Abstract
OBJECTIVE
Common variation in the FTO gene is associated with BMI and type 2 diabetes. Increased BMI is associated with diabetes risk factors, including raised insulin, glucose, and triglycerides. We aimed to test whether FTO genotype is associated with variation in these metabolic traits.
RESEARCH DESIGN AND METHODS
We tested the association between FTO genotype and 10 metabolic traits using data from 17,037 white European individuals. We compared the observed effect of FTO genotype on each trait to that expected given the FTO-BMI and BMI-trait associations.
RESULTS
Each copy of the FTO rs9939609 A allele was associated with higher fasting insulin (0.039 SD [95% CI 0.013–0.064]; P = 0.003), glucose (0.024 [0.001– 0.048]; P = 0.044), and triglycerides (0.028 [0.003– 0.052]; P = 0.025) and lower HDL cholesterol (0.032 [0.008 – 0.057]; P = 0.009). There was no evidence of these associations when adjusting for BMI. Associations with fasting alanine aminotransferase, γ-glutamyl-transferase, LDL cholesterol, A1C, and systolic and diastolic blood pressure were in the expected direction but did not reach P < 0.05. For all metabolic traits, effect sizes were consistent with those expected for the per allele change in BMI. FTO genotype was associated with a higher odds of metabolic syndrome (odds ratio 1.17 [95% CI 1.10 –1.25]; P = 3 × 10−6).
CONCLUSIONS
FTO genotype is associated with metabolic traits to an extent entirely consistent with its effect on BMI. Sample sizes of >12,000 individuals were needed to detect associations at P < 0.05. Our findings highlight the importance of using appropriately powered studies to assess the effects of a known diabetes or obesity variant on secondary traits correlated with these conditions.
The global prevalence of obesity and overweight (defined by a BMI ≥30 and ≥25 kg/m2, respectively) is increasing rapidly (1). Obesity and overweight are key risk factors for type 2 diabetes (2). Although recent increases in obesity reflect life-style changes, genetic factors are also important in predisposing some individuals to obesity.
Common variation in the FTO (fat mass– and obesity-associated) gene is associated with higher BMI and the risk of obesity in populations of European and Hispanic ancestry (3-5). Each copy of the A allele at rs9939609 is associated with a 0.10-SD (95% CI 0.08–0.12) higher BMI, equivalent to an increase of ~0.4 kg/m2, and a 1.31-fold (1.23–1.39) higher odds of obesity (3). A study of 5,243 children showed that the effect is almost exclusively mediated by differences in fat mass (3). The FTO variant is also associated with higher odds of type 2 diabetes (per allele odds ratio [OR] ~1.25; P = 5 × 10−8), although this effect can be entirely explained by differences in BMI between case and control subjects (3,6-9).
The association between FTO genotype and type 2 diabetes suggests that the FTO alleles that raise adiposity have adverse metabolic consequences. However, the question of which metabolic phenotypes and to what degree they are altered has not been tested in large numbers. Obesity is associated with insulin resistance, nonalcoholic fatty liver disease, hyperglycemia, hypertension, and dyslipidemia in the general population (10). These associations continue throughout the BMI range and are often seen as early as childhood (11). Individually and when used together to define metabolic syndrome, these traits are important predictors of type 2 diabetes and cardiovascular disease (10,12-16). An examination of the effects of FTO variation on quantitative traits may improve our understanding of how genetic alterations to fat mass could predispose to type 2 diabetes and other obesity-related diseases.
In this study, we investigated the association between common variation in the FTO gene and metabolic traits using data from seven studies (n = 17,037). We hypothesized that the FTO variant would be associated with metabolic traits but that these associations would be mediated through the effect of the variant on adiposity. We tested whether effect sizes reflected the magnitude of the FTO-BMI association and those of associations between BMI and metabolic traits from epidemiological studies.
RESEARCH DESIGN AND METHODS
We used data from seven adult studies of white European origin: two groups of nondiabetic individuals, selected from the general population (the Exeter Family Study of Childhood Health [EFSOCH] [17] and the U.K. Type 2 Diabetes Genetics Consortium Collection [UKT2D GCC] control subjects [7]), and five population-based samples (the Northern Finland Birth Cohort of 1966 [NFBC1966] [18], the Oxford Biobank [19], the Caerphilly study [20], the British Women’s Heart and Health Study [BWHHS] [21], and the InCHIANTI study [22]). The basic characteristics of the seven studies are presented in Table 1 and by Frayling et al. (3). Further details are provided in supplementary methods, which are available in an online appendix at http://dx.doi.org/10.2337/db07-1466.
TABLE 1.
Basic characteristics of all studies
| Study name | n* | Males | Age (years) | BMI (kg/m2)† | Prevalence of metabolic syndrome (NCEP definition) |
|---|---|---|---|---|---|
| NFBC1966 | 4,435 | 48.2 | 31‡ | 24.37 (20.75–28.63) | 6.6 |
| EFSOCH | 1,196 | 74.8 | 33 (30–37) | 25.59 (21.91–29.89) | NA |
| Oxford Biobank | 1,154 | 51.0 | 42 (36–46) | 25.81 (22.06–30.19) | 14.7 |
| Caerphilly | 1,328 | 100 | 56 (53–60) | 26.36 (23.01–30.19) | 20.7 |
| UK T2D GCC controls | 4,779§ | 49.4 | 60 (50–70) | 26.56 (22.56–31.27) | 16.1 |
| BWHHS | 3,244 | 0 | 69 (64–73) | 27.15 (22.86–32.26) | 45.4 |
| InCHIANTI | 901 | 44.3 | 71 (66–77) | 26.90 (23.13–31.28) | 28.2 |
Data are n, median (interquartile range) for age, geometric mean (SD range) for BMI, and % for males and prevalence of metabolic syndrome. NA, not applicable because not all criteria were available.
Number of individuals with FTO genotype, BMI, and at least one of the metabolic traits available.
SD range: 10(log10mean–log10SD) for lower value; 10(log10mean + log10SD) for upper value.
Interquartile range is not applicable to NFBC1966 because all subjects were studied at the same age.
Although blood pressure data were available for all UKT2D GCC control subjects, the maximum number of individuals with FTO genotype and fasting biochemical data was 1,902.
Metabolic phenotypes
We studied 10 quantitative traits, for which we had data from >6,000 participants. Altered fasting insulin, glucose, triglycerides, HDL cholesterol, alanine aminotransferase (ALT), γ-glutamyl-transferase (GGT), LDL cholesterol, A1C, and systolic and diastolic blood pressure are all known from epidemiological studies to be associated with higher BMI and an increased risk of type 2 diabetes and cardiovascular disease. The liver enzymes, ALT and GGT, are markers for nonalcoholic fatty liver disease, and A1C is a measure of glycemia over the preceding 2–3 months.
We grouped individuals according to the National Cholesterol Education Program (NCEP) Adult Treatment Panel III definition of metabolic syndrome (14). Individuals were classified as having metabolic syndrome on the basis of thresholds for waist circumference (men, ≥102 cm; women, ≥88 cm), triglycerides (≥1.7 mmol/l), HDL cholesterol (men, <1.03 mmol/l; women, <1.29 mmol/l), blood pressure (systolic ≥130 mmHg or diastolic ≥85mmHg), and fasting glucose (≥5.6 mmol/l). Metabolic syndrome was defined as the crossing of any three or more thresholds. Under this definition, an individual may be classified as having metabolic syndrome even if their waist and glucose measurements fall below the thresholds. Therefore, in contrast to other definitions (12,13), the NCEP definition is more independent of waist circumference and type 2 diabetes, traits that are already known to be associated with FTO genotype (3).
Given its importance in defining the metabolic syndrome, we included analyses of waist circumference as an additional quantitative trait in the current study.
Choice of marker, genotyping, and quality control
We used the single nucleotide polymorphism (SNP) rs9939609 as a marker of the FTO risk variant. Previous studies have reported that other SNPs (for example, rs9930506, rs1421085, and rs17817449) are associated with BMI and obesity (4,5), but these are strongly correlated to each other and to rs9939609 in individuals of European ancestry, based on HapMap data (r2 for all pairwise correlations >0.8).
Genotyping of rs9939609 has been described in detail previously (3). Further genotyping had been carried out for ~1,000 additional UKT2D GCC control subjects and ~400 additional Oxford Biobank participants since the previous publication (see supplementary methods in the online appendix).
Statistical methods
Within-study analyses
All quantitative traits were skewed in most studies and were therefore log10 transformed to normalize before analysis. To facilitate comparisons between studies, Z scores were generated within each study using the sex-specific means and SDs of each log10-transformed trait. Within each study, we examined the association between each quantitative trait and BMI using linear regression of log10trait Z score against log10BMI Z score. We examined the association between FTO rs9939609 genotype and each quantitative trait (including BMI) using linear regression of log10trait Z score against genotype. We used an additive genetic model (which assumes a consistent change in trait per additional risk allele) because, using the same studies, we previously found no evidence for departure from additivity in the FTO-BMI association (3). In addition, we performed all of these analyses while correcting for BMI by including log10BMI Z score in the regression model as a covariate.
To investigate the association between FTO genotype and metabolic syndrome, we grouped individuals in each cohort according to the NCEP definition (14). We used logistic regression to assess the relationship with FTO genotype.
To assess whether the inclusion of individuals on lipid-lowering or blood pressure medication (0.1–8 and 2–37% of cohorts, respectively) or with diabetes (1–11% of the population-based cohorts) influenced our results, we performed a series of sensitivity analyses with these individuals excluded. Further details of these are given in the supplementary methods in the online appendix.
Meta-analysis
Meta-analysis statistics and plots were produced using the METAN module (23), developed for Stata (College Station, TX). We used the inverse variance method to pool summary data from the linear regression analyses performed in the individual studies. We used the I2 statistic to estimate the percentage of total variation in study estimates that is due to between-study heterogeneity (24). We combined summary statistics from the six studies with sufficient data available for metabolic syndrome using a fixed-effects Mantel-Haenszel meta-analysis model.
Calculation of expected effect sizes for the associations between FTO genotype and metabolic traits
Adjusting the FTO-trait associations for BMI may help indicate whether the associations are driven by BMI, but it does not provide an accurate way of testing whether the effect sizes seen are as expected given the FTO-BMI and BMI-trait associations. We used a triangulation approach to estimate expected effect sizes for the associations between FTO genotype and metabolic traits (Fig. 1). We hypothesized that any such associations would be mediated by BMI. Therefore, the magnitude of the FTO-BMI association (Fig. 1a) and each BMI–metabolic trait association (Fig. 1b) would determine the effect size of each FTO–metabolic trait association (Fig. 1c).
FIG. 1.
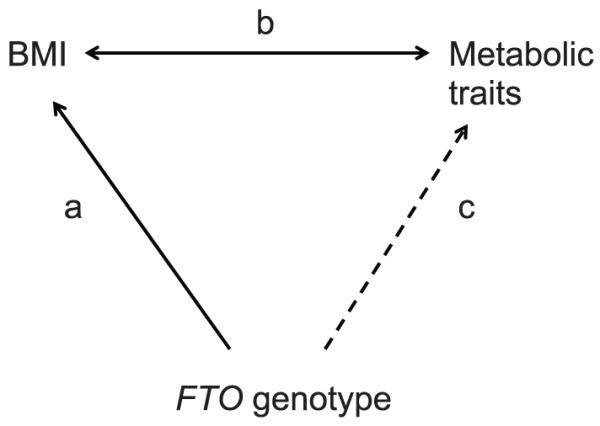
Triangulation approach used to estimate the effect size of the FTO–metabolic trait association (c) given the association between FTO and BMI (a) and the observed epidemiological associations between BMI and the traits (b). We hypothesized that associations observed between FTO genotype and metabolic traits would be mediated by BMI (i.e., c = a × b). Effect sizes would therefore be expected to reflect both the FTO-BMI association and the BMI–metabolic trait associations.
Meta-analysis of the metabolic trait–BMI effect sizes from each cohort produced an overall estimate of the SD change in each trait associated with a 1-SD increase in BMI (on the log10 scale). Because, in the current study, the per A allele effect size of FTO on BMI was 0.088 SD, we scaled down each estimate by a factor of 0.088. In this way, we were able to predict the SD change in each trait that should be associated with each additional FTO rs9939609 A allele in the genotype. For example, the change in log10(fasting insulin) Z score associated with a 1-SD increase in log10(BMI)was 0.433 SD. Multiplying this by 0.088, we calculated the expected change in log10(fasting insulin) Z score to be 0.038 SD per FTO A allele. For each metabolic trait, we computed a Z statistic (see supplementary methods in the online appendix) to assess the evidence that the observed and expected effect sizes were different. We checked that the point estimates for each BMI–metabolic trait association were similar when derived from fixed- and random-effects meta-analyses. We subjected waist circumference to the same set of analyses as the 10 metabolic traits to test the hypothesis that the known association between FTO and waist circumference is mediated through general adiposity as opposed to a specific effect on visceral adiposity.
RESULTS
Association of FTO genotype with BMI (Fig. 1a)
As described previously (3), FTO genotype was associated with BMI. In the current study, each copy of the rs9939609 A allele was associated with a 0.088-SD (95% CI 0.066–0.109) higher BMI (P = 2 × 10−15; n = 17,037). There was no detectable between-study heterogeneity, as measured by the I2 statistic (0%).
Association between BMI and metabolic traits (Fig. 1b)
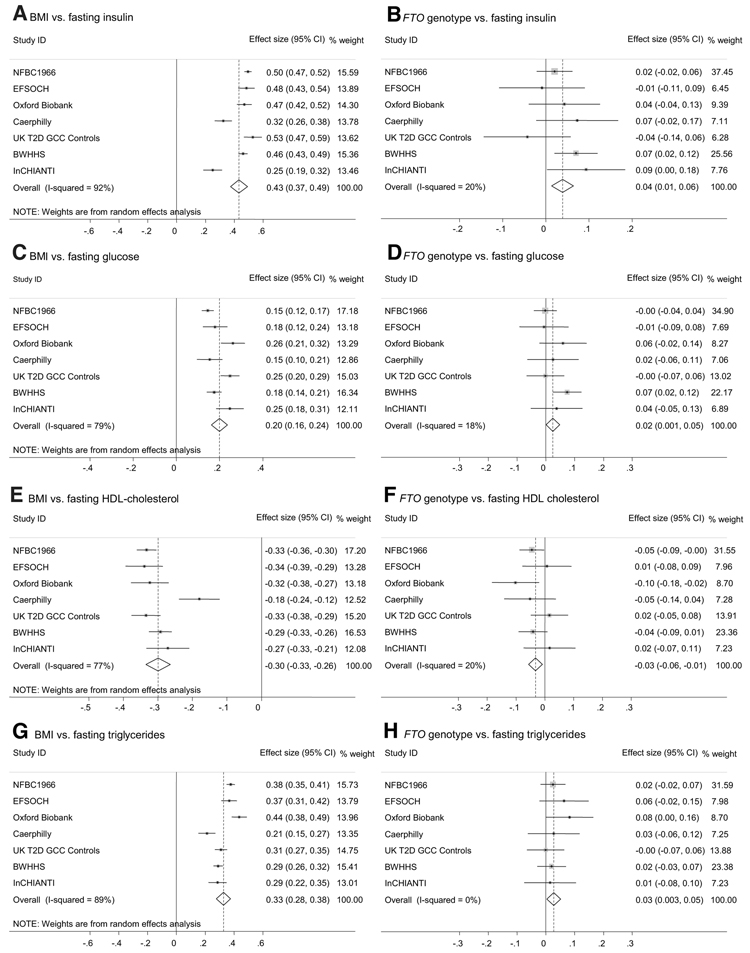
We assessed the association between BMI and 10 quantitative metabolic traits. As expected from previous epidemiological studies, all of the metabolic traits were associated with BMI (Table 2; Fig. 2A, C, E, and G; Supplementary Fig. 1). Waist circumference was highly correlated with BMI (Table 2; Supplementary Fig. 1). There was evidence of between-study heterogeneity, but the point estimates of overall effect sizes were very similar between fixed-effects models (which assume that each study comes from the same background population) and random-effects models (which account for differences between background populations; data not shown).
TABLE 2.
Meta-analysis of associations of metabolic traits with FTO rs9939609 genotype and with BMI
| Phenotype | n | Expected change in trait Z score per 0.088 SD BMI increase |
P value (BMI vs. trait)* |
Observed change in trait Z score per A allele |
P value (FTO vs. trait)† |
P value for difference between observed and expected |
Observed change in trait Z score per A allele, adjusted for BMI |
P value (FTO vs. trait)‡ |
|---|---|---|---|---|---|---|---|---|
| Fasting insulin | 12,095 | 0.038 (0.033–0.043) | 5 × 10−47 (92) | 0.039 (0.013–0.064) | 0.003 (20) | 0.95 | −0.005 (−0.027 to 0.018) | 0.69 (34) |
| Fasting glucose | 13,632 | 0.018 (0.014–0.021) | 1 × 10−25 (79) | 0.024 (0.001–0.048) | 0.044 (18) | 0.60 | 0.006 (−0.017 to 0.029) | 0.62 (22) |
| Fasting HDL cholesterol | 13,659 | −0.026 (−0.029 to −0.023) | 2 × 10−62 (77) | −0.032 (−0.057 to −0.008) | 0.009 (20) | 0.66 | −0.004 (−0.027 to 0.019) | 0.74 (44) |
| Fasting LDL cholesterol | 13,476 | 0.011 (0.004–0.018) | 0.001 (95) | 0.015 (−0.009 to 0.040) | 0.22 (0) | 0.78 | 0.001 (−0.023 to 0.026) | 0.91 (0) |
| Fasting triglycerides | 13,651 | 0.029 (0.024–0.033) | 3 × 10−39 (89) | 0.028 (0.003 to 0.052) | 0.025 (0) | 0.95 | −0.003 (−0.026 to 0.020) | 0.81 (0) |
| Systolic blood pressure | 15,624 | 0.019 (0.011–0.026) | 4 × 10−6 (97) | 0.016 (−0.007 to 0.039) | 0.16 (0) | 0.83 | 0.0004 (−0.022 to 0.022) | 0.97 (0) |
| Diastolic blood pressure | 15,619 | 0.020 (0.010–0.030) | 1 × 10−4 (98) | 0.021 (−0.002 to 0.044) | 0.067 (32) | 0.93 | 0.004 (−0.018 to 0.026) | 0.72 (15) |
| Fasting ALT | 6,171 | 0.021 (0.014–0.028) | 7 × 10−9 (89) | 0.034 (−0.003 to 0.070) | 0.069 (0) | 0.48 | 0.008 (−0.027 to 0.043) | 0.66 (0) |
| Fasting GGT | 6,596 | 0.018 (0.011–0.025) | 4 × 10−7 (90) | 0.026 (−0.009 to 0.061) | 0.15 (0) | 0.66 | 0.005 (−0.030 to 0.039) | 0.80 (0) |
| A1C | 8876 | 0.014 (0.012–0.017) | 2 × 10−33 (33) | 0.015 (−0.015 to 0.045) | 0.32 (53) | 0.97 | 0.001 (−0.029 to 0.031) | 0.95 (46) |
| Waist circumference | 16,639 | 0.075 (0.073–0.077) | < 1 × 10−100 (87) | 0.087 (0.065–0.108) | 9 × 10−15 (0) | 0.28 | 0.013 (0.001–0.024) | 0.027 (54) |
Data are means (95% CI) for observed and expected effect sizes, and I2 (%) values are given after the meta-analysis P values. All continuous traits were log10 transformed before calculation of sex-corrected Z scores. All effect sizes (95% CIs) are presented in SD units. I2 is the percentage of total variation in study estimates that is due to between-study heterogeneity (24).
P values are from random-effects meta-analysis of linear regression coefficients estimated within each study for each phenotype Z score (on the log10 scale) against BMI Z score (log10 scale).
P values are from fixed-effects meta-analysis of linear regression coefficients estimated within each study for each phenotype Z score (on the log10 scale) against rs9939609 genotype.
P values are from fixed-effects meta-analysis of within-study linear regression coefficients for each phenotype Z score (on the log10 scale) against rs9939609 genotype, with BMI Z score (log10 scale) as a covariate.
FIG. 2.
Meta-analysis plots for key quantitative traits associated with insulin resistance and the metabolic syndrome. Effect sizes for A, C, E, and G: SD change in trait (log10scale) per 1 SD higher BMI (log10 scale) (equal to the correlation coefficient between log10[trait] and log10[BMI]). Effect sizes for B, D, F, and H: SD change in trait (log10 scale) per FTO A allele.
Association between FTO genotype and metabolic traits (Fig. 1c)
Meta-analysis of the seven studies revealed evidence for association at P < 0.05 between FTO genotype and four of the metabolic traits examined: fasting insulin, glucose, triglycerides, and HDL cholesterol. Waist circumference was also strongly associated with FTO genotype, as described previously (3). There was little detectable between-study heterogeneity (Table 2). Meta-analysis plots of the associations between FTO genotype and fasting insulin, glucose, HDL cholesterol, and triglycerides are shown in Fig. 2B, D, F, and H. The effects of FTO genotype on fasting insulin, glucose, HDL cholesterol, and triglycerides are approximately equivalent to differences between homozygotes of 2 pmol/l, 0.04 mmol/l, −0.02 mmol/l, and 0.03 mmol/l, respectively. Further meta-analysis plots and data for the individual studies are provided in Supplementary Fig. 1 and Supplementary Table 1. Additional adjustment for age made little difference to the results (Supplementary Table 1).
Adjustment for BMI did change the results: The evidence for association between FTO and fasting insulin, glucose, triglycerides, and HDL cholesterol was removed, and effect size estimates for all 10 metabolic traits were reduced (Table 2). Evidence for association between FTO genotype and waist circumference was greatly reduced (P value increased from 9 × 10−15 to 0.027), and the effect size estimate was reduced from 0.09 to 0.01 SD per A allele (Table 2).
Exclusion of the 1–11% of individuals in each study with diabetes produced very similar results (Supplementary Table 2). Where information was available, excluding individuals known to be on lipid-lowering medication had little impact on the associations between FTO genotype and fasting HDL or LDL cholesterol or triglycerides, and excluding individuals known to be on medication for hypertension had little impact on the associations between FTO genotype and blood pressure (Supplementary Table 2).
Comparison of observed and expected effect sizes (Fig. 1c vs. a and b)
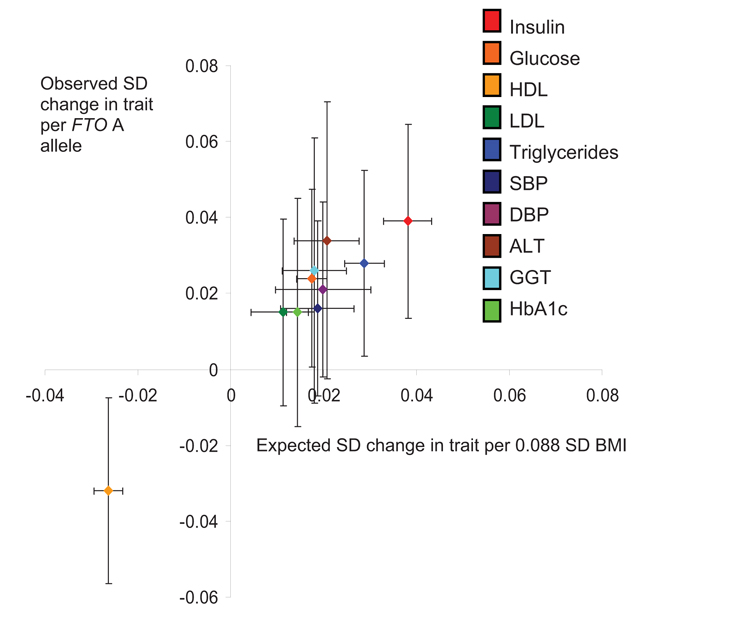
For all 10 metabolic traits, the observed per A allele change at rs9939609 was consistent with that predicted given the BMI-trait and FTO-BMI associations (Fig. 3). There was no evidence of a difference between the observed and expected effect sizes (all P > 0.25; Table 2). Observed associations remained consistent with those expected when individuals with diabetes were removed from the analyses (all P > 0.48).
FIG. 3.
Observed effect size per FTO A allele for each metabolic trait, plotted against expected effect size, given the FTO-BMI per A allele effect size estimate (0.088 SD) and the observed BMI-trait associations. Error bars represent 95% CIs.
Association between FTO genotype and metabolic syndrome
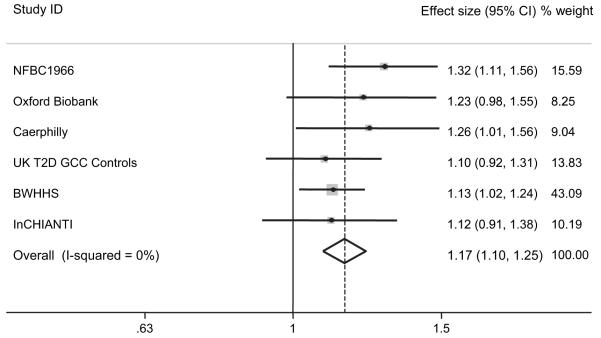
The prevalence of metabolic syndrome in each study is shown in Table 1. Meta-analysis (n = 12,555) revealed an association between FTO genotype and prevalence of the metabolic syndrome (per A allele OR 1.17 [95% CI 1.10–1.25]; P = 3 × 10−6; Fig. 4; Supplementary Table 3). Additional adjustment for age made little difference to the results (Supplementary Table 3), and exclusion of individuals with diabetes resulted in a similar effect size estimate (1.16 per allele [1.08–1.24]; P = 3 × 10−5; n = 11,965).
FIG. 4.
Meta-analysis plot of the association between the NCEP Adult Treatment Panel III definition of metabolic syndrome and FTO genotype in the six studies in which data on all criteria were available. Effect size: OR per FTO A allele.
DISCUSSION
In a large study involving >17,000 people from seven different population-based studies, we have shown that the BMI risk allele of FTO is also associated with the metabolic syndrome and its components. The sizes of the associations observed are consistent with the effect of the FTO variant on BMI and with observed epidemiological correlations between BMI and metabolic traits. This work has a number of important implications.
Further evidence that the increase in fat mass attributable to FTO genotype has a metabolic impact
The previously reported association between FTO genotype and type 2 diabetes suggested that the FTO alleles that raise adiposity have adverse metabolic consequences (3). However, the effects of FTO genotype on pre-diabetic intermediate traits were not known. The associations that we have observed between FTO genotype and metabolic traits provide further evidence that the BMI and fat mass increase attributable to FTO genotype is metabolically active. Four of the 10 associations between FTO genotype and metabolic traits (fasting insulin, glucose, triglycerides, and HDL cholesterol) reached P < 0.05. However, the observed effect sizes for all 10 traits are very similar to those expected given the FTO-BMI and BMI-trait associations. This strongly suggests that the proportion of extra fat carried by people with the FTO risk allele has a similar metabolic activity to that added by a combination of all genetic, lifestyle, and environmental factors in the general population. To explore this further, it may be informative to examine in detail the distribution of fat by genotype. Although we have shown previously that FTO genotype is associated with both skinfold thickness and waist circumference (3), precise and direct analyses of fat distribution by genotype using whole-body imaging have not been performed.
We tested the hypothesis that FTO is associated with waist circumference independently of BMI. Although a small residual association of waist circumference with FTO genotype after adjustment for BMI (P = 0.027) remained, the great reduction in effect size and the similarity of observed to expected effect sizes suggest our data are more consistent with a general effect of FTO on adiposity, which is not specifically mediated through visceral fat mass.
We observed a strong association between FTO genotype and the odds of metabolic syndrome, as defined by the NCEP Adult Treatment Panel III (14), which may be used to identify individuals at increased risk of type 2 diabetes and cardiovascular disease. Each additional A allele was associated with a 1.17-fold higher odds of metabolic syndrome (95% CI 1.10–1.25; P = 3 × 10−6). This result is not surprising given that four of the traits used to define metabolic syndrome (waist circumference, fasting glucose, fasting HDL cholesterol, and fasting triglycerides) showed individual associations with FTO genotype at P < 0.05.
Our results do not provide any further insight into how FTO genotype alters type 2 diabetes risk. Our previous data showed that each additional FTO A allele alters diabetes risk with an OR of 1.27 (95% CI 1.16–1.37) when case and control subjects are not matched for BMI (3). It seems unlikely that the small effects that we have observed in >12,000 individuals could result in this increase in diabetes risk, although lifetime exposure to these subtle differences may be expected to alter diabetes risk. Further studies are needed to test whether FTO genotype alters insulin secretion or more sophisticated measures of insulin resistance, although we note there is some evidence for association of FTO genotype with reduced whole-body insulin sensitivity (M/I; P = 0.02; n = 1,200), which is removed after adjustment for BMI (25).
The evidence is consistent with adiposity causing alterations in metabolic traits
The mechanism by which FTO alters fat mass is not known. It is therefore possible that the variant results in altered fat mass and altered metabolic traits through separate mechanisms. However, the consistency of the FTO genotype–metabolic trait effect sizes with those expected, given the FTO-BMI and BMI-trait associations, argues against this. Because FTO genotype is assigned at conception, associations between FTO alleles and traits are unlikely to be confounded. This use of genotypes proven to alter a trait to assess the causal direction of associations between that trait and others correlated with it is known as Mendelian randomization (26,27). Although it is the most widely accepted view, some have questioned whether raised adiposity is causally related to adverse metabolic and vascular outcomes (28). Our results, although not conclusive, are consistent with the view that increased adiposity causally alters metabolic traits. When the function of FTO is more fully understood, we will be able to draw firmer conclusions about how it informs this debate.
Appropriately powered studies are needed to assess the effects of known diabetes or obesity variants on secondary, correlated traits
Our findings highlight the importance of using appropriately powered studies to assess the effects of a known diabetes or obesity variant on secondary traits correlated with those conditions. Relatively modest sample sizes may be sufficient for associations between genetic variants and traits that are on the causal pathway to the associated disease, such as the type 2 diabetes–predisposing SNPs in TCF7L2 and insulin secretion (29,30). In contrast, very large numbers are likely to be needed to test for associations with traits that are secondary to the associated disease or primary quantitative trait. Here, power calculations should be informed by the association between genotype and disease and the correlation between the disease and secondary traits in a triangulation test. FTO genotype is by far the most convincing example of a common gene variant that is associated with BMI. Each additional FTO A allele is associated with a ~0.4 kg/m2 higher BMI, reflecting a difference in body fat between homozygotes of ~14% (3), and the correlations between BMI and many of the metabolic traits are strong. Despite this, our study illustrates that between 12,095 and 13,659 individuals were needed to detect associations at P < 0.05. Several traits did not reach formal significance despite the effect sizes being as expected. We estimate that for traits such as A1C and LDL cholesterol, which change only modestly with increased BMI, >70,000 individuals would be required for 80% power to detect the expected associations with FTO genotype at P < 0.05. Insufficient statistical power may help to explain why, in the first generation of genome-wide association studies, little evidence has been obtained for strong associations of disease-associated SNPs with quantitative disease-related traits (31): many of these traits will be imperfectly correlated with the disease and, therefore, require larger samples for detection. The important corollary of this point is that, given appropriate statistical power, it will be possible to improve our understanding of disease processes using gene variants known to alter a disease or trait, such as those in FTO.
Limitations
There are some limitations to our study. First, we used data from seven different studies that differed by their average age and sex distribution. This was necessary because power calculations suggested that we would need >12,000 individuals. It is likely that a single study of similar size would be more powerful. Second, the associations between BMI and metabolic traits were heterogeneous across studies. However, the point estimates were very similar in fixed- and random-effects models, which means our estimates of expected FTO–metabolic trait effect sizes are unlikely to be affected by this heterogeneity. Third, we have not taken into account the sampling error of the FTO-BMI association when calculating expected effects. To do this would require sophisticated statistical approaches such as instrumental variables analysis, which are not currently readily adaptable to meta-analyses of smaller studies. However, the narrow CIs and lack of heterogeneity in the estimate of the FTO-BMI association indicates that this is likely to result in a good approximation of the expected FTO–metabolic trait effect sizes. Fourth, we have not corrected our P values for multiple testing (10 traits). However, 4 of the 10 associations with quantitative traits reached P < 0.05, and 6 reached P < 0.1 (when we would expect only one P value <0.1 by chance); the association with metabolic syndrome, which incorporates information from several traits, reaches P = 3 × 10−6; and all of the observed effect sizes are extremely consistent with those expected. Together, this strongly suggests that our results are not false positives. Finally, our study was restricted to European white populations. Further studies are required to explore these relationships in populations of nonwhite ancestry. Associations between FTO genotype and obesity-related traits or type 2 diabetes have not been consistently observed in populations of Asian ancestry (32-35). A study of African Americans found no association between FTO genotype and obesity (5). Additional analyses of these populations using large samples will be needed to determine whether these differences are due to reduced power or reduced linkage disequilibrium between the variants tested and the putative causal variant or indicative of more fundamental heterogeneity between populations.
In summary, FTO genotype is associated with alterations in metabolic traits that are entirely consistent with its effect on BMI. The results further demonstrate that the increase in fat mass attributable to FTO genotype has an adverse metabolic impact. Our results also highlight the importance of using appropriately powered studies to assess the effects of a known diabetes or obesity variant on secondary traits correlated with these conditions.
Supplementary Material
ACKNOWLEDGMENTS
R.M.F. has received a Diabetes U.K. Research Studentship. D.A.L. has received a U.K. Department of Health Career Scientist Award. E.Z. is a Wellcome Trust Research Career Development Fellow. M.N.W. is a Vandervell Foundation Research Fellow. C.N.A.P. and A.D.M. are supported by the Scottish executive as part of the Generation Scotland Initiative. A.T.H. is a Wellcome Trust Research Leave Fellow. The work on the NFBC1966 was supported by the Academy of Finland (104781), the Medical Research Council (G0500539), and the Wellcome Trust (Project Grant GR069224). The EFSOCH was supported by National Health Service Research and Development and the Wellcome Trust. The Oxford Biobank study was supported by the British Heart Foundation. The Caerphilly study was funded by the Medical Research Council (U.K.) and the British Heart Foundation. The UKT2D GCC was supported by the Wellcome Trust (Biomedical Collections Grant GR072960). The BWHHS was funded by the U.K. Department of Health Policy Research Programme and the British Heart Foundation. The InCHIANTI study was supported by contract funding from the U.S. National Institute on Aging (NIA), and the research was supported in part by the Intramural Research Program, NIA, and National Institutes of Health. Genotyping was funded by the Wellcome Trust, Diabetes UK, the European Commission (EURODIA LSHG-CT-2004-518153), and the Peninsula Medical School.
We acknowledge the assistance of many colleagues involved in sample collection and phenotyping in all of the contributing studies. We are grateful to Prof. Leena Peltonen for providing DNA extraction for the NFBC1966 study and to Roger Harbord for helpful comments on the manuscript.
Footnotes
ALT, alanine aminotransferase; BWHHS, British Women’s Heart and Health Study; EFSOCH, Exeter Family Study of Childhood Health; GGT, γ-glutamyl-transferase; NCEP, National Cholesterol Education Program; NFBC1966, Northern Finland Birth Cohort of 1966; NIA, National Institute on Aging; SNP, single nucleotide polymorphism; UKT2D GCC, U.K. Type 2 Diabetes Genetics Consortium Collection.
Additional information for this article can be found in an online appendix at http://dx.doi.org/10.2337/db07-1466.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser. 2000;894:i–xii. 1–253. [PubMed] [Google Scholar]
- 2.Must A, Spadano J, Coakley EH, et al. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 3.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 5.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 10.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 11.Weiss R, Dziura J, Burgert TS, et al. Obesity and the metabolic syndrome in children and adolescents. N Engl J Med. 2004;350:2362–2374. doi: 10.1056/NEJMoa031049. [DOI] [PubMed] [Google Scholar]
- 12.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 13.Alberti KG, Zimmet P, Shaw J. The metabolic syndrome: a new worldwide definition. Lancet. 2005;366:1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 14.Grundy SM, Cleeman JI, Daniels SR, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 15.Sattar N, McConnachie A, Ford I, et al. Serial metabolic measurements and conversion to type 2 diabetes in the west of Scotland coronary prevention study: specific elevations in alanine aminotransferase and triglycerides suggest hepatic fat accumulation as a potential contributing factor. Diabetes. 2007;56:984–991. doi: 10.2337/db06-1256. [DOI] [PubMed] [Google Scholar]
- 16.Khaw KT, Wareham N, Luben R, et al. Glycated haemoglobin, diabetes, and mortality in men in Norfolk cohort of European prospective investigation of cancer and nutrition (EPIC-Norfolk) BMJ. 2001;322:15–18. doi: 10.1136/bmj.322.7277.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight B, Shields BM, Hattersley AT. The Exeter Family Study of Childhood Health (EFSOCH): study protocol and methodology. Paediatr Perinat Epidemiol. 2006;20:172–179. doi: 10.1111/j.1365-3016.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 18.Rantakallio P. The longitudinal study of the Northern Finland Birth Cohort of 1966. Paediatr Perinat Epidemiol. 1988;2:59–88. doi: 10.1111/j.1365-3016.1988.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 19.Tan GD, Neville MJ, Liverani E, et al. The in vivo effects of the Pro12Ala PPARgamma2 polymorphism on adipose tissue NEFA metabolism: the first use of the Oxford Biobank. Diabetologia. 2006;49:158–168. doi: 10.1007/s00125-005-0044-z. [DOI] [PubMed] [Google Scholar]
- 20.The Caerphilly and Speedwell Collaborative Group Caerphilly and Speedwell collaborative heart disease studies. J Epidemiol Community Health. 1984;38:259–262. doi: 10.1136/jech.38.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawlor DA, Bedford C, Taylor M, et al. Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women’s Heart and Health Study. J Epidemiol Community Health. 2003;57:134–140. doi: 10.1136/jech.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrucci L, Bandinelli S, Benvenuti E, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 23.Harris R, Bradburn M, Deeks J, et al. Statistical Software Components S456798. Boston College Department of Economics; Chestnut Hill, MA: [revised 19 Feb 2007]. METAN: Stata module for fixed and random effects meta-analysis. Available from http://ideas.repec.org/c/boc/bocode/s456798.html. [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pascoe L, Tura A, Patel SK, et al. Common variants of the novel type 2 diabetes genes, CDKAL1 and HHEX/IDE, are associated with decreased pancreatic β-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 26.Smith G Davey, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 27.Lawlor DA, Harbord RM, Sterne JA, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 28.Campos P, Saguy A, Ernsberger P, et al. The epidemiology of overweight and obesity: public health crisis or moral panic? Int J Epidemiol. 2006;35:55–60. doi: 10.1093/ije/dyi254. [DOI] [PubMed] [Google Scholar]
- 29.Saxena R, Gianniny L, Burtt NP, et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 30.Freathy RM, Weedon MN, Bennett A, et al. Type 2 diabetes TCF7L2 risk genotypes alter birth weight: a study of 24,053 individuals. Am J Hum Genet. 2007;80:1150–1161. doi: 10.1086/518517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nat Genet. 2007;39:813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Wu Y, Loos RJ, et al. Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57:264–268. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- 33.Horikoshi M, Hara K, Ito C, et al. Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia. 2007;50:2461–2466. doi: 10.1007/s00125-007-0827-5. [DOI] [PubMed] [Google Scholar]
- 34.Ohashi J, Naka I, Kimura R, et al. FTO polymorphisms in oceanic populations. J Hum Genet. 2007;52:1031–1035. doi: 10.1007/s10038-007-0198-2. [DOI] [PubMed] [Google Scholar]
- 35.Omori S, Tanaka Y, Takahashi A, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–795. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.