Abstract
Nonsyndromic cleft lip and palate (NSCL/P) is a complex disease resulting from failure of fusion of facial primordia, a complex developmental process that includes the epithelial-mesenchymal transition (EMT). Detection of differential gene transcription between NSCL/P patients and control individuals offers an interesting alternative for investigating pathways involved in disease manifestation. Here we compared the transcriptome of 6 dental pulp stem cell (DPSC) cultures from NSCL/P patients and 6 controls. Eighty-seven differentially expressed genes (DEGs) were identified. The most significant putative gene network comprised 13 out of 87 DEGs of which 8 encode extracellular proteins: ACAN, COL4A1, COL4A2, GDF15, IGF2, MMP1, MMP3 and PDGFa. Through clustering analyses we also observed that MMP3, ACAN, COL4A1 and COL4A2 exhibit co-regulated expression. Interestingly, it is known that MMP3 cleavages a wide range of extracellular proteins, including the collagens IV, V, IX, X, proteoglycans, fibronectin and laminin. It is also capable of activating other MMPs. Moreover, MMP3 had previously been associated with NSCL/P. The same general pattern was observed in a further sample, confirming involvement of synchronized gene expression patterns which differed between NSCL/P patients and controls. These results show the robustness of our methodology for the detection of differentially expressed genes using the RankProd method. In conclusion, DPSCs from NSCL/P patients exhibit gene expression signatures involving genes associated with mechanisms of extracellular matrix modeling and palate EMT processes which differ from those observed in controls. This comparative approach should lead to a more rapid identification of gene networks predisposing to this complex malformation syndrome than conventional gene mapping technologies.
Electronic supplementary material
The online version of this article (doi:10.1007/s12015-010-9197-3) contains supplementary material, which is available to authorized users.
Keywords: Nonsyndromic cleft lip and palate, Gene expression profile, Dental pulp, Stem cell, Epithelial-mesenchymal transition, Extracellular matrix
Introduction
Nonsyndromic cleft lip and palate (NSCL/P [MIM 119530]), a complex multifactorial disorder, is one of the most common congenital malformations, with a prevalence of 0.69 to 2.35 per 1,000 births in the Caucasian population [1]. Taking account of the complexities of this orofacial malformation and the long rehabilitation period following surgery, cleft lip and palate is considered to be a major psychosocial and economic burden for families and society. Gaining insight into the genetic causes of NSCL/P should lead to future improvement of genetic counseling, preventive and curative measures.
The development of the human face begins with migration of neural crest cells that combine with the core mesoderm and pharyngeal ectoderm, establishing the facial primordia, which in turn give rise to structures associated with upper lip and palate formation [2, 3]. The growth, differentiation and fusion of these structures are genetically determined, and it is likely that disturbances in genetic pathways orchestrating these processes result in facial abnormalities, such as cleft lip and palate [2–5]. In this context, the epithelial-mesenchymal transition (EMT) plays a central role in generating the cranial neural crest cells as well as ensuring palate and lip fusion [6–8].
The large number of genes known or suspected to be involved in clefting probably reflects the diversity of embryological events that contribute to the formation of these facial structures [4, 9–16]. Although gene mapping approaches, such as Genome-Wide Association Studies (GWAS), appeared to offer an option to identify at-risk alleles associated with NSCL/P, with better reproducibility among different studies [10, 11, 17, 18] than candidate genes, lack of progress over the last decade suggests that GWAS are still unlikely to provide sufficient information on the genetic etiology underlying the disease. However, genome-wide expression analyses based on differential gene expression associated with NSCL/P, as proposed here; present a viable and challenging alternative, since patterns of co-expression can be used to identify biological pathways or gene networks associated with disease predisposition. The current data supporting this supposition can be summarized as follows: transcriptome analysis in tissues of cleft palate (CP) patients showed a distinct gene expression signature when compared to CL/P [19]. It has been reported that a few genes coding for extracellular matrix proteins, such as TGFB3 and MMP3, are differentially expressed in fibroblasts of NSCL/P patients as compared to controls [20, 21], suggesting that the transcriptome of NSCL/P cells might exhibit a specific expression signature irrespective of origin of the cells concerned.
The use of mesenchymal stem cells (MSCs) or induced pluripotent stem cells (iPSs) has been shown to be a promising new approach to study gene function and signaling pathways in genetic disorders [22–24]. MSCs constitute a long-lived population of cells possessing self-renewal and differentiation properties [25–29]. Accordingly, these cells are a good model to study the in vitro characteristics of NSCL/P, since in addition to gene expression, they can be tested for proliferation, migration and differentiation properties, including EMT, functions that are presumed to be altered in cells of NSCL/P patients during embryonic development. MSCs were originally isolated from bone marrow, and subsequently, similar cell populations were isolated from other tissues, such as adipose tissue [27], dental pulp [28, 30], orbicularis oris muscle [26], umbilical cord blood, and umbilical cord tissue [31]. Moreover, MSCs can be easily obtained from non-invasive sources, such as exfoliated teeth, both from NSCL/P patients and control individuals [28].
The main aim of this study was to verify if there are consistent gene expression profile differences between mesenchymal stem cells from NSCL/P patients and controls. We chose to study stem cells from dental pulp as they can be obtained relatively easy from both NSCL/P patients and controls. In addition, these stem cells, as for any other cells obtained from craniofacial tissues, are derived from cranial neural crest cells which play an important role in craniofacial development, including lip and palate [32, 33]. Our results will provide a base line for further investigation and insights into genetic pathway irregularities associated with craniofacial clefts.
Materials and Methods
Sample: NSCL/P Patients and Controls
Ethical approval to obtain stem cells from dental pulp of deciduous teeth was obtained from the Biosciences Institute Research Ethics Committee (Protocol 037/2005). Samples were included only after signed informed consent by the parents or legal guardians.
Deciduous teeth from controls were obtained from odontopediatric clinics in Sao Paulo, while teeth from NSCL/P patients were excised during the exfoliation period by Dr. Bueno D.F. at Sobrapar, Campinas, Sao Paulo. An individual was classified as NSCL/P if no other malformations than clefting of both lip and palate were present.
We analyzed mRNA of dental pulp stem cell cultures (DPSC) obtained from 6 controls and 6 NSCL/P patients (Table 1, supplementary material) for microarray expression analysis and for validation of 4 genes by quantitative Real-Time PCR (qRT-PCR). Validation of the microarray expression analysis by qRT-PCR for a larger number of genes was also done in mRNA obtained from DPSC cultures of 16 additional controls and 13 NSCL/P patients.
Stem Cell Culture
Stem cell cultures obtained from DPSC of deciduous teeth were established according to previously published protocols [28]. Cells were cultured at 37°C with 5% CO2 in DMEM-F12 (Invitrogen, UK) supplemented with 15% Fetal Bovine Serum (FBS, HyClone, USA) and frozen in 40% FBS for storage. Frozen cells were thawed and grown until 80% confluency in a 75 cm2 flask and submitted to serum starvation (12 h) prior to RNA extraction. All experiments were conducted with cells between the 4th and 8th subculture.
Characterization of Mesenchymal Stem Cell Populations
Cell populations used in the microarray assays were analyzed in a flow cytometer for specific cell surface markers to evaluate homogeneity. Cells were harvested with TrypLe (Invitrogen, UK), washed with PBS, and incubated at 4°C for 30 min with the following antibodies: CD29-PE CY5, CD90 (Thy-1), CD45-FITC, CD31-PE (Becton Dickinson, USA), SH2, SH3, and SH4 (Case Western Reserve University, USA). After a second wash, samples incubated with unconjugated primary antibodies were then incubated with anti-mouse-PE secondary antibody (Guava Technologies, Hayward, CA) for an additional 15 min at 4°C. Finally, the cell suspension was washed with PBS, and signals from 105 cells were acquired with an EasyCyte Flow cytometer (Guava Technologies). Control samples for determining background noise were incubated with PBS instead of primary antibody followed by incubation with anti-mouse-PE secondary antibody. All plots generated were analyzed with Guava ExpressPlus software (Guava Technologies).
The in vitro differentiation into bone, cartilage, muscle and fat had previously been demonstrated in 2 of the NSCL/P patients (F4280, F4285) and 3 of the control (F3363, F4271, F4272) samples included in this study [28, 29]. Because of the high reproducibility of our protocols, stem cell cultures are currently characterized only with respect to their immunophenotype.
RNA Processing
Total RNA was isolated using TRIzol (Invitrogen, UK) according to the manufacturer’s protocol and purified with RNeasy Mini-kit (QIAGEN). RNA quality and concentration were assessed using Nanodrop 1000 and gel electrophoresis. Only samples with a preserved rRNA ratio (28S/18S) and no evidence of RNA degradation were used in the microarray hybridization and qRT-PCR.
Microarray Processing
Expression measurements were performed using the Affymetrix Human Gene 1.0 ST array, which interrogates 28,869 genes, following RNA labeling and hybridization protocols as recommended by the manufacturer. After array scanning, quality control was performed with GCOS software (Affymetrix, USA) according to the manufacturer’s recommendations.
Transcriptome Analysis
Gene expression values were obtained using the three-step Robust Multi-array Average (RMA) pre-processing method, implemented in the Affy package from R/Bioconductor [34].
We employed the RankProd method for the selection of differentially expressed genes (DEGs), considering a p-value cut-off of 0.05 adjusted for FDR (False Discovery Rate) [35]. RankProd is a rank-based nonparametric procedure [36]. The method has the advantage of being able to deal with few samples for identifying biologically relevant expression changes [37]. Genes selected by RankProd do not necessarily need to present homogenous expression levels across all the samples of the test and control groups. Accordingly, RankProd seems to be a good choice for identifying differential gene expression in complex diseases, in which altered expression of a given candidate gene is expected in just a subgroup of patients due to both multi-locus genetic heterogeneity and the stochastic nature of gene expression in complex systems [38].
Functional Annotation
We performed functional annotation analysis of the differentially expressed genes with the DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov) and IPA (Ingenuity Pathway Analysis, http://www.ingenuity.com) tools. In IPA, we considered the default parameter Molecules per Network = 35, Networks per Analysis = 25, only direct relationships between genes and the “Ingenuity Expert Information” Data Source, including the new option of “Ingenuity ExpertAssist Findings”, in which the information has been manually reviewed and curated from full-text scientific publications.
Validation of Microarray Expression Using Quantitative Real-Time PCR (qRT-PCR)
Initially we performed qRT-PCR for 4 genes (COL15A1, ERAP2, PPT2 and EGFL8) on the same samples used in the microarray assay. These genes were randomly selected, but they were amongst those with the highest fold change. PPT2 and EGFL8 are represented by a common probe set on the Affymetrix Human Gene 1.0 ST array and therefore both genes were tested for qRT-PCR with gene-specific primers. Next, we performed qRT-PCR for further 12 genes (ACAN, CDH6, CLDN1, CLDN11, COL4A1, COL4A2, ENPP2, HAS2, ITGA8, JAG1, MCAM and MMP3) plus the genes COL15A1, ERAP2 and PPT2 in 16 controls and 13 NSCL/P patients. Only 4 of the control and 4 of the NSCL/P patients samples were the same as those used in the microarray assay due to unavailability of RNA from the remaining 4 individuals (Table 1 in supplementary material).
One microgram of total RNA from each cell culture was reverse transcribed with Superscript II (Invitrogen, UK), according to manufacturer’s recommendations. Quantitative Real-Time PCR reactions were performed in duplicates with a final volume of 20 μl, using 20 ng of cDNA, 1X SYBR Green PCR Master Mix (Applied Biosystems) and 100–400 nM of each primer. We used ABI Prism 7500 Sequence Detection System (Applied Biosystems) with standard temperature protocol. Primers were designed with Primer Express software V.2.0 (Applied Biosystems; primers sequence in supplementary Table 2) and the amplification efficiency (E) of each primer was calculated according to the equation E = 10 (−1/slope). The expression data of the target transcripts were determined by relative quantification in comparison to a pool of RNAs (4 controls and 4 patients). GeNorm v3.4 was used to determine the most stable endogenous controls (SDHA, HPRT1 and GAPDH) and to calculate the normalization factors for each sample [39]. Expression values were calculated according to reference [40].
To compare the expression of COL15A1, ERAP2, PPT2 and EGFL8, obtained by qRT-PCR and microarray assay in the same samples, we used an unpaired t-test with Welch’s correction.
To compare the results obtained by microarray with those obtained by qRT-PCR in the novel samples, for which we do not have microarray data, we performed the following strategy: for each of the 15 genes, we calculated the average (avg) expression for controls and for NSCL/P patients obtained by both methods (Table 3 in supplementary material). The correlation between the ratios “avg_patients/avg_controls” from microarray and qRT-PCR assays was calculated using Spearman’s correlation test.
Clustering Analysis
The cluster analysis of the differentially expressed genes was performed using GEDI (Gene Expression Dynamics Inspector) software. This tool creates a 2 dimensional gene expression image for each sample in which each gene retains exactly the same position in the image of each sample and in which the gene positions are computed to give the most parsimonious gene arrangement for depicting expression level differences between the patient and control groups for all differentially expressed genes [41]. In the analysis, a 10 × 11 grid configuration of SOM (Self-Organizing Map) was used. Inspection of GEDI images allows a straightforward classification of the samples into subgroups without the aid of a clustering algorithm, but simply based on the visual differences in the patterns [42].
We also used two other conventional clustering methods: K-means and Hierarchical, both available in the MeV (MultiExperiment Viewer) software [43], with Spearman’s correlation as the distance metric. The clustering analysis of qRT-PCR data followed two criteria: a) only DEGs from network 1 (Fig. 4) and b) only DEGs that showed the same tendency of expression in the qRT-PCR and microarray assays (Fig. 2 in supplementary material).
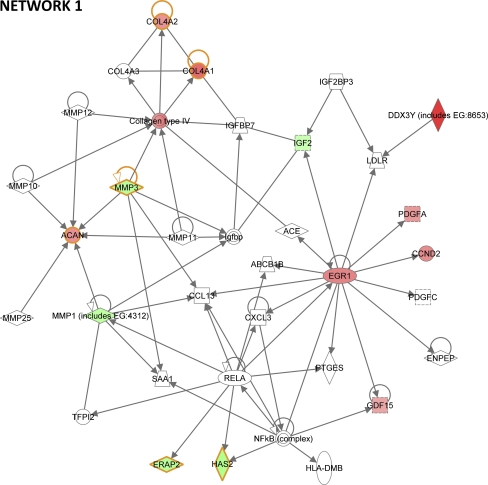
Fig. 4.
The most significant network built by IPA tool with the highest number of differentially expressed genes. The upregulated genes in NSCL/P patients are indicated in red and the downregulated genes in green. The blank symbols pertain to genes that were either not present in our array or not differentially expressed. Solid lines indicate a direct linkage among two genes. Lines with arrows indicate that one gene acts on the other, and lines without arrows indicate that the corresponding proteins interact with each other. The 6 genes circled in orange were used in clustering analysis of qRT-PCR and microarray
Results
Characterization of Mesenchymal Stem Cell Populations
The cell cultures presented positive labeling for cell adhesion (CD29, CD90) and mesenchymal stem cell markers (SH2, SH3, SH4) in most of the cells (>90%) and were negative for endothelial and hematopoietic cell markers (Fig. 2 in supplementary material). Moreover, 2 NSCL/P patients and 3 control cell cultures used in the microarray analyses had been previously shown to differentiate into bone, muscle, cartilage and fat upon in vitro induction [28, 29]. Therefore the cell populations used in this study had the main properties of stem cells.
Controls versus NSCL/P Patients
We identified a total of 87 differentially expressed genes (DEGs; 58 upregulated and 29 downregulated) in the comparison between controls and NSCL/P patients (adjusted p ≤ 0.05; Table 1).
Table 1.
List of 87 differentially expressed genes sorted out by comparing controls and NSCL/P patients with the RankProd analysis
| AFFY ID | Gene symbol | FCa | Pfpb | Cytoband |
|---|---|---|---|---|
| 8176375 | RPS4Y1 | 5.1099 | 0.0007 | Yp11.3 |
| 8176624 | DDX3Y | 4.9358 | 0.0000 | Yq11 |
| 8041206 | LBH | 4.6707 | 0.0000 | 2p23.1 |
| 8118509 | PPT2 c | 4.2212 | 0.0008 | 6p21.3 |
| 7972750 | COL4A1 d c | 3.6284 | 0.0006 | 13q34 |
| 7932254 | ITGA8 d c | 3.2852 | 0.0037 | 10p13 |
| 8125537 | HLA-DMA c | 3.2541 | 0.0005 | 6p21.3 |
| 7998927 | – | 3.0562 | 0.0053 | – |
| 8108370 | EGR1 | 3.0266 | 0.0124 | 5q31.1 |
| 8176719 | EIF1AY | 2.9797 | 0.0055 | Yq11.222 |
| 7952205 | MCAM d c | 2.9129 | 0.0123 | 11q23.3 |
| 7953200 | CCND2 | 2.7894 | 0.0089 | 12p13 |
| 8056491 | SCN9A c | 2.7724 | 0.0223 | 2q24 |
| 7964388 | NDUFA4L2 | 2.7670 | 0.0112 | 12q13.3 |
| 8113504 | C5orf13 | 2.7473 | 0.0077 | 5q22.1 |
| 8176578 | USP9Y | 2.7360 | 0.0219 | Yq11.2 |
| 8104663 | CDH6 d c | 2.7285 | 0.0263 | 5p15.1-p14 |
| 7985786 | ACAN d c | 2.6889 | 0.0042 | 15q26.1 |
| 8121838 | TPD52L1 | 2.6688 | 0.0106 | 6q22-q23 |
| 8177137 | UTY | 2.6455 | 0.0280 | Yq11 |
| 7970033 | COL4A2 d c | 2.6123 | 0.0210 | 13q34 |
| 7965573 | NTN4 c | 2.5853 | 0.0129 | 12q22-q23 |
| 8003298 | SLC7A5 c | 2.5767 | 0.0174 | 16q24.3 |
| 8156783 | COL15A1 c | 2.5452 | 0.0250 | 9q21-q22 |
| 8090565 | SNORA7B | 2.5214 | 0.0225 | 3q21.3 |
| 7954293 | PDE3A | 2.5063 | 0.0121 | 12p12 |
| 8137670 | PDGFA d c | 2.4802 | 0.0179 | 7p22 |
| 8049888 | ATG4B | 2.4474 | 0.0115 | 2q37.3 |
| 7974895 | FLJ43390 | 2.4260 | 0.0427 | 14q23.2 |
| 7958262 | TCP11L2 | 2.4172 | 0.0253 | 12q23.3 |
| 7989985 | ITGA11 c | 2.4038 | 0.0447 | 15q23 |
| 8104035 | SORBS2 | 2.3912 | 0.0291 | 4q35.1 |
| 8102800 | SLC7A11 c | 2.3697 | 0.0390 | 4q28-q32 |
| 7912157 | ERRFI1 | 2.3596 | 0.0308 | 1p36 |
| 7981728 | – | 2.3590 | 0.0407 | – |
| 7985493 | TM6SF1 | 2.3535 | 0.0442 | 15q24-q26 |
| 7981962 | SNORD116-5 | 2.3518 | 0.0419 | 15q11.2 |
| 7931977 | ITIH5 | 2.3354 | 0.0486 | 10p14 |
| 7928308 | DDIT4 | 2.3277 | 0.0467 | 10pter-q26.12 |
| 8034940 | NOTCH3 c | 2.3234 | 0.0396 | 19p13.2-p13.1 |
| 7966089 | CMKLR1 c | 2.2946 | 0.0455 | 12q24.1 |
| 8068024 | JAM2 d c | 2.2538 | 0.0397 | 21q21.2 |
| 8023152 | TCEB3CL | 2.2139 | 0.0407 | 18q21.1 |
| 8109159 | LOC728264 | 2.2134 | 0.0459 | 5q33.1 |
| 8117018 | JARID2 | 2.2051 | 0.0408 | 6p24-p23 |
| 8086752 | RNU13P3 | 2.2046 | 0.0452 | 3p21.31 |
| 7982868 | CHAC1 | 2.2041 | 0.0273 | 15q15.1 |
| 8139207 | INHBA c | 2.1839 | 0.0405 | 7p15-p13 |
| 8027352 | – | 2.1608 | 0.0399 | – |
| 8027002 | GDF15 c | 2.1594 | 0.0285 | 19p13.11 |
| 8048749 | KCNE4 c | 2.1436 | 0.0184 | 2q36.3 |
| 8007850 | LRRC37A | 2.0206 | 0.0244 | 17q21.31 |
| 8160138 | NFIB | 1.9790 | 0.0182 | 9p24.1 |
| 8131867 | – | 1.8907 | 0.0477 | – |
| 8064978 | JAG1 d c | 1.8529 | 0.0178 | 20p12.1-p11.23 |
| 8042788 | ACTG2 | 1.7905 | 0.0414 | 2p13.1 |
| 8045804 | – | 1.7730 | 0.0492 | – |
| 8092726 | CLDN1 d | 1.4872 | 0.0445 | 3q28-q29 |
| 7937772 | IGF2 | −1.8634 | 0.0165 | 11p15.5 |
| 7915592 | – | −1.8745 | 0.0195 | – |
| 8124391 | HIST1H2AB | −2.0165 | 0.0426 | 6p21.3 |
| 8163202 | SVEP1 | −2.0970 | 0.0214 | 9q32 |
| 8037240 | PSG1 c | −2.1467 | 0.0273 | 19q13.2 |
| 8044605 | LOC654433 | −2.2491 | 0.0468 | – |
| 8117594 | HIST1H2BM | −2.2734 | 0.0128 | 6p22-p21.3 |
| 7951271 | MMP1 d c | −2.3465 | 0.0026 | 11q22.3 |
| 8138888 | PDE1C | −2.4348 | 0.0263 | 7p15.1-p14.3 |
| 8140668 | SEMA3A c | −2.5040 | 0.0042 | 7p12.1 |
| 7951284 | MMP3 d c | −2.5261 | 0.0433 | 11q22.3 |
| 8107044 | ERAP2 | −2.5407 | 0.0291 | 5q15 |
| 8003667 | SERPINF1 c | −2.5688 | 0.0109 | 17p13.1 |
| 8110916 | LOC442132 | −2.5879 | 0.0258 | 5p15.31 |
| 7904293 | PTGFRN c | −2.6619 | 0.0164 | 1p13.1 |
| 8113800 | FBN2 c | −2.7034 | 0.0000 | 5q23-q31 |
| 8116418 | GFPT2 | −2.8180 | 0.0212 | 5q34-q35 |
| 8152617 | HAS2 d | −2.8501 | 0.0038 | 8q24.12 |
| 8129573 | MOXD1 c | −2.8809 | 0.0181 | 6q23.1-q23.3 |
| 7925929 | AKR1C3 | −3.1055 | 0.0165 | 10p15-p14 |
| 7917850 | ARHGAP29 | −3.1793 | 0.0031 | 1p22.1 |
| 8037251 | PSG7 c | −3.2618 | 0.0006 | 19q13.2 |
| 8180291 | – | −3.2801 | 0.0006 | |
| 8165808 | XG c | −3.6591 | 0.0065 | Xp22.33 |
| 8037272 | PSG5 // PSG5 c | −3.7775 | 0.0000 | 19q13.2 // 19q13.2 |
| 8083887 | CLDN11 d | −4.2221 | 0.0000 | 3q26.2-q26.3 |
| 7909730 | KCNK2 c | −4.7183 | 0.0000 | 1q41 |
| 8037283 | PSG4 c | −4.9049 | 0.0000 | 19q13.2 |
| 8152522 | ENPP2 d c | −5.3472 | 0.0000 | 8q24.1 |
aFC = Fold change
bPfp = estimated percentage of false positive predictions.
(c) Genes that were functionally categorized as glycoproteins by DAVID (p = 8,0E-6).
(d) Genes involved in EMT.
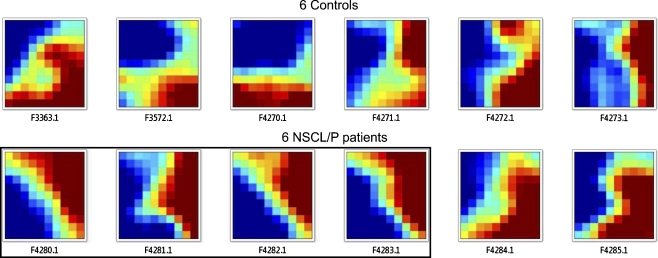
Next, in order to visualize the expression behavior of these DEGs, we performed clustering analysis with the GEDI software. An image was created reflecting the 87 DEGs’ transcriptional behavior for each individual and where the gene position was fixed to give the most parsimonious arrangement to show differential gene expression between controls and NSCL/P patients. Upon visual inspection of the GEDI images, we observed that 4 of the NSCL/P patients showed a similar expression pattern (F4280, F4281, F4282, F4283; Fig. 1).
Fig. 1.
Clustering of 87 DEGs resulted from the comparison between 6 controls and 6 NSCL/P patients. Each GEDI map (or mosaic) represents a gene expression profile of a single individual. The blue color represents the lowest expression level and red color represents the highest expression level on a scale of −4.70 to 7.98, respectively. The black frame highlights four patients with similar gene expression profile
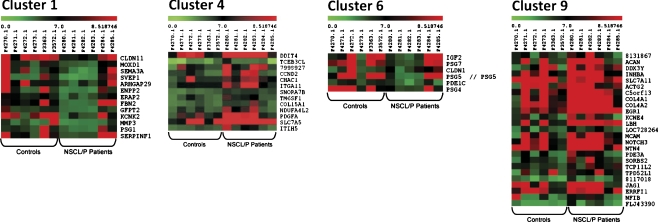
The clustering analysis with the k-means method resulted in 9 gene clusters. Four of them exhibited a similar expression profile among NSCL/P patients, most particularly in the afore-mentioned group (F4280, F4281, F4282, F4283; Fig. 2). The similarities and dissimilarities in expression levels observed between NSCL/P patients were similar for both clustering methods. The expression pattern found in 4 out of 6 NSCL/P patients illustrates the characteristic of RankProd of being capable of selecting genes with differential expression in just a subgroup of samples.
Fig. 2.
Gene clusters 1, 4, 6 and 9 resulted from k-means method (k = 9). In both clusters it is possible to observe a similar gene expression profile among 4 out of 6 patients (F4280, F4281, F4282 and F4283), indicating that many of the 87 selected DEGs are co-regulated in these 4 NSCL/P patients
Functional Annotation
Differentially expressed genes were functionally annotated and analyzed with two different tools. First, the DAVID tool led to identification of three main canonical pathways from the KEGG Database: Focal adhesion, Cell adhesion molecules and ECM-receptor interaction (Fig. 3, 4 and 5 in supplementary material). Moreover, the most relevant functional category identified through DAVID was that of Glycoproteins (p = 8.0E-6), which included 36 of the 87 DEGs (Table 1). Subsequently, the IPA tool was used to characterize the 87 DEGs regarding possible biological functions. We observed 3 relevant functions enriched with a significant number of our genes: Cellular movement (20 genes, p = 6.4E-06–2.76E-02), Cellular growth and proliferation (27 genes, p = 3.11E-05–2.68E-02) and Cellular development (27 genes, p = 3.3E-05–2.47E-02) (Table 4 in supplementary material). A putative network with the largest number of DEGs built by IPA (13 DEGs; Fig. 4) suggests functional relationship among several extracellular proteins: ACAN, COL4A1, COL4A2, GDF15, IGF2, MMP1, MMP3 and PDGFa. All of these 8 genes are DEGs.
Validation of the Microarray Analysis
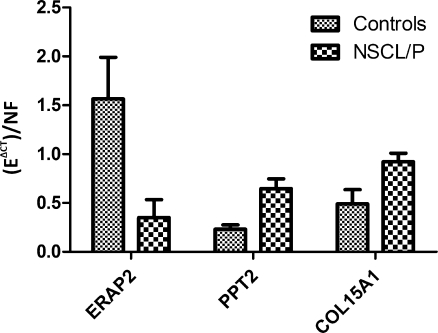
The reproducibility of gene expression assayed by Affymetrix microarrays is high [44] and 4 genes (COL15A1, ERAP2, PPT2 and EGFL8) were initially selected for validation through qRT-PCR. Except for ERAP2 (p = 0.0397), that showed lower expression levels, the other genes showed higher transcript levels in NSCL/P patients than in controls: PPT2 (p = 0.0087) and COL15A1 (p = 0.0355) (Fig. 3), which confirms the expression patterns observed in the microarray assays. EGFL8 is represented by the same probe set of PPT2. Considering that we did not find significant differential expression levels through qRT-PCR with specific primers for EGFL8 (p = 0.1781), we kept only PPT2 among our candidate genes.
Fig. 3.
Quantitative Real-Time PCR (qRT-PCR) initial analysis of NSCL/P patients and control samples for ERAP2, PPT2, COL15A1. E = primer amplification efficiency; CT = cycle threshold; delta_CT = sample’s average_CT normalized by pool’s average_CT; NF = normalization factor
For further validation of our results, in addition to COL15A1, PPT2 and ERAP2, we analyzed a further 12 other genes known to be involved with matrix remodeling in a novel sample of individuals (16 controls and 13 NSCL/P patients). By comparing the ratios of the average expression from patients/controls, we observed that ENPP2 exhibited the highest discordance between the microarray and qRT-PCR ratios (0.645 and 4.324, respectively), therefore we considered this gene expression as not validated. However, we observed a significant positive correlation between microarray and qRT-PCR expression data (p = 0.0114, r = 0.653) for the 14 genes chosen for validation (Table 3 in supplementary material). Accordingly, the analyses thus show that the data obtained from microarrays and qRT-PCR are consistent with each other, even in an enlarged novel sample, attesting to the reliability of the microarray analysis.
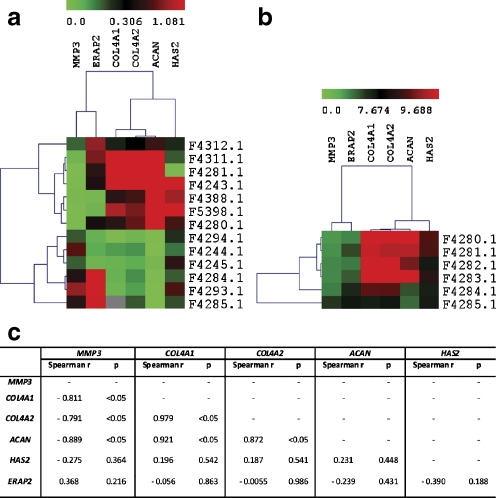
Using NSCL/P patients expression data from qRT-PCR (13 NSCL/P patients) and microarray assays (6 NSCL/P patients), we also performed a clustering analysis for the following DEGs: ACAN, COL4A1, COL4A2, ERAP2, HAS2, and MMP3. These genes belong to network 1 (Fig. 4) and exhibited the same expression tendency in both experiments (Fig. 2 in supplementary material). The hierarchical clustering performed with qRT-PCR data revealed a homogeneous cluster with 7 out of 13 NSCL/P patients (F4312, F4311, F4281, F4243, F4388, F5398 and F4280), in which MMP3 is downregulated and ACAN, COL4A1 and COL4A2 upregulated. ERAP2 and HAS2 did not exhibit a consistent expression pattern in these patients (Fig. 5a). Interestingly, the hierarchical clustering using microarray data showed the same transcriptional behavior among these genes in 4 out of 6 NSCL/P patients (F4280, F4281, F4282 and F4283), although in this case, ERAP2 and HAS2 seem to be co-regulated with MMP3 and collagens (Fig. 5b). Spearman’s correlation test (Fig. 5c) calculated for each relationship among the genes from Fig. 5a confirmed that MMP3 and ACAN are negatively correlated (r = −0.889; p < 0.05) while ACAN exhibited positive correlation with COL4A1 and COL4A2 (r = 0.921 and r = 0.872, respectively; p < 0.05), corroborating our findings thus far. On the other hand, we did not achieve significance for ERAP2 and HAS2, confirming the lack of a well-defined expression pattern for these genes, as observed in Fig. 5a.
Fig. 5.
Hierarchical clustering of all the patients analyzed by qRT-PCR and microarray, considering only the genes from IPA network 1 (Fig. 4). a Clustering of expression values obtained by qRT-PCR. b Clustering of expression values obtained by microarray. c Correlations (Spearman’s correlation test, r and p-values) between each co-regulated gene from qRT-PCR clustering. It is possible to observe that the gene MMP3 is significantly and inversely correlated to ACAN, COL4A1, COL4A2 genes in a subgroup of patients from qRT-PCR. This same pattern of co-regulation is also present in another group of patients analyzed by microarray, which includes two individuals (F4280 and F4281) from the mentioned qRT-PCR subgroup. In controls expression data of both assays, MMP3 is upregulated and ACAN, COL4A1 and COL4A2 downregulated
Discussion
In this study, we conducted the first comparative transcriptome analysis between dental pulp stem cells from NSCL/P patients and controls to obtain more information on the genetic etiology of this malformation and explore new possibilities to identify pathways associated with disease pathology.
Genome expression microarray analysis is a powerful tool for assessing changes in the transcription patterns of related genes and identification of signaling pathways associated with specific cell types, culture conditions or disease states [45, 46]. Considering that the cell populations from NSCL/P patients and controls were established and maintained under similar identical conditions, it is very likely that a large proportion of the DEGs identified is related to the genetic constitutional differences between cells from NSCL/P patients and controls. The observations that NSCL/P disease-specific expression profiles have also been previously reported in tissue biopsies and fibroblast cultures [19, 21] suggests that such profiles may be ubiquitous; in this respect our findings suggest that the disease-specific transcription profile is also present in mesenchymal stem cells. Accordingly, transcriptome analysis of stem cells represents an additional option to the study of the transcriptome and genetics of NSCL/P.
Of the 87 DEGs obtained in our microarray analysis, we noted that MMP3 had previously been proposed as a candidate gene for NSCL/P [47]. A further 2 genes, PDGFa and ERRFI1, had previously been indirectly associated with NSCL/P, since their receptors, respectively PDGFR and EGRF, were identified as predisposing loci for this form of clefting [48–50]. These observations provide further confirmation that the transcriptome of DPSCs from NSCL/P patients can also be used to identify genetic variations associated with the disease.
The functional annotation through DAVID showed that nearly half of the 87 DEGs correspond to glycoprotein molecules, including collagens, metalloproteinases, integrins, and adhesion proteins, which are important to orchestrate the signaling between the extracellular and intracellular compartment. Indeed, the three canonical pathways we identified through DAVID are mainly related to extracellular matrix components and signaling molecules located on the cell membrane (Fig. 3, 4 and 5 in supplementary material). Functional relationships among several extracellular proteins that are deregulated in our studies are also suggested by the putative network built by IPA (Fig. 4). These analyses suggest that a large proportion of the DEGs in the DPSC from NSCL/P patients are extracellular matrix components (ECM) involved in several cellular processes in facial development, such as extracellular remodeling during the epithelial-mesenchymal transition (EMT).
EMT is a fundamental mechanism behind palatal fusion. This process occurs through a regulated sequence of events determined both by the extracellular environment and the gene expression program of the cell, leading to loss of cell-cell adhesion, breakdown of basal laminae, and increased cell invasion and mobility [51, 52].
Fifteen genes enrolled in EMT are within the 87 DEGs (Table 1). Further, we observed the enrichment of biological functions involving cell proliferation, movement and invasion (Table 4 in supplementary material), all of which are expected phenotypic outcomes for genes involved with EMT.
In the most relevant IPA network we observed that 8 out of 13 DEGs (ACAN, COL4A1, COL4A2, GDF15, IGF2, MMP1, MMP3 and PDGFa, Fig. 4) are extracellular matrix components, suggesting that an abnormal expression behavior of these genes may affect EMT during palate development.
Clustering analyses showed that MMP3, ACAN, COL4A1 and COL4A2 transcripts are co-regulated in 4 out of 6 NSCL/P patients analyzed by microarray as well as in 7 out of 13 NSCL/P patients from qRT-PCR analysis. Therefore, it seems that in NSCL/P mesenchymal cells, the down-regulation of MMP3 is associated with up-regulation of ACAN, COL4A1 and COL4A2. Such a deregulated pathway probably has serious consequences in embryonic development, since it is known that the MMP3 protein cleaves a wide range of ECM proteins, including the collagens IV, V, IX, X, proteoglycans, fibronectin and laminin. It is also capable of activating other MMPs, as shown in network 1 (Fig. 4), and playing a key role in ECM degradation and remodeling [47]. In this context it has already been experimentally shown that lower levels of MMPs can block palatal fusion [7]. Therefore, it is possible that failure of lip and palate fusion in these groups of patients is at least partly associated with down-regulation of MMPs and up-regulation of collagens. It will be important in the near feature to identify the leading causative mechanism of altered expression of MMPs in these individuals. These results also show the robustness of RankProd to detect DEGs on a limited and heterogeneous group of samples, in contradistinction to a popular method SAM [53] which appears to require larger sample sizes. Moreover, RankProd is able to identify expression patterns in just a subgroup of affected samples, which is the ideal, considering that this is the expected to occur in a complex disease such as NSCL/P. Notwithstanding our favorable impression of RankProd, we are acutely aware of the small sample sizes employed in detecting DEGs in the initial microarray assay. Future studies must be directed towards confirming and/or expanding the pattern of DEGs using novel and much larger sample sizes.
In summary our results suggest that NSCL/P patients exhibit deregulation of genes participating in either EMT or embryonic processes that depend on extracellular modeling. Abnormal expression of some genes encoding for extracellular matrix proteins in NSCL/P fibroblasts has also been reported by others, reinforcing the concept that disease expression signatures for NSCL/P are present in various tissues and not necessarily confined to a specific cell type. Moreover, the penetrance of the phenotype can depend on exposure to environmental factors and it is of interest that a recent report claimed a positive association between nicotine exposure and the CL/P phenotype in conjunction with deregulation of gene expression involved in ECM synthesis and degradation [21].
The observation that the DEGs in NSCL/P patients are associated with ECM component signaling suggests that further analysis of these cells presents unique opportunities to study the complexity of molecular pathways and alleles involved in NSCL/P etiology. However, arguably the major advantage of DPSCs above other cell types, such as fibroblasts, will be their potential to study in vitro differentiation into muscle, bone and cartilage that are affected tissues in NSCL/P. In this context, it will be possible to integrate genomic and transcriptome analysis under different experimentally induced environmental insults on identical cell cultures. Our results open new avenues for the study of novel candidate genes for NSCL/P, since most of the 87 DEGs have not been previously associated with NSCL/P. In particular, the potential offered by our approach can be best visualized in the gene network 1 (Fig. 4), in which several of our DEGs are ECM components, suggesting that these genes might be enrolled in EMT during the development of NSCL/P phenotype.
If wide-spread differences in co-regulated gene transcription, as observed in our experiments, are indeed a primary cause of NSCL/P, then the research emphasis must move away from naively cataloging which genes are being differentially expressed to defining the central transcription factors and regulatory elements that are driving the disease specific transcription patterns. In this respect, identifying the putative gene networks involved, as in our observations, will be an initial crucial step towards defining which regulatory elements are the most important.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Flow cytometry analysis of dental pulp stem cell (DPSC) cultures. The graphs display the immunophenotype of adherent DPSC isolated from NSCL/P patients. The control assay (not labeled) is represented by a black line and the experimental population (labeled with specific antibodies) is in gray. Values represent the mean percentage of all assessed cells positively stained for the indicated antigens: SH2 (97.43%), SH3 (97.02%), SH4 (93.92%), CD29 (99.9%), CD31 (0.08%), CD45 (3.84%) and CD90 (97.96%). Abbreviations: CD, Cluster of Differentiation. (JPEG 551 kb)
Comparison between expression ratios (log10 avg_patients/avg_controls) obtained by qRT-PCR (black line) and microarray (grey line) of the 15 genes chosen for validation. (JPEG 223 kb)
Focal Adhesion pathway from the KEGG Database sorted out by DAVID tool. The red stars correspond to genes deregulated in the comparison between NSCL/P patients and controls (ECM, ITGA and CycD). (JPEG 782 kb)
Cell Adhesion Molecules pathway from the KEGG Database sorted out by DAVID tool. The red stars correspond to genes deregulated in the comparison between NSCL/P patients and controls (MHC-II, CLDN, JAM2, ITGA8 and CLDN11). (JPEG 1387 kb)
ECM-receptor interaction pathway from the KEGG Database sorted out by DAVID tool. The red stars correspond to genes deregulated in the comparison between NSCL/P patients and controls (Collagen, alpha8 and alpha12). (JPEG 783 kb)
Age, gender, and clinical status of the samples analyzed by qRT-PCR and/or microarray. (DOC 64 kb)
Primer sequences used for qRT-PCR. (DOC 41 kb)
Ratios of the average expression from patients/controls for 15 candidate genes chosen for validation. (DOC 55 kb)
Top 3 (out of 70) functional categories identified by Ingenuity Pathway Analysis as being significantly overrepresented by differentially expressed genes. (DOC 50 kb)
Acknowledgements
The authors would like to express their gratitude to: Mrs. Constancia Urbani for secretarial assistance, and the following colleagues: Rodrigo Atique, BSc, for helping us with figures; Eder Zucconi, PhD, and Natassia Vieira, BSc, for helping us with flow cytometry; Andrea Sertié, PhD, and Keith Okamoto, PhD for their helpful reviews and suggestions regarding the manuscript. FAPESP/CEPID, CNPq, MCT, FINEP sponsored this study.
Disclosures
The authors indicate no potential conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Daniela Franco Bueno and Daniele Yumi Sunaga contributed equally to this work
References
- 1.Gundlach KKH, Maus C. Epidemiological studies on the frequency of clefts in Europe and world-wide. Journal of Cranio-Maxillo-Facial Surgery. 2006;34(2):1–2. doi: 10.1016/S1010-5182(06)60001-2. [DOI] [PubMed] [Google Scholar]
- 2.Kerrigan JJ, Mansell JP, Sengupta A, Brown N, Sandy JR. Palatogenesis and potential mechanisms for clefting. Journal of the Royal College of Surgeons of Edinburgh. 2000;45(6):351–358. [PubMed] [Google Scholar]
- 3.Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Developmental Dynamics. 2006;235(5):1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Diseases. 2009;15(7):437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K, Hu D, Bustos T, et al. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate-ectodermal dysplasia. Nature Genetics. 2000;25(4):427–430. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- 6.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. The Journal of Clinical Investigation. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu W, Ruest LB, Svoboda KK. Regulation of epithelial-mesenchymal transition in palatal fusion. Experimental Biology and Medicine. 2009;234(5):483–491. doi: 10.3181/0812-MR-365. [DOI] [PubMed] [Google Scholar]
- 8.Kang P, Svoboda KK. Epithelial-mesenchymal transformation during craniofacial development. Journal of Dental Research. 2005;84(8):678–690. doi: 10.1177/154405910508400801. [DOI] [PubMed] [Google Scholar]
- 9.Juriloff DM, Harris MJ. Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Research. Part A: Clinical and Molecular Teratology. 2008;82(2):63–77. doi: 10.1002/bdra.20430. [DOI] [PubMed] [Google Scholar]
- 10.Mangold E, Ludwig KU, Birnbaum S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nature Genetics. 2010;42(1):24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 11.Mangold E, Reutter H, Birnbaum S, et al. Genome-wide linkage scan of nonsyndromic orofacial clefting in 91 families of central European origin. American Journal of Medical Genetics. Part A. 2009;149A(12):2680–2694. doi: 10.1002/ajmg.a.33136. [DOI] [PubMed] [Google Scholar]
- 12.Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nature Genetics. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 13.Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. American Journal of Human Genetics. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai J, Chen J, Liu Y, et al. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells. 2006;24(3):516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong SG, Gong TW, Shum L. Identification of markers of the midface. Journal of Dental Research. 2005;84(1):69–72. doi: 10.1177/154405910508400112. [DOI] [PubMed] [Google Scholar]
- 16.Brown NL, Knott L, Halligan E, Yarram SJ, Mansell JP, Sandy JR. Microarray analysis of murine palatogenesis: temporal expression of genes during normal palate development. Development, Growth & Differentiation. 2003;45(2):153–165. doi: 10.1034/j.1600-0854.2004.00686.x. [DOI] [PubMed] [Google Scholar]
- 17.Nikopensius T, Birnbaum S, Ludwig KU, et al. Susceptibility locus for non-syndromic cleft lip with or without cleft palate on chromosome 10q25 confers risk in Estonian patients. European Journal of Oral Sciences. 2010;118(3):317–319. doi: 10.1111/j.1600-0722.2010.00741.x. [DOI] [PubMed] [Google Scholar]
- 18.Beaty TH, Murray JC, Marazita ML, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature Genetics. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobsen LP, Borup R, Vestergaard J, et al. Expression analyses of human cleft palate tissue suggest a role for osteopontin and immune related factors in palatal development. Experimental & Molecular Medicine. 2009;41:77–85. doi: 10.3858/emm.2009.41.2.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown NL, Yarram SJ, Mansell JP, Sandy JR. Matrix metalloproteinases have a role in palatogenesis. Journal of Dental Research. 2002;81:826–830. doi: 10.1177/154405910208101206. [DOI] [PubMed] [Google Scholar]
- 21.Baroni T, Bellucci C, Lilli C, et al. Human cleft lip and palate fibroblasts and normal nicotine-treated fibroblasts show altered in vitro expressions of genes related to molecular signaling pathways and extracellular matrix metabolism. Journal of Cellular Physiology. 2010;222(3):748–756. doi: 10.1002/jcp.22006. [DOI] [PubMed] [Google Scholar]
- 22.Gunaseeli I, Doss MX, Antzelevitch C, Hescheler J, Sachinidis A. Induced pluripotent stem cells as a model for accelerated patient- and disease-specific drug discovery. Current Medicinal Chemistry. 2010;17(8):759–766. doi: 10.2174/092986710790514480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masotti C, Ornelas C, Splendore-Gordonos A, Moura R, Felix T, Nivaldo A, et al. Reduced transcription of TCOF1 in adult cells of Treacher Collins syndrome patients. BMC Medical Genetics. 2009;10(1):136. doi: 10.1186/1471-2350-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai M-S, Hwang S-M, Chen K-D, Lee Y-S, et al. Functional network analysis of the transcriptomes of mesenchymal stem cells derived from amniotic fluid, amniotic membrane, cord blood, and bone marrow. Stem Cells. 2007;25(10):2511–2523. doi: 10.1634/stemcells.2007-0023. [DOI] [PubMed] [Google Scholar]
- 25.Zucconi E, Vieira NM, Bueno DF, et al. Mesenchymal stem cells derived from canine umbilical cord vein—a novel source for cell therapy studies. Stem Cells and Development. 2010;19(3):395–402. doi: 10.1089/scd.2008.0314. [DOI] [PubMed] [Google Scholar]
- 26.Bueno DF, Kerkis I, Costa AM, et al. New source of muscle-derived stem cells with potential for alveolar bone reconstruction in cleft lip and/or palate patients. Tissue Engineering. 2009;15:427–435. doi: 10.1089/ten.tea.2007.0417. [DOI] [PubMed] [Google Scholar]
- 27.Vieira N, Bueno CR, Jr, Brandalise V, et al. Dystrophic mice express a significant amount of human muscle proteins following systemic delivery of human adipose-derived stromal cells without immunosuppression. Stem Cells. 2008;26(9):2391–2398. doi: 10.1634/stemcells.2008-0043. [DOI] [PubMed] [Google Scholar]
- 28.Costa AM, Bueno DF, Martins MT, et al. Reconstruction of large cranial defects in nonimmunosuppressed experimental design with human dental pulp stem cells. The Journal of Craniofacial Surgery. 2008;19:204–210. doi: 10.1097/scs.0b013e31815c8a54. [DOI] [PubMed] [Google Scholar]
- 29.Bueno, D. F. (2007). The use of adult stem cells to study the etiopathogeny of cleft lip and palate and tissue engineering. Phd in human genetics, Institute of Biosciences, University of Sao Paulo (USP), Rua do Matao, trav. 14, n321—Cidade Universitaria—Sao Paulo—Brazil.
- 30.Gronthos S, Brahim J, Li W, Fisher LW, et al. Stem cell properties of human dental pulp stem cells. Journal of Dental Research. 2002;81(8):531–535. doi: 10.1177/154405910208100806. [DOI] [PubMed] [Google Scholar]
- 31.Secco, M., Moreira, Y., Zucconi, E., et al. (2009). Gene expression profile of mesenchymal stem cells from paired umbilical cord units: Cord is different from blood. Stem Cell Reviews and Reports. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 32.Widera D, Zander C, Heidbreder M, et al. Adult palatum as a novel source of neural crest-related stem cells. Stem Cells. 2009;27(8):1899–1910. doi: 10.1002/stem.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grenier J, Teillet MA, Grifone R, Kelly RG, Duprez D. Relationship between neural crest cells and cranial mesoderm during head muscle development. PLoS ONE. 2009;4(2):e4381. doi: 10.1371/journal.pone.0004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 36.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. Rankprod: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22(22):2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 37.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 38.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol., 3(7), research0034.1–0034.11. [DOI] [PMC free article] [PubMed]
- 40.Pfaffl MW. A new mathematical model for quantification in real-time-RT-PCR. Nucleic Acids Research. 2001;9:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eichler GS, Huang S, Ingber DE. Gene expression dynamics inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 42.Guo, Y., Eichler, G. S., Feng, Y., Ingber, D. E., & Huang, S. (2006). Towards a holistic, yet gene-centered analysis of gene expression profiles: a case study of human lung cancers. Journal of Biomedicine and Biotechnology, 69141. [DOI] [PMC free article] [PubMed]
- 43.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 44.Bochukova, E., Soneji, S., Wall, S., & Wilkie, A. O. (2009). Scalp fibroblasts have a shared expression profile in monogenic craniosynostosis. Journal of Medical Genetics, [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 45.LaGamba D, Nawshad A, Hay ED. Microarray analysis of gene expression during epithelial-mesenchymal transformation. Developmental Dynamics. 2005;234(1):132–142. doi: 10.1002/dvdy.20489. [DOI] [PubMed] [Google Scholar]
- 46.Fanganiello RD, Sertié AL, Reis EM, et al. Apert pSer252Trp mutation in FGFR2 alters osteogenic potential and gene expression of cranial periosteal cells. Molecular Medicine. 2007;13(7,8):422–442. doi: 10.2119/2007-00027.Fanganiello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letra A, Silva RA, Menezes R, et al. MMP gene polymorphisms as contributors for cleft lip/palate: association with MMP3 but not MMP1. Archives of Oral Biology. 2007;52(10):954–960. doi: 10.1016/j.archoralbio.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Xu X, Bringas P, Jr, Soriano P, Chai Y. PDGFR-alpha signaling is critical for tooth cusp and palate morphogenesis. Developmental Dynamics. 2005;232(1):75–84. doi: 10.1002/dvdy.20197. [DOI] [PubMed] [Google Scholar]
- 49.Ferby I, Reschke M, Kudlacek O, et al. Mig6 is a negative regulator of EGF receptor–mediated skin morphogenesis and tumor formation. Nature Medicine. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- 50.Miettinen PJ, Chin JR, Shum L, Slavkin HC, Shuler CF, Derynck R, et al. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nature Genetics. 1999;22:69–73. doi: 10.1038/8773. [DOI] [PubMed] [Google Scholar]
- 51.Yu W, Ruest LB, Svoboda KK. Regulation of epithelial-mesenchymal transition in palatal fusion. Experimental Biology and Medicine. 2009;234(5):483–491. doi: 10.3181/0812-MR-365. [DOI] [PubMed] [Google Scholar]
- 52.Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Developmental Dynamics. 2009;238(3):716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry analysis of dental pulp stem cell (DPSC) cultures. The graphs display the immunophenotype of adherent DPSC isolated from NSCL/P patients. The control assay (not labeled) is represented by a black line and the experimental population (labeled with specific antibodies) is in gray. Values represent the mean percentage of all assessed cells positively stained for the indicated antigens: SH2 (97.43%), SH3 (97.02%), SH4 (93.92%), CD29 (99.9%), CD31 (0.08%), CD45 (3.84%) and CD90 (97.96%). Abbreviations: CD, Cluster of Differentiation. (JPEG 551 kb)
Comparison between expression ratios (log10 avg_patients/avg_controls) obtained by qRT-PCR (black line) and microarray (grey line) of the 15 genes chosen for validation. (JPEG 223 kb)
Focal Adhesion pathway from the KEGG Database sorted out by DAVID tool. The red stars correspond to genes deregulated in the comparison between NSCL/P patients and controls (ECM, ITGA and CycD). (JPEG 782 kb)
Cell Adhesion Molecules pathway from the KEGG Database sorted out by DAVID tool. The red stars correspond to genes deregulated in the comparison between NSCL/P patients and controls (MHC-II, CLDN, JAM2, ITGA8 and CLDN11). (JPEG 1387 kb)
ECM-receptor interaction pathway from the KEGG Database sorted out by DAVID tool. The red stars correspond to genes deregulated in the comparison between NSCL/P patients and controls (Collagen, alpha8 and alpha12). (JPEG 783 kb)
Age, gender, and clinical status of the samples analyzed by qRT-PCR and/or microarray. (DOC 64 kb)
Primer sequences used for qRT-PCR. (DOC 41 kb)
Ratios of the average expression from patients/controls for 15 candidate genes chosen for validation. (DOC 55 kb)
Top 3 (out of 70) functional categories identified by Ingenuity Pathway Analysis as being significantly overrepresented by differentially expressed genes. (DOC 50 kb)