Abstract
Many non-coding RNAs fold into complex three-dimensional structures, yet the self-assembly of RNA structure is hampered by mispairing, weak tertiary interactions, electrostatic barriers, and the frequent requirement that the 5′ and 3′ ends of the transcript interact. This rugged free energy landscape for RNA folding means that some RNA molecules in a population rapidly form their native structure, while many others become kinetically trapped in misfolded conformations. Transient binding of RNA chaperone proteins destabilize misfolded intermediates and lower the transition states between conformations, producing a smoother landscape that increases the rate of folding and the probability that a molecule will find the native structure. DEAD-box proteins couple the chemical potential of ATP hydrolysis with repetitive cycles of RNA binding and release, expanding the range of conditions under which they can refold RNA structures.
Key words: RNA folding, kinetic partitioning, ribozyme, RNA chaperone, DEAD-box
Introduction
RNA sequences encode tertiary structures that catalyze biochemical reactions and transduce regulatory signals from a variety of ligands. The molecular interactions that stabilize RNA structures are stable in physiological conditions and form spontaneously within a few seconds. However, the self-assembly of RNA tertiary structure encounters physical challenges, arising from the limited chemical complexity of polynucleotides, the stability of the secondary structure relative to the tertiary structure, and the negative electrostatic potential of the folded RNA.1–3
In response, cells express a variety of non-enzymatic RNA chaperones and ATP-dependent helicases that function in ribonucleoprotein (RNP) biogenesis, RNA processing and RNA regulatory interactions (reviewed in ref. 4–6). These chaperones and helicases speed up annealing and exchange of base pairs, unwind misfolded RNAs, and control the timing of specific conformational switches. For large macromolecular complexes such as ribosomes, spliceosomes and viruses, chaperones also link folding and assembly with intracellular transport, packaging and quality control.7–9 This review discusses the physical forces that partition RNA molecules among productive and non-productive folding pathways, and how chaperones and helicases overcome limitations to the fidelity and speed of self-assembly in RNA.
Hierarchical Assembly of RNA Structure
To understand how chaperones improve the outcome of RNA folding reactions, one must first understand how RNA structures self-assemble. One answer to this question is that self-organization proceeds from the hierarchy of the structure itself (Fig. 1).10,11 The primary sequence specifies a secondary structure, that is defined by double helices and single-stranded loops and joining segments. The double helices are then packed into a tertiary structure that is mediated by interactions with non-helical segments and specified by discrete tertiary interaction motifs.12 The tertiary structure is in turn recognized by protein subunits in the quaternary structure of ribonucleoproteins (RNPs).
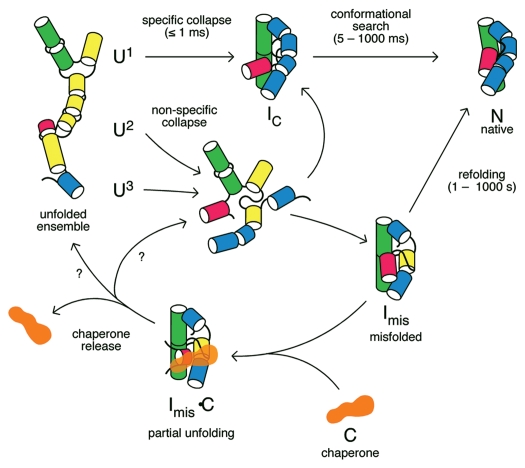
Figure 1.
RNA folding pathways and refolding by chaperones. In the absence of chaperones (top), the initial collapse transition produces an ensemble of compact intermediates (IC) that rearrange to the native structure (N). Because the unfolded RNA contains a mixture of structures, including non-native base pairs (yellow), different subpopulations (U1, U2, U3) fold through different pathways (reviewed in ref. 134). Non-specific collapse transitions lead to kinetically trapped misfolded intermediates (Imis). Chaperone proteins (C) bind and destabilize RNA folding intermediates, releasing partially unfolded RNAs that can fold again. The structure of the RNA immediately after chaperone release is unknown (question marks).
A large number of experiments on ribozymes, riboswitches, rRNA and many other structured RNAs have shown, however, that the mechanism of folding is both more complicated and more sophisticated than indicated by this hierarchical scheme (reviewed in ref. 3, 13–15). First, many RNA sequences can form more than one stable secondary structure, because of the chemical similarity of the four natural bases.1 Even 100 nt RNAs of random sequence were found to form rod-like secondary structures,16 emphasizing the degree to which base pairing is favorable even in the absence of any tertiary structure. Thus, before the correct tertiary structure can be achieved, the correct secondary structure must be selected from among many energetically similar alternatives.
Second, SAXS, NMR, fluorescence spectroscopy and gel electrophoresis experiments have shown that RNAs initially collapse into compact intermediates before they reach their native states (reviewed in ref. 15). This collapse transition correlates with assembly of the core helices, and is promoted by transient tertiary interactions between double helices that stabilize the compact structures.17–20 Thus, helix assembly and tertiary folding are at least partially, if not strongly, coupled with each other. The formation of tertiary interactions during the initial collapse transition can make the helix assembly more accurate, by favoring those secondary structures that can also form tertiary interactions.15 In the Azoarcus group I ribozyme, loss of a tetraloop-helix interaction made base pairing of the core helices less cooperative, and caused more of the RNA population to become kinetically trapped in misfolded intermediates.21
Third, the early compact intermediates must go through further conformational rearrangements to reach the native tertiary structure. Many RNA tertiary motifs are context dependent, and in some cases, the bases only take on the correct configuration when the interacting partner is present.12,22 Consequently, the formation of stable tertiary interfaces can require the sacrifice of energetically favorable base pairs. This final step of tertiary folding depends on the stability of the native structure, and can be driven by association of the RNA with Mg2+ ions, small molecules (riboswitches) or proteins (RNPs).
The search for the native conformation can involve local reorganization of base pairing and base stacking, or much larger changes to the RNA secondary structure (Fig. 1). For example, stable docking of two helical domain in the hairpin ribozyme requires restructuring of internal loops in both domains.23,24 Similarly, RNA stem-loops distort their helix geometry and rearrange base pairing and stacking to accommodate peptide ligands.25 Examples of more dramatic secondary structure rearrangements that are coupled to tertiary folding transitions are found in the P5abc domain of the Tetrahymena group I ribozyme,26 the Varkud satellite (VS) ribozyme,27 and a pseudoknot in the E. coli α operon mRNA.28
From these examples, it is apparent that the hierarchical levels of RNA structure are intimately linked with each other during the folding process. The conformations populated at any stage of folding depend simultaneously on the base pairing potential of the sequence, the stability of tertiary interactions (which depends on Mg2+ concentration), and the availability of RNA binding proteins to stabilize the folded RNA. Chaperone proteins must engage these ensembles of RNA conformations to alter the speed or efficiency of the folding process.
RNA Folding Free Energy Landscapes
While unfolded RNA can take on many different conformations, the number of possible structures narrows as the RNA chain begins to interact with itself (Fig. 2). The probabilities of forming specific conformations can be described by “free energy landscapes”, that depend on the relative energies of specific conformations and the kinetic barriers between them (Fig. 2) (reviewed in ref. 29 and 30). Single molecule experiments and simulations are beginning to describe the free energy landscapes for folding small RNAs in great detail.31–35
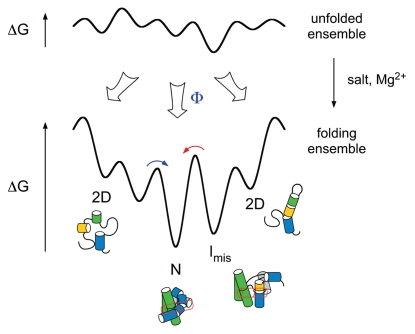
Figure 2.
Free energy landscapes for RNA folding. When conditions favor the unfolded state (e.g., low salt), RNA interactions are weak and molecules can adopt many different structures (top). When the RNA population is jumped to native conditions (e.g., Mg2+), RNA interactions are stable and the molecules seek out low free energy structures such as the native state (N) (bottom). Free energy landscapes for RNA folding are rough, because non-native structures are stable, corresponding to local minima on the energy landscape (Imis).29,41,42 A fraction of the population Φ takes a direct route to N and folds rapidly.29 Molecules that are initially trapped in misfolded structures must cross high transition states (red arrow), and thus fold slowly.
As RNA structures are stabilized by many weak, non-covalent interactions, self-assembly of the native structure should proceed smoothly as the RNA chain makes more and more energetically favorable intramolecular interactions.11 In reality, all RNA sequences encode molecular interactions that are incompatible with the native structure, “frustrating” the search for the native state.29,36 Even when equilibrium conditions favor the correct structure, folding is impeded by non-native interactions, delaying the acquisition of native structure.
In RNA, this topological frustration is acute, because short range interactions are very stable and form rapidly. For example, short hairpins can close in 10–100 µs, depending on the size of the loop.34,37,38 In contrast, tertiary interactions that shape the global structure of the RNA are enthalpically less stable than Watson-Crick base pairs, and form more slowly (10 ms–1 s).37,39,40 The tertiary interactions are also entropically disfavored because they occur between residues far apart in the sequence, and require co-localization of metal ions. Consequently, locally stable but incorrect structures form easily in RNA, and the high free energy barriers for unraveling incorrect conformations kinetically trap misfolded RNAs in local minima on the free energy landscape.29,41,42
As individual unfolded RNAs can have different initial structures, they can follow different paths toward the native structure, partitioning the population into fast-folding and slow-folding subpopulations (Fig. 2).29 The kinetic partitioning of RNA populations among parallel folding pathways has been observed in many single molecule FRET and force denaturation experiments, demonstrating that large and small RNA structures have heterogeneous folding dynamics (reviewed in ref. 14, 43 and 44). A handful of smaller RNAs, such as tRNA, RNase P C domain, the Azoarcus group I and VS ribozymes, refold into their native tertiary structure in less than 50 ms,37,45–47 virtually ensuring that the RNA is functional soon after it is transcribed. However, larger RNAs can require minutes or even hours to refold in vitro, because the non-native intermediates formed during the collapse transition require extensive reorganization (reviewed in ref. 48), and few molecules in the population bypass these kinetically trapped states.
Kinetic Partitioning of the Tetrahymena Ribozyme
The Tetrahymena group I ribozyme presents a well-studied example of kinetic partitioning in RNA folding.29 Although the majority of in vitro transcripts become kinetically trapped in inactive conformations, due to mispairing of the P3 pseudoknot in the ribozyme core, 5–10% of molecules reach the native structure in a few seconds.49–51 Single molecule FRET experiments directly showed that ∼10% of ribozyme molecules form a native-like structure in a single transition, while the remaining 90% fold through an intermediate.52 Tertiary interactions in the P4–P6 domain and peripheral domains form much more rapidly than the P3–P9 domain,40,53 and stabilize the misfolded intermediates as well as the native state.40,54–57
In addition to the potential for mispairing, the sequence separation of interacting residues (contact order) and the stability of the tertiary structure contribute to the propensity to misfold in vitro (reviewed in ref. 14). In the Tetrahymena ribozyme, the P3 pseudoknot mispairs more frequently because its strands are 100 nt apart in the ribozyme sequence; when the sequences are brought closer by a permutation of the sequence, mispairing is reduced.58,59 Moreover, the imbalance between stable peripheral interactions, which trap misfolded intermediates, and weak tertiary interactions, which diminish the accuracy of helix assembly, exacerbate the potential for misfolding.60 More recently, it has been proposed that P3 mispairing leads to an inactive but very stable topological isomer,61 that might explain extremely slow rates of ribozyme reactivation in some conditions.62 In other RNAs, unusual ribose puckers63 and local interactions have been proposed to trap RNAs in non-native structures.64
Kinetic Partitioning and Molecular Chaperones
If a certain fraction of the RNA population folds correctly, as predicted by the kinetic partitioning model, the yield of native RNA can be increased by giving misfolded molecules additional chances to fold correctly (reviewed in ref. 1, 2, 65 and 66) (Fig. 1). Both ATP-dependent RNA helicases and non-ATP utilizing RNA chaperones have been shown to accelerate RNA folding by destabilizing double helices and partially unfolding the RNA.67–70 In a similar way, chemical denaturants speed up RNA folding reactions by raising the free energy of the misfolded intermediates, thereby lowering the kinetic barriers between the misfolded state and the native state.49,62,71
In the classical model, the chaperone is not needed to stabilize the final structure of the RNA and refolding occurs as the chaperone dissociates (Fig. 1). Consequently, the folding pathways and partitioning among alternative folding pathways are specified by the RNA sequence and refolding in the presence of the chaperone should closely ressemble the folding pathway in the absence of the chaperone. If the RNA or protein substrate has a low probability of folding correctly (low partition factor), many rounds of chaperone-assisted folding may be necessary to yield a sufficient number of native molecules.
The basic tenets of this “partitioning” model for molecular chaperones have been borne out by experiments on non-enzymatic RNA chaperones and members of the DEAD-box family of RNA-dependent ATPases. In an early demonstration of the chaperone activity in RNA binding proteins, a fragment of the hnRNP A1 protein was shown to assist renaturation of tRNAs.72 Other prominent examples of RNA binding proteins that facilitate RNA refolding including HIV-1 nucleocapsid protein (NC), which facilitates dimerization of the genomic RNA and accelerates priming and strand transfer reactions during retroviral replication (see also reviews of retroviral NC proteins in this issue).73,74 By destabilizing RNA helices and promoting product release, HIV NC and hnRNP A1 were able to stimulate turnover of the hammerhead ribozyme.75,76 The E. coli StpA protein was found to reduce misfolding of the phage T4 td group I intron in vitro and in vivo, via non-specific interactions with the intron RNA (see also review of StpA by M. Doetsch, T. Gstrein, R. Schroeder and B. Fürtig in this issue).77–79 Other examples of RNA binding proteins that non-specifically unfold RNA structures include ribosomal proteins,80,81 yeast La and human Ro proteins,82–84 and the bacterial Sm-like protein Hfq (see also review of RNA folding in living cells by G. Zemora and C. Waldsich in this issue).85–88
Recent work has shown that the DEAD-box proteins can rescue misfolded ribozymes through repeated rounds of RNA unfolding, giving misfolded RNAs another chance to partition between productive and unproductive folding pathways (see review of DEAD-box proteins by C. Pan and R. Russell in this issue).89–93 Using the Tetrahymena ribozyme as a substrate, Russell and colleagues found that CYT19 accelerated conversion of the misfolded ribozyme to the native state in the presence of ATP.70,91 In agreement with the model above, kinetic partitioning of the RNA population between misfolded and correct folding pathways is the same in the presence and absence of CYT19 and the rate constants for refolding are also unchanged.91 CYT19 binds the Tetrahymena riboyzme non-specifically as expected,70 can thus also unfold the native riboyzme.
RNA Unwinding by Perturbation of Free Energy Landscapes
Although non-enzymatic and ATP-dependent RNA chaperones both destabilize RNA duplexes, non-enzymatic RNA binding proteins must facilitate RNA refolding by exploiting thermodynamic differences between native and intermediate states (Fig. 3). RNA chaperones typically promote unfolding by binding single-stranded RNA more tightly than double-stranded RNA, and consequently must dissociate from the RNA for the RNA to refold.1,5 The more tightly the chaperone binds partially or fully unfolded forms of the RNA (U) compared to the folding intermediates (I), the greater capacity it will have to unfold the I states (ΔGfC; Fig. 3A). On the other hand, if the protein binds too tightly, the RNA cannot refold. The native RNA (N) must remain thermodynamically more stable than the chaperone-RNA complex (U•C) (ΔGnet; Fig. 3B). Thus, RNA chaperones must dissociate rapidly from the RNA to avoid denaturing functional molecules and ensure fast refolding kinetics.5 Consistent with this idea, the dissociation rate of RNA binding proteins has been found to correlate with their degree of chaperone activity.5,94,95
Figure 3.
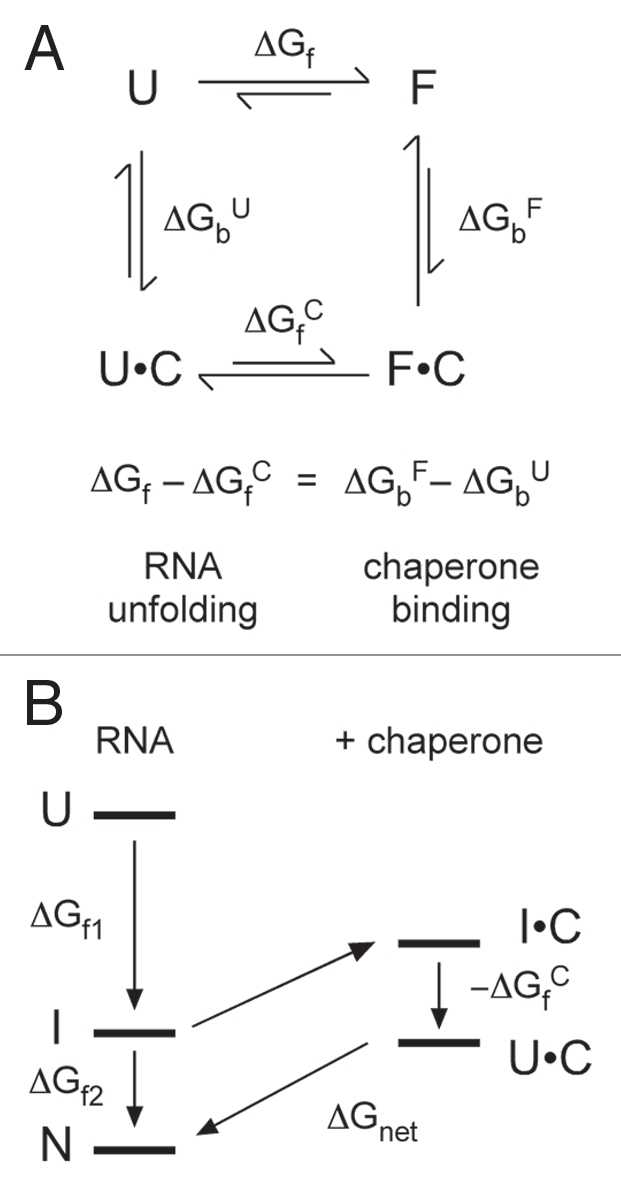
Thermodynamics of RNA unfolding by non-enzymatic chaperones. (A) Thermodynamic cycle for chaperone (C) binding to unfolded (U) and folded (F) RNA, in which the folded RNA can be an intermediate (I) or native (N) form. The difference in the free energy of chaperone binding to U and F (ΔGbF − ΔGbU) is equal to the perturbation of the folding free energy by the chaperone (ΔGf − ΔGfC). (B) Chaperones that interact tightly with unfolded RNA lower the free energy of U relative to I and N. If the unfolded RNA-chaperone complex (U•C) becomes too stable, the energy gap ΔGnet between the U•C complex and N is small, and folding is less favorable overall.
For the same reasons, RNA chaperones are likely to be most effective when the energy gap between the I and N states is large, and the I states have a more open structure that permits easy loading of the chaperone protein.67 For example, StpA protein was found to act more effectively on wild type td group I intron than on mutant forms in which the native structure was less stable.79,96 By contrast, StpA was ineffective against the stable and highly structured folding intermediates of the Tetrahymena ribozyme (Nikolcheva T and Woodson SA, unpublished data). Greater thermal fluctuations in misfolded structures, which are often less tightly packed than native structures, may allow proteins to transiently bind single-stranded segments.70
The arguments above suggest that non-enzymatic RNA chaperones must operate in a narrow thermodynamic window between the stabilities of the misfolded and native structures. How can chaperone proteins unwind RNAs efficiently, yet allow the correct structure to form? First, cycles of unwinding can be coupled to ATP hydrolysis, providing greater control over the timing of the RNA-chaperone interactions, as discussed below. Next, I discuss growing evidence that enzymes and non-enzymatic chaperones stimulate RNA folding by altering the dynamics of RNA structures, and not just by reducing its thermodynamic stability.
Active RNA Unwinding
In contrast to “passive” chaperones, DEAD-box ATPases use the chemical potential of ATP hydrolysis to drive repetitive cycles of RNA binding and release from the enzyme (reviewed in ref. 97 and 98). Interactions with the DEAD-box protein locally destabilize interactions in the RNA, and members of this protein family have been shown to act in helix unwinding and protein dissociation,99,100 RNP remodeling101,102 and strand annealing.103 Since interactions with RNA substrates are coupled to ATP binding and hydrolysis, this class of proteins has the potential to stimulate RNA folding over a wider range of conditions than non-enzymatic RNA binding proteins. It also allows the timing of conformational changes in the RNA to be regulated, a feature that is important in complex processes such as ribosome assembly and pre-mRNA splicing.7,101,104,105
Work with model substrates and ribozymes suggests that DEAD-box proteins do not require ATP hydrolysis to bind and unwind the RNA, but rather use ATP hydrolysis to cycle between states with high and low RNA affinity (reviewed in ref. 93 and 106). Many DEAD-box proteins, including CYT19, have a low intrinsic RNA unwinding activity in the absence of ATP, and the first round of RNA unwinding can be observed in the presence of non-hydrolyzable ATP analogs, suggesting that the RNA is passively unwound when the DEAD-box protein binds.92,107 However, ATP is needed for multiple turnover unwinding, supporting the conclusion that enzyme recycling depends on ATP hydrolysis.93,108
If ATP is used to control protein-RNA affinity, then how is the RNA unfolded? DEAD-box RNA helicases contain a binding site for single-stranded RNA109 that straddles the two RecA-like ATPase domains.110 As for other members of the SF2 superfamily of helicases, the orientation of the RecA-like domains changes during ATP binding and phosphate release, and these motions appear coupled to the capture and release of single-stranded RNA segments.97,98 The backbone of the bound RNA is kinked in several structures of DEAD-box complexes, which may contribute to helix unwinding.110–112 The low processivity of non-viral DEAD-box proteins, and the observation that long helices are unwound more slowly than short helices,66,98 lead to the idea that loosely structured regions of an RNA tertiary structure are targeted for unwinding because they are most accessible to the enzyme. In the Tetrahymena ribozyme, for example, CYT19 can efficiently unwind the P1 helix, which lies on the surface of the ribozyme and is less tightly docked in the misfolded state than in the native state.70
How local unfolding of RNA helices stimulates refolding of large RNA tertiary structures remains an open question. Mutational and single-molecule FRET studies showed that peripheral tertiary interactions in the Tetrahymena ribozyme break when misfolded intermediates transition to the native state, showing that the folding transition state lies high on the free energy landscape.54,56,57 Force denaturation experiments have shown that even minimal tertiary structure such as a kissing loop greatly increases the amount of force needed to rupture RNA base pairing interactions.113 Therefore, to be effective against RNAs with tertiary structure, a chaperone or helicase must unfold large regions of the RNA simultaneously. Alternatively, the destabilizing effect of local unwinding may propagate through the RNA tertiary structure more easily than currently thought. Finally, many DEAD-box chaperones contain auxiliary RNA binding domains that tether the protein to the RNA.99,114 These domains may directly increase the dynamics of the RNA through the electrostatic mechanisms discussed below.
Electrostatics and RNA Dynamics
RNA chaperone proteins are rich in basic amino acids, and electrostatic interactions with their substrates may directly stimulate RNA refolding. Owing to the negative charge on every phosphate, counterions and electrostatic interactions strongly influence the stability and dynamics of RNA structures.3,10,115 RNA tertiary structures are particularly sensitive to the presence of multivalent cations, because the folded structure creates pockets of strongly negative electrostatic potential.116 In general, multivalent cations stabilize the folded RNA more efficiently than monovalent cations, because multivalent ions interact more strongly with the electrostatic potential of the RNA, fewer multivalent ions must be localized around the nucleic acid to offset its charge, and multivalent metal ions such as Mg2+ can coordinate specific sites within the folded RNA.
For multivalent counterions, the density of positive charge in the counterion also affects the dynamics of the system (Fig. 4A). Bulky cations such as polyamines, in which the positive charges are separated by neutral carbon atoms, pack less tightly around the folded RNA and compact the RNA less efficiently than smaller metal cations such as Mg2+ or [Co(NH3)6]3+.117–119 SAXS experiments show that folding intermediates are less compact (on average) in spermidine and Ba2+ than in Mg2+ or [Co(NH3)6]3+.120 This correlates with faster refolding rates in low charge density ions such as polyamines and monovalent salts, compared with multivalent metal ions such as Mg2+.121–124 Folding intermediates are less stable in polyamines than in Mg2+ and the transition states for folding are broader and more plastic.122 Both of these effects produce a smoother free energy landscape for folding and faster folding kinetics.
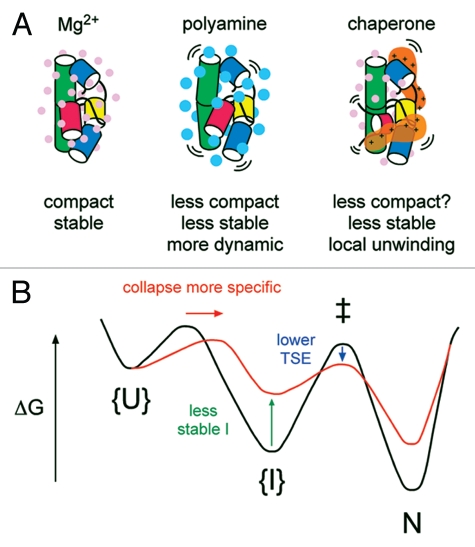
Figure 4.
Counterion charge density and RNA folding dynamics. (A) The valence and size or charge density, of counterions influences the stability of RNA structure. Small, multivalent ions (Mg2+, left) stabilize RNA tertiary structures more than larger ions (polyamines, middle).119 Basic polypeptides in RNA chaperones may destabilize RNA structures by lowering the density of positive charge around the RNA (right). (B) Chaperones can smooth free energy landscapes for RNA folding by destabilizing I states (green arrow) and stabilizing transition state ensembles (TSE; blue arrow). As the landscape becomes smoother, the transition state for collapse moves toward N, increasing the probability Φ an RNA will fold correctly and bypass non-native I's.122
In an analogous way, basic proteins, in which the positive charges are separated by polar and apolar groups, can buffer the electrostatic interactions between the RNA and its surrounding metal ions, smoothing out the free energy landscape so that the most stable configuration can be achieved in a shorter time (Fig. 4A). Non-specific electrostatic interactions between proteins and RNA have the potential to stabilize transitions states for strand exchange and annealing.1,125,126 The electrostatic environment is critical to the strength of the RNA-protein interactions, and in the case of HIV NC, can modulate the balance between its strand exchange and strand annealing activities.127
In many RNA binding proteins with RNA chaperone activity, such as HIV NC, ribosomal protein S12 and Hfq, basic amino acids are located in disordered or flexible regions that can interact with different RNA structures (reviewed in ref. 5). In ribosomal protein S1, hnRNP A1 and yeast Prp24, basic RNA binding domains are connected by flexible linkers. This flexibility is likely key to their chaperone activity, allowing the chaperone to remain in contact with the RNA as its structure is remodeled.128 Disordered domains rich in arginines, glycines and serines are also found in non-specific DEAD-box RNA chaperones such as CYT19 and Mss116,111,114 and may contribute to RNA unwinding by those enzymes (reviewed in ref. 98).
Chaperones and the Accuracy of RNA Folding
As discussed above, molecular chaperones not only improve RNA folding by unwinding misfolded intermediates, they can also lower the electrostatic barriers to folding while in complex with the protein. Both actions smooth the free energy landscapes for folding, as local minima become less pronounced while the free energy barriers between conformations are reduced (Fig. 4B). An important consequence of a smoother free energy landscape is that the propensity of macromolecules to misfold in the first place is lessened—that is to say, more molecules partition into fast, “trap-free” folding pathways when the landscape is smooth than when it is rough.129
Why does the smoothness of the landscape correlate with the population of fast folding molecules? First, partitioning among different folding pathways depends on the structures present at the start of the folding reaction. In several studies, more RNA partitioned into direct (fast) folding pathways when the RNA was prefolded in salt before Mg2+ was added.130–133 Partial denaturation of the unfolded Azoarcus ribozyme reduced the heterogeneity of the initial collapse kinetics, allowing more RNA to collapse in ≤1 ms.134
Second, more RNA molecules can avoid misfolding if the forces driving helix assembly and collapse are lessened, because the RNA has more time to search for its most stable structure before the transition state for folding is crossed.2,129 Moreover, when conditions barely favor the native state, a larger number of energetically favorable interactions must accumulate to push the RNA over the folding transition state. Nucleation of the collapse transition becomes more specific, and the transition state itself occurs closer the native state, in accord with the Hammond postulate122 (Fig. 4B, red arrow).
The relationship between the specificity of folding and the force (or change in chemical potential) was validated by mechanical denaturation of tethered RNA hairpins (reviewed in refs. 135 and 136). When the force changed very slowly, unfolding and refolding occured reversibly, with a high yield of native hairpin in each cycle.137–139 When the force was ramped quickly relative to the relaxation rate of the hairpin, the molecule unfolded and refolded irreversibly along different pathways, populating new metastable states on the refolding trajectory.138 Similarly, relaxation times in temperature jump experiments are shortest near the melting point, when the folding equilibrium is near one.
The arguments above suggest that chaperones may do more than rescue misfolded RNA; they may improve the accuracy of RNA folding by improving the specificity of structural nucleation. Evidence that proteins can stimulate large changes in RNA structure without having to dissociate from the RNA comes from single-molecule FRET experiments on RNA bound to the yeast CBP2 splicing protein140 and changes in RNA backbone accessibility during ribosome assembly.141 More work will be needed to understand the nature of non-specific interfaces in RNA-protein complexes, and the extent to which such interactions can alter RNA folding trajectories.
Protein-Facilitation of Intermolecular Base Pairing
Further evidence that proteins can change the transition states for RNA folding reactions comes from RNA “annealers” that increase the rate at which two separate RNAs come together. Although unstructured RNA oligonucleotides base pair rapidly (τ ∼ 1 s when the local strand concentration is 1 µM), proteins are required to facilitate the association of natural antisense RNAs, which must reorganize their secondary structures to form the antisense complex.142 An important function of HIV NC is to promote dimerization of the genomic RNA.74 Similarly, a major function of bacterial Hfq protein is to promote the association of bacterial sRNAs with their targets.85
One way that RNA chaperones drive the annealing of two RNA strands is to simply destabilize secondary structures that compete with the intermolecular complex.74,81 For example, E. coli Hfq protein remodels the sodB mRNA so that it is more accessible to its complementary partner,143 and destabilize an inhibitory secondary structure in the rpoS mRNA that is the target of several sRNAs.144,145 On the other hand, E. coli Hfq has a potent annealing activity on short RNA oligomers.88,144 Similarly, the unstructured N-terminal region of NC is associated with its potent strand aggregation activity, which can stimulate strand association 100 to 105 fold.146,147
A common feature of proteins with strong RNA annealing activity is the presence of more than one RNA binding surface, either in separate domains (e.g., NC, StpA,81 yeast Prp24,148) or on different surfaces of a ring-like structure (e.g., Hfq,149 human Ro150). How the ability to bind different RNA strands simultaneously promotes annealing is not understood (reviewed in ref. 5 and 151). One possibility is that the annealer simply increases local strand concentration. Alternatively, the different RNA binding surfaces may allow intra- and inter-molecular RNA base pairs to exchange along an energetically neutral path. Regardless of the mechanism, the annealing reaction is reversible and the net outcome of the annealing reaction depends on whether the intermolecular RNA complex is more stable than the individual strands.88,144,147,152
Long-Range Interactions and Co-transcriptional Folding
Although the energy landscape picture described above was first based on the refolding of proteins and RNAs in vitro, a number of studies suggest that RNA tertiary interactions and double helices have similar stabilities in the cell, as long as differences in salt and Mg2+ concentrations are accounted for.153–155 An analysis of splicing and turnover rates of the Tetrahymena pre-RNAs expressed in E. coli or yeast showed that while most transcripts fold and self-splice quickly, a minority of transcripts partition into an inactive pool that is ultimately degraded.156,157 Thus, kinetic partitioning between misfolded and correctly folded structures also occurs in vivo, and is sensitive to mutations that increase RNA misfolding.157,158
In cells, nucleotides at the 5′ end of the RNA have an opportunity to fold during transcription, before nucleotides at the 3′ end are synthesized. Therefore, the free energy landscape for folding evolves as the RNA is elongated. Experiments using the hammerhead ribozyme showed that RNA helices exchange more rapidly in yeast than in vitro, perhaps due to chaperones, but that ≥10 bp helices resist thermodynamic equilibration with downstream structures during transcription.159 Experiments on RNase P RNA showed that the locations of transcriptional pauses have a strong effect on the co-transcriptional RNA folding pathway, because transcriptional pauses allow time for upstream sequences to fold.160,161
Metastable structures formed during RNA synthesis are exploited in a large variety of regulatory schemes, including phage coat protein translation, tRNA response elements (T-boxes) and many riboswitches.162 The considerations above suggest that co-transcriptional folding is usually disadvantageous for RNAs containing long-distance interactions, because the downstream partner is not immediately available, allowing other metastable structures to form.163 For long RNAs in which critical base pairing interactions are separated by more than a few hundred base pairs, chaperones are presumably required to ensure the formation of long-distance contacts.
The biogenesis of ribosomal subunits provides a wonderful example of how transcription, RNA folding, protein recognition, processing and RNA chaperones must be coordinated to ensure robust assembly (reviewed in 104, 105, 164 and 165). In the small and large subunit ribosomal RNAs, structural domains are pinched off by long-distance base pairs separated by several hundred to several thousand nucleotides. In the small subunit rRNA, these long-distance helices are joined by a pseudoknot that is made late during assembly,166 and that is sensitive to the presence of extra-ribosomal assembly factors. In bacteria, the 5′ spacer makes metastable interactions with sequences of the pseudoknot, perhaps acting as a temporary “place holder”, until the proper native interaction with downstream residues can be made.167,168 In eukaryotes, U3 snoRNA, which is required for 5′ processing of the 18S rRNA and formation of the pseudoknot (reviewed in ref. 169), pairs with the 5′ and 3′ end of the 18S sequence.170,171 U3 and other snoRNAs may guide folding of the pre-rRNA by providing an intermolecular metastable structure that can be replaced with the mature intramolecular structure later in assembly (reviewed in ref. 165).
Especially in eukaryotes, conformational changes in the pre-rRNA are both facilitated and regulated by assembly factors, including DEAD-box helicases and GTPases.105,172,173 These assembly factors, which are themselves regulated through a variety of pathways, link RNA folding and ribosomal protein binding with maturation of the rRNA, nuclear export of the complexes and with quality control pathways. Thus, for very long and structurally complex RNAs, specific metastable interactions and their associated chaperones may have evolved to gain greater control over the intrinsically stochastic nature of the RNA folding landscape.
Conclusion
In summary, non-coding RNAs can fold into elaborate and beautiful structures, yet the self-assembly of such structures is compromised by strong mispairing and weak tertiary interactions. Long natural transcripts are at a further disadvantage, as interactions between the 5′ and 3′ ends of the sequence cannot form until the entire sequence is synthesized. Chaperones can unwind RNA structures by destabilizing base pairs, allowing the RNA a chance to refold. However, the brittle nature of RNA tertiary structures and the delicate task of destabilizing misfolded RNAs without denaturing functional molecules means that a thermodynamic strategy is likely insufficient for robust chaperone function. Instead, emerging data suggest that chaperones also act by raising the dynamics of the RNA, lowering or altering transition states for refolding.
Additional structures of chaperones in complex with natural substrates and information on the motions of the RNA and protein, will provide new insight into how proteins alter RNA folding landscapes. Other tantalizing questions for the future are how chaperone-assisted refolding of the RNA is coupled with specific protein recognition and downstream processing reactions, and at what stage misassembled RNPs are designated as being beyond repair.
Acknowledgements
The author thanks members of her research group, D. Thirumalai, G. Storz and S. Gottesman and for helpful discussions and the NIGMS for support of research in the author's laboratory.
Abbreviations
- RNP
ribonucleoprotein
- NC
nucleocapsid
- NMR
nuclear magnetic resonance
- SAXS
small angle X-ray scattering
- VS
varkud satellite
Footnotes
Previously published online www.landesbioscience.com/journals/rnabiology/article/13615
References
- 1.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:77–79. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 2.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 3.Chen SJ. RNA folding: conformational statistics, folding kinetics and ion electrostatics. Annu Rev Biophys. 2008;37:197–214. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 5.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 6.Wolin SL, Wurtmann EJ. Molecular chaperones and quality control in noncoding RNA biogenesis. Cold Spring Harbor Symp Quant Biol. 2006;71:505–511. doi: 10.1101/sqb.2006.71.051. [DOI] [PubMed] [Google Scholar]
- 7.Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuniga S, Sola I, Cruz JL, Enjuanes L. Role of RNA chaperones in virus replication. Virus Res. 2009;139:253–266. doi: 10.1016/j.virusres.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafontaine DL. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 11.Tinoco IJ, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 12.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shcherbakova I, Mitra S, Laederach A, Brenowitz M. Energy barriers, pathways and dynamics during folding of large, multidomain RNAs. Curr Opin Chem Biol. 2008;12:655–666. doi: 10.1016/j.cbpa.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sosnick TR. Kinetic barriers and the role of topology in protein and RNA folding. Protein Sci. 2008;17:1308–1318. doi: 10.1110/ps.036319.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodson SA. Compact intermediates in RNA folding. Annu Rev Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat Struct Mol Biol. 2005 doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- 17.Buchmueller KL, Weeks KM. Near native structure in an RNA collapsed state. Biochemistry. 2003;42:13869–13878. doi: 10.1021/bi035476k. [DOI] [PubMed] [Google Scholar]
- 18.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 19.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, et al. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol. 2004;343:1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 20.Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, et al. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J Mol Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 24.Pljevaljcic G, Klostermeier D, Millar DP. The tertiary structure of the hairpin ribozyme is formed through a slow conformational search. Biochemistry. 2005;44:4870–4876. doi: 10.1021/bi047772i. [DOI] [PubMed] [Google Scholar]
- 25.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 26.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen AA, Collins RA. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc Natl Acad Sci USA. 2001;98:7730–7735. doi: 10.1073/pnas.141039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gluick TC, Gerstner RB, Draper DE. Effects of Mg2+, K+ and H+ on an equilibrium between alternative conformations of an RNA pseudoknot. J Mol Biol. 1997;270:451–463. doi: 10.1006/jmbi.1997.1119. [DOI] [PubMed] [Google Scholar]
- 29.Thirumalai D, Woodson SA. Kinetics of folding of protein and RNA. Acc Chem Res. 1996;29:433–439. [Google Scholar]
- 30.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 31.Liphardt J, Onoa B, Smith SB, Tinoco IJ, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 32.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Chen SJ. RNA hairpin-folding kinetics. Proc Natl Acad Sci USA. 2002;99:1931–1936. doi: 10.1073/pnas.032443099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. Exploring the energy landscape of a small RNA hairpin. J Am Chem Soc. 2006;128:1523–1530. doi: 10.1021/ja0553856. [DOI] [PubMed] [Google Scholar]
- 35.Hyeon C, Dima RI, Thirumalai D. Size, shape and flexibility of RNA structures. J Chem Phys. 2006;125:194905. doi: 10.1063/1.2364190. [DOI] [PubMed] [Google Scholar]
- 36.Go N. Theoretical studies of protein folding. Annu Rev Biophys Bioeng. 1983;12:183–210. doi: 10.1146/annurev.bb.12.060183.001151. [DOI] [PubMed] [Google Scholar]
- 37.Crothers DM, Cole PE, Hilbers CW, Shulman RG. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974;87:63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- 38.Kuznetsov SV, Ren CC, Woodson SA, Ansari A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008;36:1098–1112. doi: 10.1093/nar/gkm1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarrinkar PP, Williamson JR. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat Struct Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
- 40.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 41.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 42.Chen SJ, Dill KA. RNA folding energy landscapes. Proc Natl Acad Sci USA. 2000;97:646–651. doi: 10.1073/pnas.97.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bokinsky G, Zhuang X. Single-molecule RNA folding. Acc Chem Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- 44.Aleman EA, Lamichhane R, Rueda D. Exploring RNA folding one molecule at a time. Curr Opin Chem Biol. 2008;12:647–654. doi: 10.1016/j.cbpa.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Fang XW, Pan T, Sosnick TR. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat Struct Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- 46.Hiley SL, Collins RA. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodson SA. Recent insights on RNA folding mechanisms from catalytic RNA. Cell Mol Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan J, Thirumalai D, Woodson SA. Folding of RNA involves parallel pathways. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 50.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 51.Pan J, Deras ML, Woodson SA. Fast folding of a ribozyme by stabilizing core interactions: evidence for multiple folding pathways in RNA. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 53.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 54.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 55.Treiber DK, Williamson JR. Concerted kinetic folding of a multidomain ribozyme with a disrupted loopreceptor interaction. J Mol Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 56.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, et al. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan J, Woodson SA. The effect of long-range loop-loop interactions on folding of the Tetrahymena self-splicing RNA. J Mol Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 58.Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–733. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lease RA, Adilakshmi T, Heilman-Miller S, Woodson SA. Communication between RNA folding domains revealed by folding of circularly permuted ribozymes. J Mol Biol. 2007;373:197–210. doi: 10.1016/j.jmb.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thirumalai D, Woodson SA. Maximizing RNA folding rates: a balancing act. RNA. 2000;6:790–794. doi: 10.1017/s1355838200000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 62.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 63.Mortimer SA, Weeks KM. C2′-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci USA. 2009;106:15622–15627. doi: 10.1073/pnas.0901319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008;36:7088–7099. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 66.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karpel RL, Miller NS, Fresco JR. Mechanistic studies of ribonucleic acid renaturation by a helix-destabilizing protein. Biochemistry. 1982;21:2102–2108. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 68.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci USA. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat Struct Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 72.Karpel RL, Swistel DG, Miller NS, Geroch ME, Lu C, Fresco JR. Acceleration of RNA renaturation by nucleic acid unwinding proteins. Brookhaven Symp Biol. 1975:165–174. [PubMed] [Google Scholar]
- 73.Thomas JA, Gorelick RJ. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 75.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 76.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang A, Derbyshire V, Salvo JL, Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 78.Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 81.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belisova A, Semrad K, Mayer O, Kocian G, Waigmann E, Schroeder R, et al. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 86.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, et al. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 87.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 90.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 91.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J Mol Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 96.Grossberger R, Mayer O, Waldsich C, Semrad K, Urschitz S, Schroeder R. Influence of RNA structural stability on the RNA chaperone activity of the Escherichia coli protein StpA. Nucleic Acids Res. 2005;33:2280–2289. doi: 10.1093/nar/gki515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 98.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 99.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 101.Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 104.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 106.Banroques J, Doere M, Dreyfus M, Linder P, Tanner NK. Motif III in superfamily 2 “helicases” helps convert the binding energy of ATP into a high-affinity RNA binding site in the yeast DEAD-box protein Ded1. J Mol Biol. 2010;396:949–966. doi: 10.1016/j.jmb.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 107.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 110.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 111.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 2009;37:4464–4471. doi: 10.1093/nar/gkp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li PT, Bustamante C, Tinoco I., Jr Unusual mechanical stability of a minimal RNA kissing complex. Proc Natl Acad Sci USA. 2006;103:15847–15852. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, et al. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 116.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 117.Thomas TJ, Bloomfield VA. Ionic and structural effects on the thermal helix-coil transition of DNA complexed with natural and synthetic polyamines. Biopolymers. 1984;23:1295–1306. doi: 10.1002/bip.360230713. [DOI] [PubMed] [Google Scholar]
- 118.Rouzina I, Bloomfield VA. Influence of ligand spatial organization on competitive electrostatic binding to DNA. J Phys Chem. 1996;100:4305–4313. [Google Scholar]
- 119.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge density of divalent metal cations determines RNA stability. J Am Chem Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, Thirumalai D, et al. Metal ion dependence of cooperative collapse transitions in RNA. J Mol Biol. 2009;393:753–764. doi: 10.1016/j.jmb.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heilman-Miller SL, Pan J, Thirumalai D, Woodson SA. Counterion condensation in folding of the Tetrahymena ribozyme. II. Counterion dependence of folding kinetics. J Mol Biol. 2001;309:57–68. doi: 10.1006/jmbi.2001.4660. [DOI] [PubMed] [Google Scholar]
- 122.Koculi E, Thirumalai D, Woodson SA. Counterion charge density determines the position and plasticity of RNA folding transition states. J Mol Biol. 2006;359:446–454. doi: 10.1016/j.jmb.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 123.Fang XW, Thiyagarajan P, Sosnick TR, Pan T. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc Natl Acad Sci USA. 2002;99:8518–8523. doi: 10.1073/pnas.142288399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brown TS, Chadalavada DM, Bevilacqua PC. Design of a highly reactive HDV ribozyme sequence uncovers facilitation of RNA folding by alternative pairings and physiological ionic strength. J Mol Biol. 2004;341:695–712. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- 125.Record MT, Jr, Anderson CF, Lohman TM. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening and ion effects on water activity. Q Rev Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 126.Draper DE. Themes in RNA-protein recognition. J Mol Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 127.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J Mol Biol. 2009;386:773–788. doi: 10.1016/j.jmb.2008.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tompa P, Kovacs D. Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol. 2010;88:167–174. doi: 10.1139/o09-163. [DOI] [PubMed] [Google Scholar]
- 129.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu Rev Phys Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 130.Heilman-Miller SL, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J Mol Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 131.Webb AE, Weeks KM. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat Struct Biol. 2001;8:135–140. doi: 10.1038/84124. [DOI] [PubMed] [Google Scholar]
- 132.Su LJ, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J Mol Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- 133.Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. Distinct contribution of electrostatics, initial conformational ensemble and macromolecular stability in RNA folding. Proc Natl Acad Sci USA. 2007;104:7045–7050. doi: 10.1073/pnas.0608765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Roh JH, Guo L, Kilburn JD, Briber RM, Irving T, Woodson SA. Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering. J Am Chem Soc. 2010;132:10148–10154. doi: 10.1021/ja103867p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Woodside MT, Garcia-Garcia C, Block SM. Folding and unfolding single RNA molecules under tension. Curr Opin Chem Biol. 2008;12:640–646. doi: 10.1016/j.cbpa.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li PT, Vieregg J, Tinoco I., Jr How RNA unfolds and refolds. Annu Rev Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 137.Li PT, Collin D, Smith SB, Bustamante C, Tinoco I., Jr Probing the mechanical folding kinetics of TAR RNA by hopping, force-jump and force-ramp methods. Biophys J. 2006;90:250–260. doi: 10.1529/biophysj.105.068049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li PT, Bustamante C, Tinoco I., Jr Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci USA. 2007;104:7039–7044. doi: 10.1073/pnas.0702137104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hyeon C, Thirumalai D. Mechanical unfolding of RNA: from hairpins to structures with internal multiloops. Biophys J. 2007;92:731–743. doi: 10.1529/biophysj.106.093062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bokinsky G, Nivon LG, Liu S, Chai G, Hong M, Weeks KM, et al. Two distinct binding modes of a protein cofactor with its target RNA. J Mol Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 143.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 147.Zeng Y, Liu HW, Landes CF, Kim YJ, Ma X, Zhu Y, et al. Probing nucleation, reverse annealing and chaperone function along the reaction path of HIV-1 single-strand transfer. Proc Natl Acad Sci USA. 2007;104:12651–12656. doi: 10.1073/pnas.0700350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bae E, Reiter NJ, Bingman CA, Kwan SS, Lee D, Phillips GN, Jr, et al. Structure and interactions of the first three RNA recognition motifs of splicing factor prp24. J Mol Biol. 2007;367:1447–1458. doi: 10.1016/j.jmb.2007.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat Struct Mol Biol. 2006;13:1002–1009. doi: 10.1038/nsmb1156. [DOI] [PubMed] [Google Scholar]
- 151.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 152.Heilman-Miller SL, Wu T, Levin JG. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J Biol Chem. 2004;279:44154–44465. doi: 10.1074/jbc.M401646200. [DOI] [PubMed] [Google Scholar]
- 153.Nikolcheva T, Woodson SA. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J Mol Biol. 1999;292:557–567. doi: 10.1006/jmbi.1999.3083. [DOI] [PubMed] [Google Scholar]
- 154.Brion P, Schroede rR, Michel F, Westhof E. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: A comparative study. RNA. 1999;5:947–958. doi: 10.1017/s1355838299990477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 156.Koduvayur SP, Woodson SA. Intracellular folding of the Tetrahymena group I intron depends on exon sequence and promoter choice. RNA. 2004;10:1526–1532. doi: 10.1261/rna.7880404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jackson SA, Koduvayur S, Woodson SA. Self-splicing of a group I intron reveals partitioning of native and misfolded RNA populations in yeast. RNA. 2006;12:2149–2159. doi: 10.1261/rna.184206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nikolcheva T, Woodson SA. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J Mol Biol. 1999;292:557–567. doi: 10.1006/jmbi.1999.3083. [DOI] [PubMed] [Google Scholar]
- 159.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci USA. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci USA. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grundy FJ, Henkin TM. Regulation of gene expression by effectors that bind to RNA. Curr Opin Microbiol. 2004;7:126–131. doi: 10.1016/j.mib.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 163.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 164.Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17:281–286. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 165.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- 167.Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 168.Pardon B, Wagner R. The Escherichia coli ribosomal RNA leader nut region interacts specifically with mature 16S RNA. Nucleic Acids Res. 1995;23:932–941. doi: 10.1093/nar/23.6.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 170.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Borovjagin AV, Gerbi SA. Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA. 2004;10:942–953. doi: 10.1261/rna.5256704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Granneman S, Tollervey D. Building ribosomes: even more expensive than expected? Curr Biol. 2007;17:415–417. doi: 10.1016/j.cub.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 173.Karbstein K. The role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. [DOI] [PubMed] [Google Scholar]