Abstract
Hepatocellular carcinoma (HCC), the primary form of human adult liver malignancy, is a highly aggressive tumor with average survival rates that are currently less than a year following diagnosis. Most patients with HCC are diagnosed at an advanced stage, and no efficient marker exists for predicting prognosis and/or response(s) to therapy. We previously reported a high level of [1-13C] alanine in an orthotopic HCC using single-voxel hyperpolarized [1-13C] pyruvate magnetic resonance spectroscopy (MRS). In the present study, we implemented a 3D-magnetic resonance spectroscopic imaging (MRSI) sequence to investigate this potential hallmark of cellular metabolism in rat livers bearing HCC (N=7 buffalo rats). In addition, quantitative real-time PCR was used to determine the mRNA levels of lactate dehydrogenase A, NAD(P)H dehydrogenase quinone 1, and alanine transaminase. The enzymes levels were significantly higher in the tumor than the normal liver tissues within each rat, which is associated with the in vivo MRSI signal of [1-13C]alanine and [1-13C]lactate after a bolus intravenous injection of [1-13C]pyruvate. Histopathological analysis of these tumors confirms successful growth of HCC as a nodule in buffalo rats’ livers revealing malignancy and hyper-vascular architecture. More importantly, the results demonstrate that the metabolic fate of [1-13C]pyruvate conversion to [1-13C]alanine significantly supersedes that of [1-13C]pyruvate conversion to [1-13C]lactate potentially serving as a marker of HCC tumors.
Keywords: hyperpolarized 13C 3D MRSI, [1-13C]pyruvate, hepatocellular carcinoma, alanine transaminase
INTRODUCTION
The incidence rates of hepatocellular carcinoma (HCC) tripled in the United States from 1975 through 2005 across all ethnic groups, with marked recent increases among middle-aged black, Hispanic, and white males (1). Most patients with HCC are diagnosed when the disease is already at an advanced stage, thereby limiting therapeutic options and leading to a dismal one-year cause-specific survival rate (1). This health challenge warrants efforts to effectively manage and treat the disease. Research on alterations of gene and protein expressions of HCC could facilitate identification of molecular hallmarks for effective therapeutic strategies. Therefore, suitable animal models of orthotopic HCC that permit the control of genetic and environmental conditions in longitudinal studies will recapitulate all phases of the disease, facilitate the development of diagnostic or prognostic biomarkers, develop robust tumor imaging approaches, and provide evaluation of potential therapeutic strategies.
A characteristic feature of cancer cells is the alteration of their central carbon metabolism. It is generally acknowledged that cancer cells preferentially utilize glycolysis rather than oxidative phosphorylation for energy production irrespective of oxygen supply (2). However, mechanistic explanations for this glycolytic phenotype are controversial, perhaps ATP production for energy, biosynthesis for cell growth, or anaplerotic flux for the tricarboxylic acid (TCA) cycle (3–5). As technological improvements increase the feasibility of studying cancer metabolism, growing number of reports have investigated the molecular connections between malignant transformation and cell metabolism. Recently, a state-of-the-art metabolome analysis tool based on capillary electrophoresis coupled to mass spectrometry (6) quantified the various levels of metabolites involved in central carbon metabolism in human tumor tissues and globally mapped the glycolysis, pentose phosphate, and TCA pathways. In addition, the liquid state preservation of polarized nuclear spins from dynamic nuclear polarization (DNP) (7) has advanced 13C magnetic resonance spectroscopic imaging (MRSI) and enabled the implementation of in vivo tumor metabolic imaging with hyperpolarized [1-13C]pyruvate (8–14), 13C-labelled bicarbonate (15), [2-13C]fructose (16), [1,4-13C2]fumarate (17) or [1-13C] ketoisocaproate (18) to investigate local changes in the carbon metabolic pathways after intravenous administration of the hyperpolarized substrate. Detection of these substrates and their metabolic products provide crucial information about multiple transporters and enzymes involved in carbon metabolism. Due to the short lifetime of the hyperpolarized signal (~ 60 s), complete investigation of carbon metabolism is not feasible.
Hyperpolarized [1-13C]pyruvate MRSI was previously used to demonstrate changes in metabolism of fasted rat liver where the [1-13C]lactate to [1-13C]alanine ratios increased as compared to normal rat liver (19). Another study demonstrated an increased lactate production rate in rat liver when [1-13C]pyruvate was co-administered with ethanol (20). This finding was attributed to increased nicotinamide adenine dinucleotide (NADH) in relation to ethanol metabolism in the rat liver. More recently, it was reported that a fasted rat bearing orthotopic HCC showed increased [1-13C]lactate and [1-13C]alanine levels after a bolus intravenous injection of hyperpolarized [1-13C]pyruvate (14). Unlike most [1-13C]pyruvate studies, this single-voxel MRS study revealed a marked increase in [1-13C]alanine above that from [1-13C]lactate in tumors. These contributions are significant because they may characterize inherent biomarkers of HCC that can provide new insights into the progression of unresectable hepatomas. Studies have previously investigated the pattern of glycolytic enzymes in buffalo rat hepatomas using invasive tissue assay analyses (21,22). With the exceptions of glucokinase, phosphofructokinase and pyruvate kinase, the activities of the enzymes of the main glycolytic pathway are generally similar in rat liver and hepatomas (21). The activities of these three enzymes, glucokinase, phosphofructokinase and pyruvate kinase, reflect the growth potential of the tumors that is consistently being highest in the more rapidly growing tumors and gradually decreasing from slowly growing tumors to normal rat liver. Notwithstanding, the enzyme patterns of the rat hepatomas also showed distinctive changes indirectly related to glycolysis at branched points that involve alternate pathway to the main glycolytic pathway. One such pathway is via lactate dehydrogenase (LDH). The ratio of LDH to glycerolphosphate dehydrogenase activities was highest in the most rapidly growing tumors and the lowest in the slowly growing tumors as compared to normal liver, thereby suggesting a correlation to the rate of aerobic glycolysis of malignant tissues. Also, it has been noted that total tyrosine aminotransferase in many host livers and hepatomas were slightly elevated in rats fed a vitamin B6-deficient diet (22). These results could point out some distinguishable metabolic markers of HCC that may be identifiable in vivo. In light of these findings, little or no investigation has reported enzyme levels in connection with metabolites produced from in vivo HCC rat models or even in humans.
We postulated, on the basis of preliminary data, that in vivo 13C MRS measurement of the conversion of administered hyperpolarized [1-13C]pyruvate to the downstream metabolites of [1-13C]alanine and [1-13C]lactate will produce surrogate markers concomitant with associated enzymes to characterize orthotopic HCC in rat. In this study, we implemented a three-dimensional double-spin-echo echo-planar spectroscopic imaging (3D DSE-EPSI) pulse sequence to investigate potential hallmarks of cellular carbon metabolism in rat liver bearing orthotopic HCC. In addition, quantitative real-time polymerase chain reaction (PCR) was performed to measure the mRNA levels of lactate dehydrogenase A [LDH-A, EC 1.1.1.27], NAD(P)H dehydrogenase quinone 1 [NQO1, EC 1.6.5.2], and alanine transaminase [ALT, EC 2.6.1.2] in our rat hepatoma model. We show that differentially expressed enzymes in pyruvate metabolism could be exploited to distinguish HCC tissues from normal liver tissues. Our imaging data based on metabolite profiles may also be used as molecular hallmarks for improved diagnosis and prognosis of HCC, and for monitoring of response to drug treatment.
EXPERIMENTAL
In Vivo Hyperpolarized 13C MRS Imaging
A total of 7 male buffalo rats (Charles River Laboratories International Inc., Wilmington, MA) were used in this study. All rats were studied under a protocol approved by Stanford University Administrative Panel on Laboratory Animal Care (APLAC). During single orthotopic implantation, a subxiphoid incision was made and a total of 1×106 Morris hepatoma McA-RH7777 cells (American Type Culture Collection, Manassas, VA) suspended in 100 μL of phosphate-buffered saline (PBS) were injected slowly (typically over 15–30 seconds) beneath the parenchyma of the middle lobe. A sterile cotton stick was applied for 1–2 minutes over the needle insertion site followed by application of about 100 μL of 70% isopropyl alcohol to the peritoneal cavity to prevent extrahepatic tumor formation. The incision was then closed in layers with absorbable surgical suture. During tumor growth, the tumor volume was estimated by measuring its dimensions on three orthogonal MR imaging planes (a, b, c) and using the formula: (π/6)× a×b×c (23). At 12 to 14 days post-implantation, tumor volume of 100 mm3 to 200 mm3 was considered appropriate for subsequent MRS analysis. Each rat was fed ad libitum with water and rodent formula 5P04 from LabDiet® (Brentwood, MO) up to the time of setup in the magnet. Experiments were performed on rats (~300 g) anesthetized with 2% isoflurane in oxygen at a flow rate of 1.5 L/min. An MR-compatible small animal monitoring and gating system (Model 1025, SA Instruments Inc., Stony Brook, NY) was used to measure and record physiological parameters of each rat, specifically respiration (66±5 breath/min), temperature (37.2±.1°C), pulse rate (396±30 beats/min), and oxygen saturation (96±3%). A custom-built dual-tuned (1H/13C) quadrature rat coil was used for both radiofrequency (RF) transmission and reception in a 3.0 Tesla clinical system (Signa™ MR Scanner, GE Healthcare, Waukesha, WI). An 8 molar 13C-urea phantom was placed alongside each rat for experimental calibrations. A mixture of [1-13C]pyruvic acid and trityl radical was polarized to approximately 23.7±1.7% using a DNP polarizer (HyperSense™, Oxford Instruments Molecular Biotools, Oxford, UK). After the mixture was polarized for 1 hour, it was rapidly dissolved using a Tris/EDTA NaOH buffer (pH=7.5±0.1). A 3 mL bolus injection of hyperpolarized [1-13C]pyruvate (80 mM) was administered via the tail vein over a 14 s interval followed by a 4 s saline flush to clear the 0.5 mL catheter dead volume. Based on calculation, the bolus injection resulted in a peak blood concentration of approximately 8 mM. Acquisition of MRSI data began 20 s after the start of bolus injection.
A 3D DSE-EPSI sequence was utilized, which allows a maximum in-plane spatial resolution of 3.4 mm with spectral bandwidth (BW) of 500 Hz to include the metabolite peaks. A 30 mm axial slab was excited to cover the rat liver. The EPSI gradient was applied in the right-left direction. Eighteen phase encoding steps were applied in the anterior-posterior (A/P) direction and eight in the superior-inferior (S/I) direction. The acquisition parameters were: matrix size = 26 × 18 × 8, FOV = 90 mm × 60 mm × 40 mm, spectral BW = 500 Hz, time points = 64 (spectral resolution = 7.8 Hz), flip angle = 8, TR = 245 ms, and total scan time = 35 s. Details of the 3D DSE-EPSI pulse sequence were previously published by Yen et al (24). Data reconstruction and analysis were performed using custom software written in Matlab (The MathWorks, Inc., Natick, MA, USA). The symmetric EPSI waveform was used as a weighting function in a gridding algorithm to effectively correct for phase among sample points in kx vs. time domain. Apodizations were performed in kx and ky with a Hamming filter and in time domain with a line broadening of 15 Hz. Then the data were zero-filled to 64 × 42 × 8 and fast Fourier transform was applied to reconstruct the spectroscopic images. B0 correction was performed for spectral peaks above noise threshold to align the spectral peaks across all voxels. After reconstruction, metabolite maps of [1-13C]pyruvate, [1-13C]lactate, and [1-13C]alanine were computed by integrating the area under respective peaks of each voxel’s magnitude (complex modulus) spectrum and overlay on T1-weighted proton images.
Quantitative Real-Time PCR Analysis
Following the MRSI examinations of 4 rats, rat liver tissues were harvested and immediately snap-frozen in cold acetone and stored at −80°C. About 30 mg of snap-frozen tissue was used from each rat specimen to extract mRNA. Total RNA enriched with mRNA was purified with RNeasy Mini Kit (Qiagen, Valencia, CA). All procedures were performed according to the manufacturers’ recommendations. Following a 1:50 dilution of samples, aliquots were pipetted into microplate wells and absorbance at 260 nm was measured using a PowerWave™ XS Microplate Spectrophotometer (BioTek Instruments, Winooski, VT). The quantity of RNA (ng/μL) was calculated using the Beer-Lambert equation. Reverse transcription (RT) using 500 ng of total RNA in a 10 μL reaction was performed using the Applied Biosystems RT Kit (Applied Biosystems, Foster City, CA) according to the instruction manual. Synthesized cDNA was then maintained at 37°C for 1 h, followed by 10 min at 95°C. cDNA was PCR-amplified in a 20-μL reaction using the TaqMan® Gene Expression Assay Kit and Master Mix Kit (Applied Biosystems, Forster City, CA) according to the instruction manual. Briefly, thermal cycling was performed using the Mx3000P real-time PCR system (Stratagene®, Santa Clara, CA), amplification rates were measured automatically, and the number of cycles needed to cross the threshold (Ct) determined. Each sample was assayed in triplicates for levels of LDH-A (Assay number Rn00820751_gl), ALT (Assay number Rn01538341_ml) and NQO1 (Assay number Rn00566528_ml). The mRNA from Morris hepatoma McA-RH7777 cells, used for tumor implantation, was used to generate standard curves for each enzyme assay. Ct values of respective enzymes were normalized to a reference/housekeeping gene (18S rRNA) as endogenous control. Negative controls included a sample without enzyme in the RT reaction and another sample without template during the PCR.
Immunohistochemistry
For the remaining 3 rats, liver tissues were harvested upon completion of the MRSI examinations and immediately fixed in 10% neutral buffered formalin, paraffin embedded and sectioned into 5-μm slices. Multiple histologic stains of Hematoxylin & Eosin (H&E), anti-CD31, and anti-Caspase-3, were performed according to standard protocols (Histo-tec Laboratory, Hayward, CA). The stained slides were submitted to a pathologist for histopathological evaluations. Digital images of the histological slides were acquired with a 20× objective lens at a light microscope, Nikon DXM1200F digital camera (Nikon, Japan).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 4.0a for Mac OS X (GraphPad Software, San Diego, California, USA). A two-tailed unpaired student’s t test was performed to verify the statistical significance of the means in the compared groups. A p-value ≤ 0.05 was considered statistically significant. All results are displayed as mean±SEM (standard error of mean) of measures in groups.
RESULTS
In Vivo Hyperpolarized 13C MRS Imaging revealed elevated levels of [1-13C]-lactate and [1-13C]-alanine in HCC
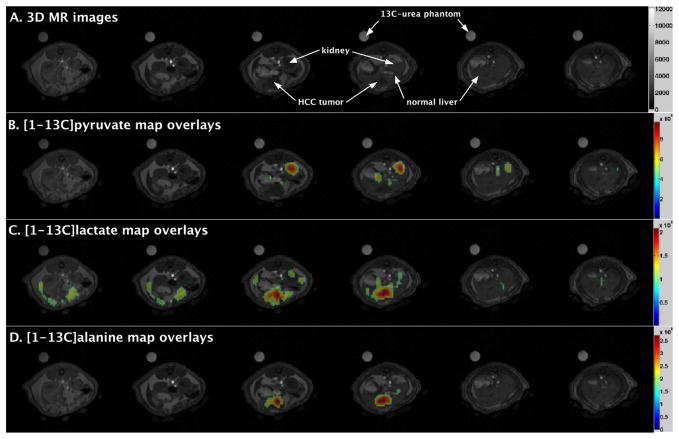
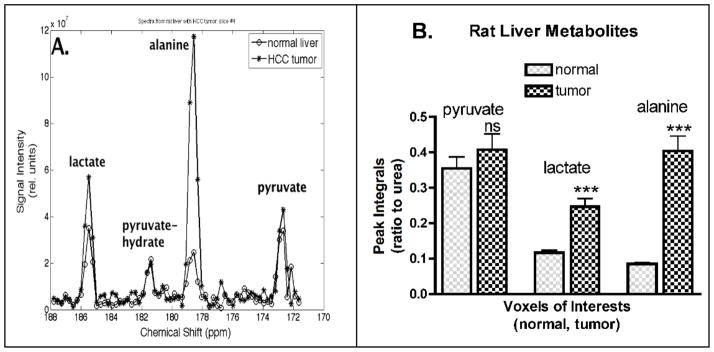
Metabolic maps of [1-13C]pyruvate, [1-13C]lactate, and [1-13C]alanine from a representative rat are shown in Figure 1. These maps are color-coded with values ranging from minimum (blue) to maximum (red) and normalized (0 to 1). The maps were set at a threshold value of 0.4 on a scale of 0 to 1. It is notable that both [1-13C]lactate and [1-13C]alanine maps are co-localized with the HCC tumor, and the tumor voxel values are greater than those of normal liver (Fig. 1). Spectra from reconstructed voxels (1.4×1.4×5 mm3) in tumor and normal liver are shown (Fig. 2A). The integral values of four reconstructed voxels (1.4×1.4×5 mm3) per region (tumor or normal) were tabulated across all 7 rats for statistical comparisons (Fig. 2B). Each metabolite peak integral was expressed as a ratio to the reference 13C-urea phantom, always placed alongside each rat. Difference in the means of normal liver and tumor were statistically significant for [1-13C]lactate (p < 0.0001) and for [1-13C]alanine (p < 0.0001), but not significant for [1-13C]pyruvate (p = 0.3428). Such comparable levels of [1-13C]pyruvate in tumor and normal liver may indicate that both tissues were well perfused with [1-13C]pyruvate. Also, the sum of [1-13C]pyruvate, [1-13C]lactate and [1-13C]alanine, representative of the total 13C-carbon signal was found to be significantly higher (p < 0.0001) in tumors (4.86±0.29 × 108) than normal rat liver (2.74±0.18 × 108).
Figure 1.
Representative metabolite maps of a rat liver bearing hepatocellular carcinoma (HCC) tumors showing [1-13C]lactate and [1-13C]alanine production after a bolus injection of hyperpolarized [1-13C]pyruvate. Metabolite maps were computed from the integral of metabolite peak for each voxel’s spectrum from 3D DSE-EPSI data. [A] 3D axial magnetic resonance images of a rat abdomen with HCC tumors in the liver, [B] [1-13C]pyruvate metabolic map overlays, [C] [1-13C]lactate metabolic map overlays, and [D] [1-13C]alanine metabolic map overlays.
Figure 2.
[A] Representations of in vivo hyperpolarized 13C MRS spectra of normal liver and HCC tumor obtained within the same rat. [B] Quantitative measures of 13C-metabolites peaks integrals plotted as bar graphs for 7 rats. All bar graphs are displayed as mean±SEM, and the p-values were evaluated by npaired t test (nsp-value > 0.05, *p-value ≤.05, ** p-value ≤ 0.01, *** p-value ≤ 0.001).
Quantitative Real-Time PCR Analysis indicated higher gene expressions of LDH-A, NQO1, and ALT in HCC tissues
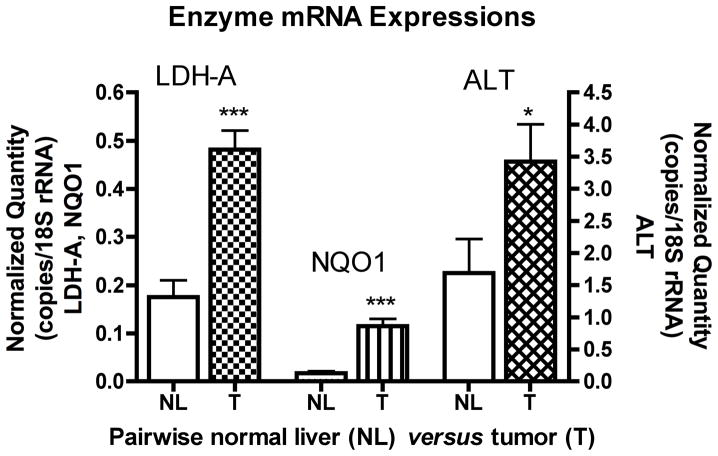
To account for the observed elevated levels of metabolites in HCC tumors, we used quantitative real-time PCR to measure the expression levels of putative enzymes (LDH-A, NQO1, and ALT) associated with the metabolism of [1-13C]pyruvate. Measurements were made in triplicates for each specimen (tumor or normal liver) from four rats. All three enzymes, LDH-A (p < 0.0001), NQO1 (p < 0.0001), and ALT (p = 0.0173) were significantly elevated (p ≤ 0.05) in HCC tumors compare to normal liver tissues (Fig. 3). We anticipated the levels of NQO1 to be in accordance with LDH-A, because NQO1 is an enzyme that produce NADH, and NADH is used in the LDH-A catalyzed reaction of [1-13C]pyruvate to [1-13C]lactate. Although the levels of ALT (Fig. 3) were significantly greater in tumor than normal liver (p = 0.0173), the degree of significance is less than that of the [1-13C]-alanine metabolite (p < 0.0001) (Fig. 2).
Figure 3.
Pairwise bar graphs of enzyme mRNA expression from normal liver and tumor tissues in control and treated cohorts. All bar graphs are displayed as mean±SEM, and the p-values were evaluated by unpaired t-test (*p-value ≤ 0.05, ** p-value ≤ 0.01, *** p-value ≤ 0.001). LDH-A, lactate dehydrogenase A; NQO1, NAD(P)H dehydrogenase quinone 1; ALT, alanine transaminase; NL, normal liver; T, tumor.
Immunohistochemistry confirms a nodular HCC displaying malignant characteristics
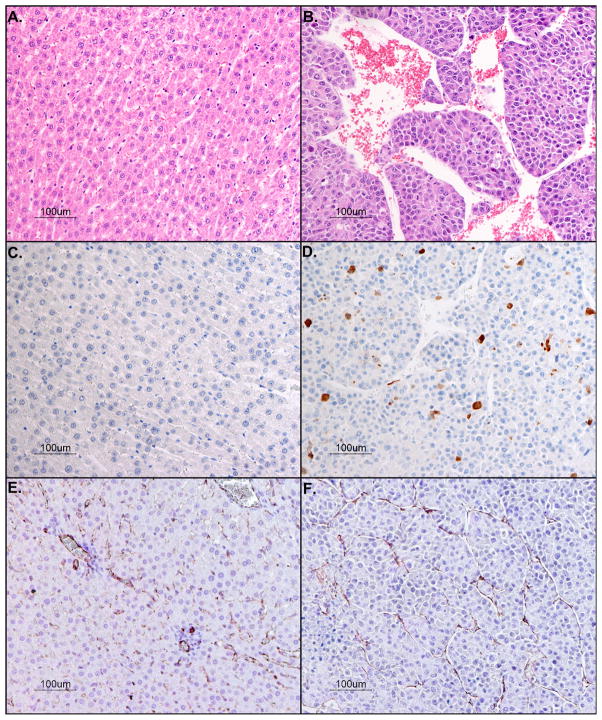
Histopathologic examinations were performed on H&E stained liver tissues (Fig. 4B). Multiple sections of liver are examined, revealing multiple demarcated nodular masses within the middle lobe of the liver. The masses are comprised of haphazard, 2- to 8-cell thick trabeculae and solid lobules of cells with scant vascular stroma. The cells are polygonal with defined cell borders enclosing small amounts of eosinophilic cytoplasm. The nuclei are irregularly round with dispersed stippled chromatin and single, prominent nucleoli. An average of 7 mitoses per 400X field (range from 4 to 12) were observed in tumors. Anisocytosis and anisokaryosis are markedly noticeable (> 3-fold each). Few apoptotic bodies are noted scattered throughout the masses. Multifocal, random, discrete areas of coagulation necrosis are noted throughout the nodules. There are few but distinct areas of local invasion by the nodules into surrounding hepatic parenchyma, and discrete solid aggregates of a few of the cells are noted within adjacent vascular spaces.
Figure 4.
Histopathologic analysis with hematoxylin and eosin stains of rat liver bearing hepatocellular carcinoma [A] normal liver and [B] tumor (20-fold magnification. Photomicrograph of anti-Caspase-3 positive stain for apoptotic cells [C] normal liver and [D] tumor (20-fold magnification). Immunostain of endothelial cells lining vessel walls with anti-CD31 (brown) and counterstained with hematoxylin for cell nuclei (blue) [E] normal liver and [F] tumor (20-fold magnification).
Sections immunostained with anti-Caspase-3 antibody (Fig. 4D) reveal strong (3+) specific brown cytoplasmic immunoreactivity in some of the cells of the nodular masses, at an average rate of 9 immunostained cells per 100 cells in tumors. Anti-Caspase-3 immunoreactivity is not noted in any cells in the normal liver (Fig. 4C) outside the nodular masses.
The vascular architecture of rat liver tissue was examined with anti-CD31 antibody (Fig. 4E-F). Sections immunostained with anti-CD31 antibody reveal presence of normal endothelial cells outside the nodular masses (Fig. 4E), as well as normal endothelial cells within the nodular masses (Fig. 4F). These endothelial cells are extremely flattened and elongated, displaying strong (3+) specific brown cytoplasmic immunoreactivity. The tumors exhibit more vascularization (as determined by CD31 immunostaining) (Fig. 4F) than normal liver (Fig. 4E).
DISCUSSION
We sought to investigate potential molecular hallmarks of an in vivo orthotopic HCC model. Our data suggest that the conversion of exogenous [1-13C]pyruvate to [1-13C]lactate and [1-13C]alanine is a characteristic marker of HCC in vivo. Coupled to this finding, the associated enzymes (LDH-A, NQO1, and ALT) are significantly elevated in HCC tumor as compared to normal liver. Histopathologic analysis of liver tissues revealed a nodular tumor with malignant features.
Previous reports of in vivo hyperpolarized 13C MRSI of tumors have implicated constitutive increase in [1-13C]lactate after a bolus injection of [1-13C]pyruvate(8–14). Similarly, we observed that [1-13C]lactate levels were significantly elevated and co-localized within the HCC tumor. The observed elevated levels of lactate may be attributed to exchange of [1-13C]pyruvate labeling with preexisting lactate pools, a net flux of [1-13C]pyruvate to [1-13C]lactate, or a combination of both. Regardless of whether this technique measures isotopic exchange or net flux, the enzyme responsible for the conversion is LDH-A. Our results indicate significantly higher levels of LDH-A mRNA in HCC tumor as compared to normal liver, suggesting that the elevated levels of lactate in the HCC tumor is ascribed to the increased LDH-A enzyme levels.
While [1-13C]-lactate might be a main product of aerobic glycolysis, the significant [1-13C]alanine production observed in our study is consistent with recent report of human colon and stomach cancers (6) and rat HCC tumors (14), even though metabolic imaging was not implemented at the time. We determined that these elevated metabolites in HCC tumor could be ascribed to the density of enzymes associated with the metabolism of [1-13C]pyruvate. Despite incomplete understanding of the underlying in vivo mechanisms by which the [1-13C]pyruvate to [1-13C]alanine metabolism supports HCC growth, our study identified metabolic signatures in HCC and their relation to putative enzymes levels. Furthermore, extended investigations into these metabolic signatures may help to unravel the mechanisms by which they support HCC growth and to identify promising targets for intervention.
We measured mRNA levels of enzymes involved in pyruvate metabolism in our HCC tissues, so as to infer enzyme activities based on their expression levels. Our results indicate that the levels of LDH-A, NQO1, and ALT are consistently elevated in orthotopic HCC and that the [1-13C]pyruvate metabolism of these tumors appears concomitant with the in vivo changes in the glycolytic pathway connected to the production of [1-13C]lactate and [1-13C]alanine. These crucial enzymes, LDH-A, NQO1, and ALT, are highly elevated in the orthotopic HCC tissues, and the glycolytic capacities of HCC tumors are likely dependent on the up-regulation of these enzymes. Our observed elevated LDH-A level is in concordance with increased LDH activity in mouse tumors and cancer cells (3,10,12). This increased level can be linked to the elevated lactate levels in this orthotopic model, which was similarly demonstrated in cancer cells (3,12). NQO1 is a prevalent cytosolic flavoenzyme that catalyzes two-electron reduction of various quinones, with NADH as an electron donor (25,26). This NQO1-mediated reduction mechanism may be responsible for scavenging superoxides anions generated during oxidative stress as a cellular defense against various toxic quinones. Significant expressions of NQO1 have been reported in response to liver damage or primary biliary cirrhosis in human liver (27) or rat liver (28), and NQO1 expression is altered in a plethora of cancers cells (29). Also, NQO1 plays a key role in regulating the p53 tumor suppressor gene, a labile protein that induces either growth arrest or controlled apoptosis (30,31). Histologically, the HCCs in rats revealed higher mitotic activity and apoptotic rate. Since NQO1 regulates the p53 gene (30,31), the apoptotic features only localized to the HCC nodule may be attributed to increased NQO1 levels in these tumors. Thus, NQO1 may play varied and important roles in cancers, and may have diagnostic and/or therapeutic potential.
ALT, mostly found in liver, is specific for catalyzing an exchange between pyruvate and alanine, and between 2-oxoglutarate and glutamate. Pyruvate conversion to alanine mostly occurs in the cytoplasm, because ALT expression in rat hepatoma cells is found predominantly in the cytoplasm with a small fraction in the mitochondria (32). We observed a significantly greater level of ALT in rat HCC tumor as compared to normal liver. This finding may be due to a prolific induction or transcription of ALT in HCC tumors. It has been noted that total tyrosine aminotransferase in many host livers and hepatomas were slightly elevated in rats fed a vitamin B6-deficient diet (22). During fasting, pyruvate dehydrogenase is inactivated in rat liver bearing hepatoma (33). This could be a potential reason that excess pyruvate gets shuttled through alternative metabolic pathways, like lactate or alanine. The inhibition of pyruvate dehydrogenase in our HCC model is unlikely, because all rats were fed ad libitum during the hyperpolarized 13C MRSI experiments.
This hyperpolarized 13C 3D MRSI technique is currently limited to only preclinical studies. Our study may have overestimated the physiological levels of metabolites because we administered an exogenous metabolic substrate that surpasses physiological levels. However, advanced techniques in mass spectrometry have demonstrated elevated levels of lactate and alanine in ex vivo human tumors at physiological levels (6). Despite these limitations, our findings strongly support the advent of novel in vivo imaging technologies that hold promise for improved diagnosis and prognosis of HCC.
In summary, HCC exhibits a characteristic increase in lactate and alanine production in vivo following a bolus infusion of hyperpolarized [1-13C]pyruvate that can be detected with non-invasive imaging. Concomitant up-regulation of the enzymes (LDH-A, ALT, NQO1) may explain the observed increases in metabolic products in HCC. Hyperpolarized 13C 3D MRSI is a potential diagnostic tool for detection of HCC and may become an important new imaging tool to measure surrogate markers or endpoints for drug treatment.
Acknowledgments
Financial Support: CA-09695, RR-09784, AA-018681, EB-009070, T.S. Kwok Foundation, AA-005965
The authors express special appreciation to Mr. Oliver Hsu and Mr. Evan Nunez for their superb caretaking of animals during the hyperpolarized 13C 3D MRSI experiments, and thank Dr. Richard Luong for his expertise in histopathologic analysis.
List of Abbreviations
- HCC
hepatocellular carcinoma
- TCA
tricarboxylic acid
- MRSI
magnetic resonance spectroscopic imaging
- NADH
nicotinamide adenine dinucleotide
- LDH
lactate dehydrogenase
- 3D DSE-EPSI
three-dimensional double-spin-echo echo-planar spectroscopic imaging
- PCR
polymerase chain reaction
- NQO1
NAD(P)H dehydrogenase quinone 1
- EC
enzyme commission
- ALT
alanine transaminase
- APLAC
Stanford University Administrative Panel on Laboratory Animal Care
- PBS
phosphate-buffered saline
- 1H/13C
dual-tuned proton and carbon
- RF
radiofrequency
- EDTA
ethylenediaminetetraacetic acid
- NaOH
sodium hydroxide
- FOV
field-of-view
- BW
spectral bandwidth
- mRNA
messenger ribonucleic acid
- rRNA
ribosomal ribonucleic acid
- cDNA
complementary deoxyribonucleic acid
- Ct
cycles needed to cross the threshold
- H&E
Hematoxylin & Eosin
- CD31
glycoprotein expressed on endothelial cells and in platelets
- SEM
standard error of mean
References
- 1.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular Carcinoma Incidence, Mortality, and Survival Trends in the United States From 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburg O, Posener K, Negelein E. Uber den Stoffwechsel der Carcinomzelle. Biochem Z. 1924;152:309–344. [Google Scholar]
- 3.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ. Is cancer a disease of abnormal cellular metabolism? New angles on a old idea. Genet Med. 2008;10(11):767–777. doi: 10.1097/GIM.0b013e31818b0d9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plathow C, Weber WA. Tumor Cell Metabolism Imaging. J Nucl Med. 2008;49(6):43S–63S. doi: 10.2967/jnumed.107.045930. [DOI] [PubMed] [Google Scholar]
- 6.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative Metabolome Profiling of Colon and Stomach Cancer Microenvironment by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Cancer Res. 2009;69(11):4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 7.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. PNAS. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golman K, in ’t Zandt R, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic Imaging by Hyperpolarized 13C Magnetic Resonance Imaging for In vivo Tumor Diagnosis. Cancer Res. 2006;66(22):10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 9.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen Y-F, Hurd RE, Tropp J, Bok R, Pauly JM, Nelson SJ, Kurhanewicz J, Vigneron DB. Hyperpolarized C-13 Spectroscopic Imaging of the TRAMP Mouse at 3T-Initial Experience. Magn Reson Med. 2007;58:1099–1106. doi: 10.1002/mrm.21256. [DOI] [PubMed] [Google Scholar]
- 10.Day SE, Kettunen MI, Gallagher FA, Hu D-E, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13(11):1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 11.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen Y-F, Nelson SJ, Vigneron DB, Kurhanewicz J. Hyperpolarized 13C Lactate, Pyruvate, and Alanine: Noninvasive Biomarkers for Prostate Cancer Detection and Grading. Cancer Res. 2008;68(20):8607–8615. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward CS, Venkatesh HS, Chuameil MM, Brandes AH, Criekinge MV, Dafni H, Sukumar S, Nelson SJ, Vigneron DB, Kurhanewicz J, James CD, Haas-Kogan DA, Ronen SM. Noninvasive Detection of Target Modulation following Phosphatidylinositol 3-Kinase Inhibition Using Hyperpolarized 13C Magnetic Resonance Spectroscopy. Cancer Res. 2010;70(4):1296–1305. doi: 10.1158/0008-5472.CAN-09-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kettunen MI, Hu D-E, Witney TH, McLaughlin R, Gallagher FA, Bohndiek SE, Day SE, Brindle KM. Magnetization Transfer Measurements of Exchange Between Hyperpolarized [1-13C]Pyruvate and [1-13C]Lactate in a Murine Lymphoma. Magn Reson Med. 2010;63:872–880. doi: 10.1002/mrm.22276. [DOI] [PubMed] [Google Scholar]
- 14.Yen Y-F, Le Roux P, Mayer D, King R, Spielman D, Tropp J, Pauly KB, Pfefferbaum A, Vasanawala S, Hurd R. T2 relaxation times of 13C metabolites in a rat hepatocellular carcinoma model measured in vivo using 13C-MRS of [1-13C]pyruvate. NMR Biomed. 2010;23(4):414–423. doi: 10.1002/nbm.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher FA, Kettunen MI, Day SE, Hu D-E, Ardenkjaer-Larsen JH, in’t Zandt R, Jensen PR, Karlsson M, Golman K, Lerche MH, Brindle KM. Magnetic resonance imagin of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 16.Keshari KR, Wilson DM, Chen AP, Bok R, Larson PEZ, Hu S, Criekinge MV, Macdonald JM, Vigneron DB, Kurhanewicz J. Hyperpolarized [2-13C]Fructose: A Hemiketal DNP Substrate for In Vivo Metabolic Imaging. J Am Chem Soc. 2009;131(48):17591–17596. doi: 10.1021/ja9049355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher FA, Kettunen MI, Hu D-E, Jensen PR, in ’t Zandt R, Karlsson M, Gisselsson A, Nelson SK, Witney TH, Bohndiek SE, Hansson G, Peitersen T, Lerche MH, Brindle KM. Production of hyperpolarized [1,4-13C2]malate from [1,4-13C2]fumarate is a marker of cell necrosis and treatment response in tumors. PNAS. 2009;106(47):19801–19806. doi: 10.1073/pnas.0911447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson M, Jensen PR, in ’t Zandt R, Gisselsson A, Hansson G, Duus JO, Meier S, Lerche MH. Imaging of Branched Chain Amino Acid Metabolism in Tumors with Hyperpolarized 13C Ketoisocaproate. Int J Cancer. 2010;127(3):729–736. doi: 10.1002/ijc.25072. [DOI] [PubMed] [Google Scholar]
- 19.Hu S, Chen AP, Zierhut ML, Bok R, Yen Y-F, Schroeder MA, Hurd RE, Nelson SJ, Kurhanewicz J, Vigneron DB. In Vivo Carbon-13 Dynamic MRS and MRSI of Normal and Fasted Rat Liver with Hyperpolarized 13C-Pyruvate. Mol Imag Biol. 2009;11(6):399–407. doi: 10.1007/s11307-009-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spielman DM, Mayer D, Yen Y-F, Tropp J, Hurd RE, Pfefferbaum A. In Vivo Measurement of Ethanol Metabolism in the Rat Liver Using Magnetic Resonance Spectroscopy of Hyperpolarized [1-13C]Pyruvate. Magn Reson Med. 2009;62:307–313. doi: 10.1002/mrm.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shonk CE, Morris HP, Boxer GE. Patterns of Glycolytic Enzymes in Rat Liver and Hepatoma. Cancer Res. 1965;25:671–676. [PubMed] [Google Scholar]
- 22.Reynolds RD, Morris HP. Effects of Dietary Vitamin B6 on the In Vitro Inactivation of Rat Tyrosine Aminotransferase in Host and Morris Hepatomas. Cancer Res. 1979;39:2988–2994. [PubMed] [Google Scholar]
- 23.Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor Measurement in the Nude Mouse. J Surg Oncol. 1986;31(4):229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 24.Yen Y-F, Kohler SJ, Chen AP, Tropp J, Bok R, Wolber J, Albers MJ, Gram KA, Zierhut ML, Park I, Zhang V, Hu S, Nelson SJ, Vigneron DB, Kurhanewicz J, Dirven HAAM, Hurd RE. Imaging Considerations for In Vivo13C Metabolic Mapping Using Hyperpolarized 13C-Pyruvate. Magn Reson Med. 2009;62:1–10. doi: 10.1002/mrm.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C, Simard-Duquesne N, Ernster L, Hoberman H. Stereochemistry of hydrogen-transfer in the energy-linked pyridine nucleotide transhydrogenase and related reactions. Biochim Biophys Acta. 1965;105(3):397–409. doi: 10.1016/s0926-6593(65)80226-0. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Bianchet MA, Talalay P, Amzel LM. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc Nat Acad Sci USA. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aleksunes LM, Goedken M, Manautou JE. Up-regulation of NAD(P)H quinone oxidoreductase 1 during human liver injury. World J Gastroenterol. 2006;12(12):1937–1940. doi: 10.3748/wjg.v12.i12.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arizono K, Sugiura S, Miyazato S, Takiguchi M, Ariyoshi T. DT-Diaphorase Induction by Lead Acetate inthe Liver of Rats. Bull Environ Contam Toxicol. 1996;57:41–46. doi: 10.1007/s001289900153. [DOI] [PubMed] [Google Scholar]
- 29.Winski SL, Koutalos Y, Bentley DL, Ross D. Subcellular Localization of NAD(P)H:quinone Oxidoreductase 1 in Human Cancer Cells. Cancer Res. 2002;62:1420–1424. [PubMed] [Google Scholar]
- 30.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. PNAS. 2001;98(3):1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsvetkov P, Asher G, Reiss V, Shaul Y, Sachs L, Lotem J. Inbition of NAD(P)H:quinone oxidoreductase 1 activity and induction of p53 degradation by the natural phenolic compound curcumin. PNAS. 2005;102(15):5535–5540. doi: 10.1073/pnas.0501828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparkes RS, Weiss MC. Expression of Differentiated Functions in Hepatoma Cell Hybrids: Alanine Aminotransferase. Proc Nat Acad Sci USA. 1973;70(2):377–381. doi: 10.1073/pnas.70.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fields ALA, Wolman SL, Cheema-Dhadli S, Morris HP, Halperin ML. Regulation of Energy Metabolism in Morris Hepatoma 7777 and 7800. Cancer Res. 1981;41:2762–2766. [PubMed] [Google Scholar]