.1021/bi00053a008. [DOI] [PubMed] [Google Scholar]
- Esteban J. A., Salas M., Blanco L. Activation of S1 nuclease at neutral pH. Nucleic Acids Res. 1992 Sep 25;20(18):4932–4932. doi: 10.1093/nar/20.18.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney A. J., Victor K. D., Vu K., Nadel B., Chukwuocha R. U. Influence of the V(D)J recombination mechanism on the formation of the primary T and B cell repertoires. Semin Immunol. 1994 Jun;6(3):155–163. doi: 10.1006/smim.1994.1021. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., Lundberg P., Vogel H. J., van de Sande J. H. Perturbation of DNA hairpins containing the EcoRI recognition site by hairpin loops of varying size and composition: physical (NMR and UV) and enzymatic (EcoRI) studies. Nucleic Acids Res. 1990 Mar 25;18(6):1489–1498. doi: 10.1093/nar/18.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G., García A. E., Hiriyanna K. T. Sampling of the conformations of the d(CGCTGCGGC) hairpin in solution by two-dimensional nuclear magnetic resonance and theoretical methods. Biochemistry. 1993 Jan 26;32(3):948–960. doi: 10.1021/bi00054a028. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Haasnoot C. A., de Bruin S. H., Joordens J. J., van der Marel G. A., van Boom J. H. Hairpin formation in synthetic oligonucleotides. Biochimie. 1985 Jul-Aug;67(7-8):685–695. doi: 10.1016/s0300-9084(85)80156-5. [DOI] [PubMed] [Google Scholar]
- Hirao I., Kawai G., Yoshizawa S., Nishimura Y., Ishido Y., Watanabe K., Miura K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994 Feb 25;22(4):576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard F. B., Chen C. Q., Ross P. D., Miles H. T. Hairpin formation in the self-complementary dodecamer d-GGTACGCGTACC and derivatives containing GA and IA mispairs. Biochemistry. 1991 Jan 22;30(3):779–782. doi: 10.1021/bi00217a030. [DOI] [PubMed] [Google Scholar]
- Johnson P. H., Laskowski M., Sr Mung bean nuclease I. II. Resistance of double stranded deoxyribonucleic acid and susceptibility of regions rich in adenosine and thymidine to enzymatic hydrolysis. J Biol Chem. 1970 Feb 25;245(4):891–898. [PubMed] [Google Scholar]
- Kallick D. A., Wemmer D. E. 1H NMR of 5'CGCGTATATACGCG3', a duplex and a four-membered loop. Nucleic Acids Res. 1991 Nov 11;19(21):6041–6046. doi: 10.1093/nar/19.21.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner C. U., Patil C. K., Evans J. W., Cuomo C. A., Fried L. M., Carter T., Oettinger M. A., Brown J. M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995 Feb 24;267(5201):1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- Kroeker W. D., Kowalski D., Laskowski M., Sr Mung bean nuclease I. Terminally directed hydrolysis of native DNA. Biochemistry. 1976 Oct 5;15(20):4463–4467. doi: 10.1021/bi00665a020. [DOI] [PubMed] [Google Scholar]
- Lafaille J. J., DeCloux A., Bonneville M., Takagaki Y., Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989 Dec 1;59(5):859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- Lewis S. M. P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc Natl Acad Sci U S A. 1994 Feb 15;91(4):1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- Lieber M. R. Site-specific recombination in the immune system. FASEB J. 1991 Nov;5(14):2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- Mazumder A., Engelman A., Craigie R., Fesen M., Pommier Y. Intermolecular disintegration and intramolecular strand transfer activities of wild-type and mutant HIV-1 integrase. Nucleic Acids Res. 1994 Mar 25;22(6):1037–1043. doi: 10.1093/nar/22.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack W. T., Tjoelker L. W., Carlson L. M., Petryniak B., Barth C. F., Humphries E. H., Thompson C. B. Chicken IgL gene rearrangement involves deletion of a circular episome and addition of single nonrandom nucleotides to both coding segments. Cell. 1989 Mar 10;56(5):785–791. doi: 10.1016/0092-8674(89)90683-1. [DOI] [PubMed] [Google Scholar]
- Meier J. T., Lewis S. M. P nucleotides in V(D)J recombination: a fine-structure analysis. Mol Cell Biol. 1993 Feb;13(2):1078–1092. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Leon L., Huang L. C., Umlauf S. W., Cox M. M., Inman R. B. Holliday intermediates and reaction by-products in FLP protein-promoted site-specific recombination. Mol Cell Biol. 1988 Sep;8(9):3784–3796. doi: 10.1128/mcb.8.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989 Nov;8(11):3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. An NMR study of polymorphous behaviour of the mismatched DNA octamer d(m5C-G-m5C-G-A-G-m5C-G) in solution. The B-duplex and hairpin forms. Eur J Biochem. 1987 Dec 30;170(1-2):225–239. doi: 10.1111/j.1432-1033.1987.tb13690.x. [DOI] [PubMed] [Google Scholar]
- Orbons L. P., van der Marel G. A., van Boom J. H., Altona C. Hairpin and duplex formation of the DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. An NMR study. Nucleic Acids Res. 1986 May 27;14(10):4187–4196. doi: 10.1093/nar/14.10.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergola F., Zdzienicka M. Z., Lieber M. R. V(D)J recombination in mammalian cell mutants defective in DNA double-strand break repair. Mol Cell Biol. 1993 Jun;13(6):3464–3471. doi: 10.1128/mcb.13.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. M., de Vroom E., van der Marel G. A., van Boom J. H., Koning T. M., Kaptein R., Altona C. Hairpin structures in DNA containing arabinofuranosylcytosine. A combination of nuclear magnetic resonance and molecular dynamics. Biochemistry. 1990 Jan 23;29(3):788–799. doi: 10.1021/bi00455a029. [DOI] [PubMed] [Google Scholar]
- Reddy M. K., Bauer W. R. Activation of the vaccinia virus nicking-joining enzyme by trypsinization. J Biol Chem. 1989 Jan 5;264(1):443–449. [PubMed] [Google Scholar]
- Roth D. B., Lindahl T., Gellert M. Repair and recombination. How to make ends meet. Curr Biol. 1995 May 1;5(5):496–499. doi: 10.1016/s0960-9822(95)00101-1. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Menetski J. P., Nakajima P. B., Bosma M. J., Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992 Sep 18;70(6):983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- Taccioli G. E., Gottlieb T. M., Blunt T., Priestley A., Demengeot J., Mizuta R., Lehmann A. R., Alt F. W., Jackson S. P., Jeggo P. A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994 Sep 2;265(5177):1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- Taccioli G. E., Rathbun G., Oltz E., Stamato T., Jeggo P. A., Alt F. W. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993 Apr 9;260(5105):207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- Van de Ven F. J., Hilbers C. W. Nucleic acids and nuclear magnetic resonance. Eur J Biochem. 1988 Dec 1;178(1):1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- Weaver D. T. V(D)J recombination and double-strand break repair. Adv Immunol. 1995;58:29–85. doi: 10.1016/s0065-2776(08)60619-7. [DOI] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G., van Boom J. DNA hairpin loops in solution. Correlation between primary structure, thermostability and reactivity with single-strand-specific nuclease from mung bean. Nucleic Acids Res. 1991 Apr 11;19(7):1505–1511. doi: 10.1093/nar/19.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Roth D. B. Characterization of coding ends in thymocytes of scid mice: implications for the mechanism of V(D)J recombination. Immunity. 1995 Jan;2(1):101–112. doi: 10.1016/1074-7613(95)90082-9. [DOI] [PubMed] [Google Scholar]
- van Gent D. C., McBlane J. F., Ramsden D. A., Sadofsky M. J., Hesse J. E., Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995 Jun 16;81(6):925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- van der Ende A., Langeveld S. A., Teertstra R., van Arkel G. A., Weisbeek P. J. Enzymatic properties of the bacteriophage phi X174 A protein on superhelical phi X174 DNA: a model for the termination of the rolling circle DNA replication. Nucleic Acids Res. 1981 May 11;9(9):2037–2053. doi: 10.1093/nar/9.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]