Abstract
The main mediator of the lipopolysaccharide (LPS) response in macrophages is activation of Toll-like receptor 4 (TLR4). This generates interferon-beta (INFbeta) production that stimulates increased expression of the RNA editing enzyme ADAR1. To determine if there is an increase in RNA editing in mature miRNA in response to TLR4 activation upon Salmonella infection of macrophages we analyzed small RNA deep sequencing data. Interestingly, we found that direct infection of macrophage cell lines with Salmonella does not result in an increase of edited mature miRNA. Thus, despite elevated levels of ADAR1 during TLR4 activation of macrophages mediated by Salmonella infection, ADAR1 does not result in redirection of miRNA. The most common consequence of ADAR activity on miRNA is a reduction in the mature miRNA level due to interference with miRNA processing of pri-miRNA. However, we found very few miRNAs with reductions in level, and no significant difference between miRNAs previously reported to be edited and those reported to be not edited. In particular, we did not see significant decrease in mir-22 and mir-142, nor editing of pri-mir-22 or pri-mir-142 in infected RAW macrophages. Thus, ADAR1 has very little, if any, effect on the miRNA machinery following TL4 activation by Salmonella infection.
Key words: RNA editing, miRNA, ADAR, deep sequencing
Introduction
In mammals, ADAR1 and ADAR2, members of the adenosine deaminase family that act on double-stranded RNA (dsRNA), have been demonstrated to deaminate specific adenosines in miRNAs.1–6 This deamination usually results in a reduction in the mature miRNA levels as RNA editing and binding of ADARs can inhibit processing of pri-miRNA.7 RNA editing has also been shown to redirect miRNA activity when the editing event is located in the mature miRNA sequence.3
Intuitively, when ADAR activity increases there is an expectation of more frequent editing of miRNAs. It is well documented that ADAR1 is a ubiquitously expressed 110 kDa protein that is predominantly nuclear. However upon interferon induction a larger isoform of 150 kDa is expressed that shuttles between nucleus and cytoplasm (reviewed in ref. 8) and mice that are null for ADAR1 exhibit high levels of interferon suggesting that there may even be a feedback loop between ADAR1 and interferon levels.
In this study we wanted to determine if an increase in ADAR1 levels due to interferon would affect mature miRNAs. We chose to study Salmonella infection as Samuels and colleagues have shown that mice infected with Salmonella have a large induction of ADAR1 transcript levels.9 The increase is due to the induction of an interferon inducible isoform of ADAR1.10 Furthermore, Rabinovici and colleagues demonstrated that editing activity of ADAR1 rises dramatically upon alveolar macrophage activation by LPS.11 Thus, we wanted to ascertain if there was an increase in editing of mature miRNA upon Salmonella infection of macrophages. We also wanted to determine whether the consequences would be redirection or inhibition of pri-miRNA processing.
This work also addresses the role ADAR1 may have in innate immunity. In many physiological pathways miRNAs are prominent players, and evidence is accumulating that miRNAs are regulated as part of the innate immune response. Notably, LPS simulation of macrophages alters the levels of mir-146a,12 mir-155,13,14 and mir-125b.14 An increased level of mir-146 has been directly linked to a reduction of IRAK1 and TRAF6, two important members of the TLR signaling pathway.12 Conversely, a decrease in mir-125b was hypothesized to be linked to changes in TNFalpha levels, as TNFalpha is a target gene for mir-125b,14 indicating that decreases in miRNA levels can also be important. These studies demonstrate the important role miRNAs have in the regulation of the innate immune response and motivate the examination of how interferon induction of ADAR1 may contribute miRNA regulation.
Finally, Salmonella enterica serovar Typhimurium (henceforth Salmonella) is a model pathogen that invades macrophages and activates a TLR4-dependent host response.15 Mutation of the SPI-1 virulence region renders the bacteria unable to invade the host cell.16 Thus, by comparing infection with the SPI-1 mutant and wild-type Salmonella, we can determine if mature miRNA editing events are a result of contact with Salmonella or are infection dependent.
Due to the expansion in sequencing libraries, much effort is being directed towards how to correctly measure the sequence content of these libraries, including how to measure editing of small RNAs. The deamination of adenosine produces inosine, which base-pairs like guanosine, and is read by reverse transcriptase as guanosine.17 Thus, in sequencing small RNA libraries a replacement of adenosine with guanosine can be evidence of ADAR activity. Most analysis approaches utilize a modified blast algorithm, with the goal to match as many reads as possible to the genome. However, to map the largest number of reads possible the stringency of sequence homology must be reduced. This can lead to artifacts in the assignment of reads to genomic loci that are interpreted as editing events.18 Therefore we use a perfect matching approach to ascertain the level of editing in mature miRNA in Salmonella infection of macrophages, where correct identification (specificity) is more critical then a large number of matched reads (sensitivity).
In this study we analyzed small RNA libraries produced by infection of RAW, HeLa, THP-1, J774 and HEK cell lines with different strains of Salmonella. The editing levels of mature miRNA were determined with our simplified matching algorithm. Our study revealed that potential A to I editing levels do not rise in mature miRNA in macrophages, despite induction of ADAR1 by Salmonella infection. Further, we examined two miRNA transcripts reported to be edited by ADAR1 and did not observe an increase in editing. Overall, our results support the conclusion that inosine is not a common modification present in mature miRNA. This suggests that the miRNA machinery is resistant to increased ADAR activity caused by TLR4 activation.
Results
Sequence analysis of miRNAs from infected and non infected macrophage cell lines.
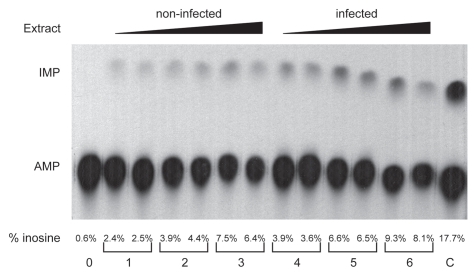
Initially we wanted to verify that ADAR1 was present in RAW macrophage cells and that it was induced in RAW macrophages infected with Salmonella. We choose a 24 h time point as it has been reported that ADAR1 induction peaks approximately 5 h after injection of mice with LPS,19 so we reasoned that if miRNAs were edited then 24 h post infection would be the optimum time to analyze them. We found that 24 h post infection the ADAR1 RNA level increased approximately 2 fold, ADAR1 protein level determined by SILAC increased approximately 3.7 fold20 (data not shown) whereas non-specific editing activity increased approximately 1.5 fold (Fig. 1).
Figure 1.
dsRNA deaminase activity assay. The lanes are 0 extract, (1) 5 µg extract from non-infected cells, (2) 10 µg extract from non-infected cells, (3) 20 µg extract from non-infected cells, (4) 5 µg extract from infected cells, (5) 10 µg extract from infected cells, (6) 20 µg extract from infected cells, (C) positive control with recombinant hADAR2. The upper limit of editing was 10 percent of the input dsRNA signal. Quantization was performed by measuring the signal of the inosine and adenosine spots. Each sample was done in duplicate and the experiment performed twice with similar result. There was more editing activity in the infected RAW macrophages than in the control non-infected macrophages.
Next we examined seventeen small RNA libraries of several macrophage cell lines infected with mutant and wild-type strains of Salmonella. The SP1 Salmonella mutant is known to block entry of the bacteria into macrophages. Thus, using the SP1 mutant we can separate the effect on the host in terms of bacterial contact and infection. The macrophage cell types used were, RAW, THP1 and J774, each has distinct origins. RAW macrophages were derived from mice infected with Ableson murine leukemia virus.21 THP1 cells are a human acute monocytic leukemia cell line from a patient tumor.22 Whereas, J774 are a murine macrophage cell line established from a tumor.23
Examinations of the libraries yielded on average a surprising 40% mapping of reads to perfect matches of the mature miRNA annotations in mirBase (Table 1 summary of RAW and HeLa cell lines. Suppl. Tables provide individual miRNA results. Very similar results were obtained for THP1 and J774 and HEK 293, data not shown). Extending the counts of matched reads to include potential A to G, G to A, C to U, U to C and U to A base changes, slightly increased the number of reads identified as miRNA. A to G mismatches between the miRNA sequence database and the sequences are interpreted as evidence for adenosine deamination, as inosine is recognized as guanosine by polymerases.
Table 1.
Summary of analysis of RAW and Hela sequence libraries using simplified perfect match algorithm
| RAW | Total | Perfect match | Reads with A to G | Reads with G to A | Reads with C to U | Reads with U to C | Reads with U to A | Library information |
| 0 hr control non-infected | 15325 | 12702 | 80 | 2431 | 11 | 3 | 98 | control non-infected |
| 24 hr control non-infected | 15137 | 12775 | 69 | 2158 | 2 | 3 | 130 | control 24 hours |
| 24 hr SPI1 mutant | 16461 | 13728 | 339* | 2302 | 21 | 2 | 69 | 24 hr infection with SPI-1 mutant |
| 24 hr WT infection | 13772 | 11329 | 381* | 1956 | 6 | 1 | 99 | 24 hr infection with wild-type |
| Hela | Total | Perfect match | Reads with A to G | Reads with G to A | Reads with C to U | Reads with U to C | Reads with U to A | Library information |
| 0 hr control non-infected | 21106 | 19044 | 126 | 1894 | 11 | 2 | 29 | control non-infected |
| 24 hr control non-infected | 23135 | 20800 | 70 | 2196 | 34 | 9 | 26 | control 24 hours |
| 24 hr WT infection | 18088 | 16478 | 61 | 1499 | 18 | 7 | 25 | 24 hr infection with wild-type |
Number in box is the number of reads.
The percent of A to G reads for each miRNA does not change, the increase in number of A to G reads is due to an increase in mir-155.
Initial observations indicate that only one miRNA contained potential A to I editing events; mir-155. This miRNA dramatically increased in RAW cells upon exposure to the SP1 and wild-type strains of Salmonella. The response to the SP1 mutant is particularly revealing as this mutant cannot invade macrophages but does contain lipopolysaccharides in its membrane. Initially, we observed that 14% of reads mapping to mir-155 were potentially A to I edited at 24 h post RAW cell exposure to Salmonella. The number of reads for mir-155 was 337, therefore the number of edited mature mir-155 reads would be a significant contributor to the miRNA pool. Supplementary Tables 1 and 2 contain the number of reads for individual miRNA. As can be seen 337 reads would place edited mir-155 at levels comparable to the eight most abundant miRNA for this library.
However, when we re-examined the potential G to A transversions, we observed that the nucleotide just 3′ of the A to G change in mir-155 had a significant number of G to A mismatches as well. There is no known enzyme that can catalyze a G to A transversion in RNA however RNA polymerase stuttering has been shown to insert more than one G when there are multiple Gs.24 Further, when we examined the percent of A to G reads for mir-155 across the RAW cell libraries we found that the percent does not change in response to infection. Indeed, the percent editing of mir-155 is the same in each of the libraries associated with a specific cell type. Additionally, we found that the primary mir-155 transcript is not edited either in non-infected nor infected macrophages (data not shown). Though the possibility exits that pre-mir-155 is edited, we conclude that the potential editing of mir-155 is a systematic error or artifact, in sequencing, dependent on the sequence context. Therefore we found no evidence of A to I editing in mature miRNA in the libraries.
ADAR1 does not inhibit processing of miRNAs.
The general lack of A to I editing indicates that Salmonella infection did not result in ADAR1 editing of mature miRNA, and hence redirection of miRNA. To explore this result further, we examined the potential for ADAR1 to be involved in reduction of miRNA levels by inhibiting processing. Comparing the levels of miRNA before and after infection with Salmonella revealed that very few miRNA had a reduction of even 2-fold (Fold Changes are in Suppl. Tables 1 and 2). Further, we analyzed sixty-five miRNA previously reported to be edited and there is no significant difference between alterations in the levels of miRNA reported to be ADAR substrates and miRNA not known to be ADAR substrates. Figure 2 illustrates this point by showing that the absolute fold-changes in miRNA abundance are low and that both the average and spread of the fold-changes is similar. This indicates that ADAR1 is not participating in a global reduction in miRNA levels, and suggests that miRNA previously reported to be edited are no more likely to be reduced in level than those reported not to be edited.
Figure 2.
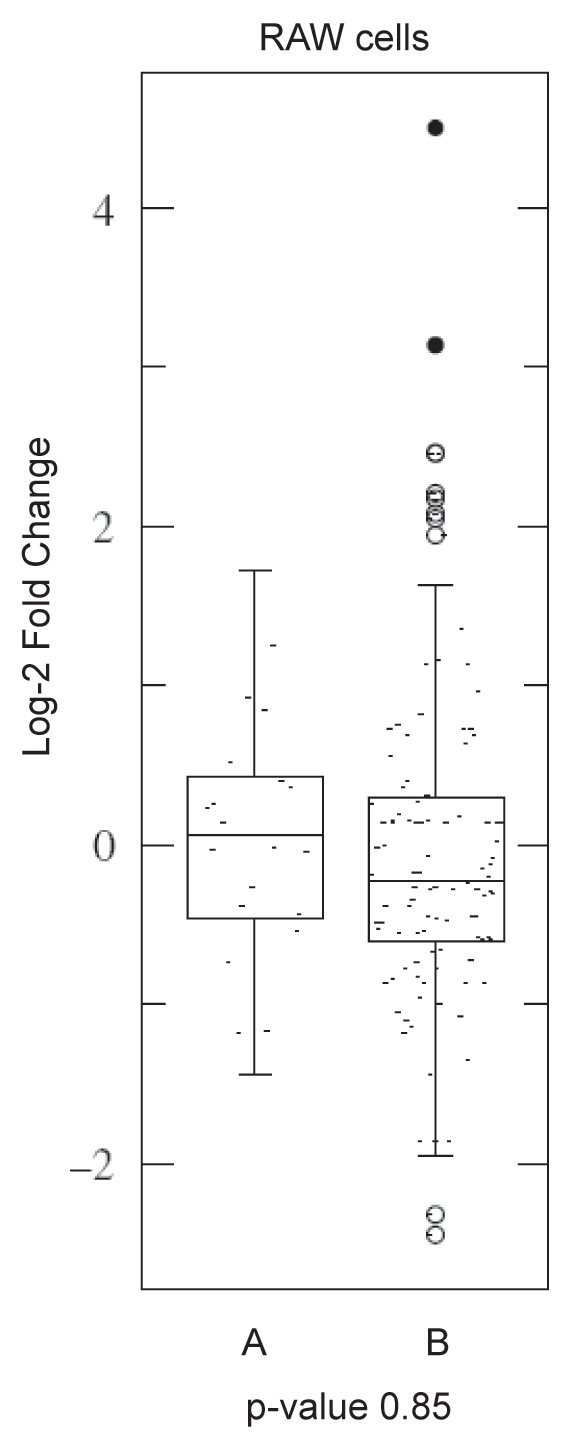
Data Scatter Box plots showing similarity of mature miRNA levels between miRNA reported to be editing substrates and those not edited. The p-value indicates the similarity between the distribution of the miRNA previously reported to be edited (marked “A”) and the miRNA not reported to be edited (marked “B”). THP1 and J774 macrophage cell lines exhibited similar trends (with p-values of 0.44 and 0.17 respectively). Very few miRNA show a two-fold decrease in levels and there is no significant difference between the edited and non-edited miRNA. This indicates that the miRNA previously found to be edited are no more likely to have a reduced level than non-edited miRNAs.
Additionally, we examined pri-miRNA sequences for two well-documented ADAR substrates, mir-22 and mir-142.1,25 We did not find evidence of upregulation of editing of the pri-miRNA for mir-22 and mir-142 after Salmonella infection, and the mature miRNA levels do not decrease significantly in RAW macrophages. The lack of editing of the pri-miRNA in RAW macrophages indicates that alteration of mir-142 is not dependent on ADAR activity on the pri-miRNA, and the increase in mir-22 further supports the assertion that induction of ADAR1 after infection with Salmonella does not lead to a strong negative effect of ADAR1 on miRNA biogenesis, even for known editing substrates.
Discussion
Given that ADAR1 expression and activity is inducted by interferon, it is surprising that Salmonella infection did not result in an increase of edited mature miRNA. As part of the innate immune response TLR4 is stimulated by LPS.26 Recently it was shown that TLR4′;s MyD88 dependent and independent signaling pathways can be separated into membrane bound and cytoplasmic TLR4 signaling.27 The MyD88 independent signaling pathway results in the activation of IRF-3 and production of β-interferon and it has been demonstrated that many effects attributed to the MyD88 independent pathway are due to the autocrine action of β-interferon. Similarly, TLR3, 7 and 9 induce production of β-interferon.27 ADAR1 has an interferon responsive promoter that responds to the presence of β-interferon. Importantly, an increase in ADAR1 activity was observed in alveolar macrophage stimulated by LPS11 directly linking ADAR1 activity with TLR4 activation. Here we observed that RAW macrophages also have increased dsRNA deaminase activity after infection with Salmonella. Interestingly, this did not lead to increased production of A to I edited mature miRNA. Thus, given that the number of mature miRNA with potential A to I editing remains constant, we propose that increased activity of ADAR1 due to TLR mediated innate immune response does not involve redirecting miRNA.
A possible explanation is that A to I editing of miRNA inhibits the biogenesis of mature miRNA. In fact, one of the most common features of A to I editing of miRNA is inhibition of processing by editing of the primary miRNA transcript. However, in our study few miRNA have decreased levels of mature miRNA upon infection. This suggests that reduction in mature miRNA levels is not a general feature of Salamonella infection of RAW macrophages. Significantly, we did not see editing of pri-mir-22 and pri-mir-142, two miRNA shown to be edited by ADAR1.1,25 Primir-22 has been found to be edited in various tissues including lung and brain. However, mir-142 is edited in T-cells, although no physiological role could be found for this editing. This highlights the potential for cell-type specific editing, which could explain the lack of editing in macrophages. Our results support the conclusion that ADAR1 does not interfere with the production of these prominent, edited-miRNA in macrophages. Thus, the increase in ADAR1 in response to interferon is for some other purpose than increasing the interactions between ADAR1 and miRNA.
Materials and Methods
Salmonella infection and culture of macrophages.
RAW264.7, HeLa, THP-1, J774 and HEK293 cells were grown in RPMI 1640 (Gibco) supplemented with 10% foetal calf serum (Biochrom), 2 mM L-glutamine (Gibco), 1 mM sodium-pyrovate (GIBCO) and 0.5% β-mercapto-ethanol (Gibco) in a 5% CO2, humidified atmosphere, at 37°C. Salmonella enterica serovar Typhimurium strain SL1344 was used as wild type throughout this study. The ΔSPI1 (JVS-0405) or ΔSPI2 (JVS-1103) strains with deletions of Salmonella Pathogenicity Islands 1 or 2 were provided by S. Pätzold28 or Karsten Tedin, respectively.29 The ΔSPI1/ΔSPI2 strain (JVS-3614) was constructed by phage P22 transduction of strain JVS-0405 with a lysate of strain JVS-1103.
One day prior to infection, 4 × 105 of host cells were seeded into 6-well plates. Overnight cultures of bacteria were diluted 1:100 in fresh L-broth medium and grown aerobically until an OD of 2. Bacteria were harvested by centrifugation and resuspended in complete RPMI medium. RAW 264.7 cells were infected at MOI of 1, and HeLa cells at MOI of 10. After addition of bacteria, the cells were centrifuged at 250x g for 10 min at room temperature followed by 20 min incubation in 5% CO2, humidified atmosphere, at 37°C. Medium was then replaced for complete RPMI containing gentamicin (50 µg/ml) to kill extracellular bacteria. Following 30 min incubation, cells were supplied with new complete RPMI containing 10 µg/ml gentamicin for the remainder of the experiment.
RNA library preparation.
The RNA libraries were prepared as previously described.30
Sequencing library editing analysis.
A PERL script was written to implement a simple matching approach to determine the percentage of editing in reads corresponding to a given miRNA (The script is available upon request from B. Heale; Email: bheale@gmail.com).
In most applications, sophisticated probabilistic alignment programs have better performance scores than perfect matching approaches. This is because, when using small data sets, the former use more of the available sequences than the latter. Thus, probabilistic alignment programs can include sequence artifacts produced by the experimental methodology (including base changing and insertion/deletion of nucleotides), and they use a model where multiple biologically relevant sequence alterations occur in the same sequence. In contrast, utilizing perfect matching implicitly ignores the majority of methodological artifacts, and thereby reduces greatly the number of false positive results. Thus, perfect matching is well-suited to very large data-sets.
We wanted to use large data sets generated by 454-sequencing and avoid artifacts produced by sequencing errors. Thus, we employ a perfect matching approach due to its extremely high stringency. For a sequence from the library to be used, it must have perfect, contiguous (20–23 nt) match to sequence annotations for mature miRNA from mirBase. As we are interested only in A to G changes, the only alteration allowed was potential A to G changes. In doing so, we are assuming that A to G changes do not co-occur with other sequence alterations as there is no data to suggest that A to G changes occur with other modifications in mature miRNA.
Our sequence analysis method begins by finding perfect matches between the mirBase annotations of mature miRNA,31–33 counting them and removing them from the library of reads. Next, a pattern is constructed where each edited base is replaced by a qualifier that allows specific mismatching. For example, when looking for the A to I editing of the sequence GCC ATT GAG G, the pattern GCC(A or G)TTG(A or G)GG is constructed. The pattern is then used to find reads that are potential editing reads of the particular miRNA. These reads are counted. When the entire library had been investigated, the output is sent to two files (comma-space-delimited for readability in Excel).
The results file contains the miRNA name, number of total matches and number of edited matches. The output file contains the sequence of the miRNA and the sequence of any edited reads along with the number of reads representing the sequence.
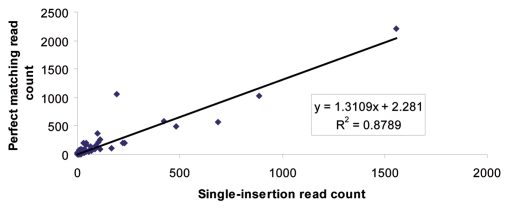
Sequence analysis that permits single mismatches, deletions and insertions locates a higher number of reads than matching approaches. To explore our sequencing libraries further, we added an additional level of analysis to consider the reads not mapped by our perfect matching methodology. As part of the methodology of Berninger and colleagues,34 used to produce the mappings for the mammalian miRNA expression atlas,35 single nucleotide insertions were allowed in mapping sequencing library reads. Thus, we looked at the number of additional sequences obtained by allowing single nucleotide insertions. When single nucleotide insertions were allowed the number of reads mapped to mature miRNA for control RAW cells and 24 h WT Salmonella infected RAW cells rose by 17% and 16% respectively. However the numbers of edited sequences did not rise. Strikingly, the number of additional reads mapped to each miRNA did not change the relative level of a particular miRNA in comparison with other miRNA. This is illustrated by the good correlation between the numbers of reads mapped to each miRNA (Fig. 3). This means that the relative level of each miRNA remains constant and is independent of the inclusion of reads with potential single-nucleotide insertions. Thus, the perfect matching approach is sufficient to determine relative miRNA levels.
Figure 3.
Correlation between the numbers of perfect match reads mapped to mature miRNA and the number of reads mapped by allowing single-nucleotide insertions. The read number for the insertions excludes the read number for perfect matches. The strong correlation in reads mapped to specific miRNA indicates that the perfect matching approach is sufficient to capture the rank and relative abundance of mature miRNA levels.
The number of reads matched was also analyzed by using shortened versions of the mature miRNA sequences. The motivation behind using shortened sequences is that it has been observed that Drosha and Dicer sometimes cleave +1 or −1 to the annotations in mirBase. Thus, we trimmed the first and last nucleotides of the mirBase annotations and searched for matches among the reads not matched to perfect matches nor to single-nucleotide insertions. This resulted in an increase in the number of reads mapping to mature miRNA. For example, the control RAW cell library had an additional 6% more reads and the 24 h WT infected RAW cell library had an additional 10% more reads mapped to mature miRNA.
However, as expected, when the shorter sequences were used to query the sequencing reads, a decrease in stringency was observed. The numbers of reads mapping to G to A, U to C and U to A mismatches were exceedingly increased, rising from levels of one or two to nearly a hundred. Therefore, if one takes G to A and U to C as sequencing errors then the use of the shortened mature miRNA, while allowing a larger number of reads to be mapped with homology to mature miRNA sequences, was more prone to artifacts that appeared as potential RNA editing events. Thus, we recommend caution when reducing the length of homology between a read and mapping site. Overall, the additional reads did not reveal an increase potential A to I editing events, outside the 3′ most nucleotide, again supporting the result of the perfect matching approach.
The importance of assigning reads to family members for analysis of editing was recently demonstrated by deHoon and colleagues.18 The authors demonstrated that due to similarity among mature miRNA families it is necessary to examine perfect matching of miRNA prior to using a probabilistic approach. Here we began with finding and removing, reads with perfect matches to all known miRNA. Next we looked for potential editing events. Thus, we were not therefore concerned about spurious matches due to homology between family members as each family member had already been analyzed separately for its occurrence.
A summary of the results of the number of mapped reads for each library is presented in Table 1 and Supplementary Table 1–2 contain the number of reads mapped to particular miRNA, including potential A to G, G to A, C to U, U to C and U to A editing. Across the libraries there was a definite lack of reads mapping to A to G editing events or C to U editing events. Strikingly there was a large number of G to A changes. When these are examined closely the G to A changes are located at the last nucleotide of the miRNA. For example, in the library for RAW cells infected with wild-type Salmonella for 24 hours, there are 1,956 reads with G to A mismatches. 1,693 of these are in the last nucleotide of the mature miRNA (87%). These probably represent artifacts of the sequencing methodology as addition of a poly-A tail was used during library preparation.
miRNA read level analysis.
Prior to comparison, read numbers were normalized between libraries of the same cell line by average normalization. Supplementary Tables 1 and 2 shows the fold-change and the Log-2 fold-change between the levels of miRNA 24 h after infection and control cells. Students-t test and data box plots reported in Figure 2. Sixty-five miRNA previously reported to be edited came from.1,4,5,25,35 For analysis, only those miRNA with at least one read in the control library were used.
Deaminase activity assay.
Non-specific deaminase assay was performed as previously reported.36 The extract was made from ten million RAW macrophage cells that had been either infected or not infected for 24 h with wild-type Salmonella. The extract was prepared by homogenizing 10 × 10-6 cells in 50 mM Tris pH7.9, 5 mM EDTA, 200 mM KCl, 10% glycerol, 1 mM DTT, 1 mM PMSF, 0.7 µg of pepstatin per ml, 0.4 µg of leupeptin per ml. The cells were homogenized for one minute on ice, followed by three minute incubation on ice, a second round of one minute homogenization was then performed. The extract was centrifuged and the supernatant was removed and the protein quantified with Bradford solution. The editing assay was performed twice and samples done in duplicate.
Primary miRNA (pri-miRNA) editing analysis.
As reported previously,7 primers were chosen to anneal 50 to 100 nts upstream (forward) or downstream (reverse) of the mirBase pre-miRNA annotation. PCR was used to amplify the pri-miRNA. Traditional sequencing was performed with the forward primers and confirmed using the reverse primers. Chromatograms were analyzed to determine if editing occurred.
Acknowledgements
M.O.C. was supported by funding from the Medical Research Council (U1275.1.5.1.1). B.H. was supported by a Fellowship from the Marie Curie Foundation.
Abbreviations
- LPS
lipopolysaccharide
- TLR4
toll-like receptor 4
- INFbeta
interferon-beta
- ADAR
adenosine deaminase that act on RNA
- dsRNA
double-stranded RNA
- salmonella
Salmonella enterica serovar typhimurium
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/13269
Supplementary Material
References
- 1.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. Rna. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, et al. RNA editing of human microRNAs. Genome Biol. 2006;7:27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohman M. A-to-I editing challenger or ally to the microRNA process. Biochimie. 2007;89:1171–1176. doi: 10.1016/j.biochi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heale BSE, O'Connell MA. Biological Roles of ADARs. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin: Landes Bioscience; 2009. pp. 243–258. [Google Scholar]
- 9.Shtrichman R, Heithoff DM, Mahan MJ, Samuel CE. Tissue selectivity of interferon-stimulated gene expression in mice infected with Dam(+) versus Dam(−) Salmonella enterica serovar Typhimurium strains. Infect Immun. 2002;70:5579–5588. doi: 10.1128/IAI.70.10.5579-5588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George CX, Wagner MV, Samuel CE. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J Biol Chem. 2005;280:15020–15028. doi: 10.1074/jbc.M500476200. [DOI] [PubMed] [Google Scholar]
- 11.Rabinovici R, Kabir K, Chen M, Su Y, Zhang D, Luo X, et al. ADAR1 is involved in the development of microvascular lung injury. Circ Res. 2001;88:1066–1071. doi: 10.1161/hh1001.090877. [DOI] [PubMed] [Google Scholar]
- 12.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NFkappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNFalpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 15.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 16.Monack DM, Raupach B, Hromockyj AE, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, et al. Crossmapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JH, Luo X, Nie Y, Su Y, Zhao Q, Kabir K, et al. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 21.Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 23.Ralph P, Nakoinz I. Phagocytosis and cytolysis by a macrophage tumour and its cloned cell line. Nature. 1975;257:393–394. doi: 10.1038/257393a0. [DOI] [PubMed] [Google Scholar]
- 24.Vidal S, Curran J, Kolakofsky D. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 27.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paetzold S, Lourido S, Raupach B, Zychlinsky A. Shigella flexneri phagosomal escape is independent of invasion. Infect Immun. 2007;75:4826–4830. doi: 10.1128/IAI.00454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen-Wester I, Chakravortty D, Hensel M. Functional transfer of Salmonella pathogenicity island 2 to Salmonella bongori and Escherichia coli. Infect Immun. 2004;72:2879–2888. doi: 10.1128/IAI.72.5.2879-2888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berezikov E, Thuemmler F, van Laak LW, Kondova I, Bontrop R, Cuppen E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Computational analysis of small RNA cloning data. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connell MA, Keller W. Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc Natl Acad Sci USA. 1994;91:10596–10600. doi: 10.1073/pnas.91.22.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.