SUMMARY
Background
The development of the germline in Caenorhabditis elegans is a complex process involving the regulation of thousands of genes in a coordinated manner. Several genes required for small RNA biogenesis and function are among those required for the proper organization of the germline. EGO-1 is a putative RNA-directed RNA polymerase (RdRP) that is required for multiple aspects of C. elegans germline development and efficient RNAi of germline-expressed genes. RdRPs have been proposed to act through a variety of mechanisms including the post-transcriptional targeting of specific mRNAs as well as through a direct interaction with chromatin. Despite extensive investigation, the molecular role of EGO-1 has remained enigmatic.
Results
Here we use high-throughput small RNA and messenger RNA sequencing to investigate EGO-1 function. We found that EGO-1 is required to produce a distinct pool of small RNAs antisense to a number of germline-expressed mRNAs through several developmental stages. These potential mRNA targets fall into distinct classes, including genes required for kinetochore and nuclear pore assembly, histone-modifying activities and centromeric proteins. We also found several RNAi-related genes to be targets of EGO-1. Finally, we show a strong association between the loss of small RNAs and the rise of mRNA levels in ego-1(−) animals.
Conclusions
Our data support the conclusion that EGO-1 produces triphosphorylated small RNAs derived from mRNA templates and that these small RNAs modulate gene expression through the targeting of their cognate mRNAs.
INTRODUCTION
Several processes, including RNA interference (RNAi) in C. elegans, quelling in Neurospora and posttranscriptional gene silencing (PTGS) in plants have been shown to be related in both their method of action and their required components [1–3]. A fundamental link between these processes is their use of double-stranded RNA (dsRNA) and/or short interfering RNAs (21–25nt) as key effector molecules and reaction intermediates [4–7].
In addition to these exogenous silencing mechanisms, there is growing evidence for endogenous small RNAs playing a critical role in development. In C. elegans, one well-defined small RNA-based regulatory pathway involves a two-step process for endogenous small RNA production. The primary step in this pathway involves the RNA-directed RNA polymerase (RdRP) RRF-3, the dsRNA-binding protein RDE-4 and the RNase-III like enzyme DCR-1 in producing a small initial pool of 26 nt, 5′-monophosphorylated small RNAs [8–11]. The secondary step involves the activity of the RdRP RRF-1 in the soma and involves the production of a much larger pool of approximately 22 nt, 5′-triphosphorylated small RNAs which are essential for effective gene silencing [8–11].
The properties of small RNAs produced during exogenously-triggered RNAi in C. elegans provide further evidence for an amplification step in small RNA production and function. Initial RNAi experiments with exogenous long dsRNA (>500 nt) showed that as little as a few molecules per cell led to silencing [5]. Additionally, small RNAs have been found to map both upstream and downstream of the initial dsRNA trigger [12, 13]. Finally, the majority of small RNAs present during an RNAi response, are triphosphorylated at their 5′ ends and map antisense to exonic sequences [13]. Small RNAs that carry triphosphorylated 5′ termini are likely the direct products of RdRP initiation (unlike DCR-1 cleavage products that have been shown to carry 5′ monophosphates).
EGO-1 – a germline RdRP
The RdRP EGO-1 is related RRF-1 [14] and is a candidate to perform the secondary step in small RNA production in the germline. ego-1(−) animals are inefficient in exogenous RNAi against germline-expressed genes [14]. Moreover, EGO-1 is important in multiple aspects of germline development [14–17].
ego-1 appears to belong to a functional group of at least four loci with germline roles. Mutations in ego-1, csr-1, drh-3, and ekl-1 all exhibit defects in heterochromatin assembly on unpaired DNA [17, 18] and are enhancers of lethality in ksr-1, an important component of the Ras-ERK signaling pathway [19]. CSR-1 is a member of the large C. elegans Argonaute family and has been shown to bind small RNAs [20–22]. CSR-1 is also required for transgene-mediated cosupression [23] and efficient RNAi of germline-expressed genes [24]. DRH-3 is a DEAH/D-box helicase that associates with the Dicer protein, DCR-1 and is also required efficient RNAi in the germline and for the production of endogenous small RNAs [25]. EKL-1 is a Tudor domain-containing protein that has also been shown to be required for efficient germline RNAi, transgene silencing, and cosuppresion [26–28]. Antibodies to these four proteins have been reported to stain structures associated with DAPI-stained chromosomes undergoing mitosis in fixed C. elegans embryos. However, in adult germline tissue, where EGO-1 function is essential, EGO-1 staining is not evident on chromosomes. Rather, antibody staining of adult germline tissue suggests EGO-1 associates with perinuclear RNA-containing granules [21].
EGO-1 function
While much is known about the physical morphology of ego-1(−) mutant animals, little is known about the molecular phenotype in these animals. This has left several questions unanswered: What is EGO-1 doing to promote the proper development of the germline and specifically, what genes are being misregulated in the absence of EGO-1?
RESULTS
To better understand the role of EGO-1 in germline development and RNAi, we used an RNAseq approach to track small RNA (sRNA) and messenger RNA (mRNA) levels from ego-1(om84) and control animals. To avoid complications of variable embryonic development, we used strains in which the temperature-sensitive allele fem-1(hc17ts) had been introduced into both mutant and control animals. Using fem-1(hc17ts), all animals were feminized via growth at the restrictive temperature of 25°C (all library data summarized in Supp Methods Table 1).
EGO-1-dependent small RNA production
To determine what populations of small RNAs are dependent on EGO-1 activity we sequenced small RNA libraries from staged L3, L4, and adult animals. Previous work has indicated that a majority of RdRP-produced sRNAs are triphosphorylated at their 5′ ends [9, 13]. In order to capture all putative EGO-1-produced small RNAs we first applied a 5′-phosphate independent protocol for sequencing [8].
We sequenced multiple independent libraries of 5′-phosphate-independent sRNAs from L3 (2 independent libraries), L4 (2 independent libraries), and adult (3 independent libraries)-staged experimental and control animals. Data from replicates at each stage was examined independently. To test the reproducibility of library preparation and sequencing, we compared count numbers between replicate samples on a gene-by-gene basis (Figure 1, Supp Figure 1). These comparisons show a strong reproducibility of differential expression results from our library preparation and sequencing.
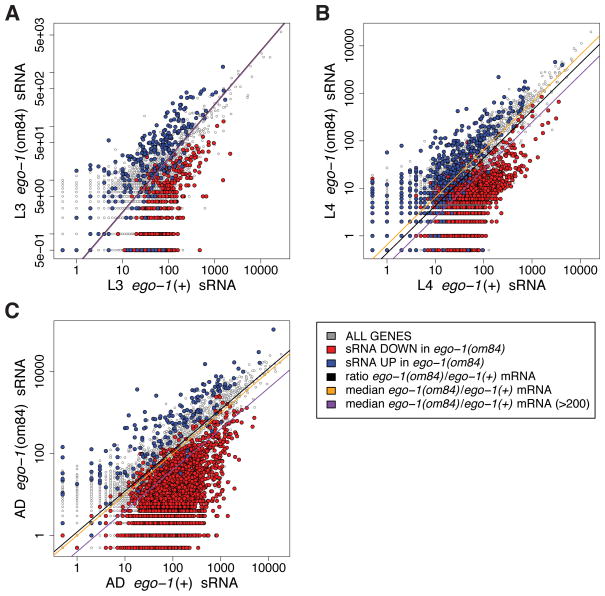
Figure 1. Stage-by-stage small RNA replicates.
Scatter plots depict a gene-by-gene comparison of small RNA abundance. Each plot compares data from ego-1(+) and ego-1(om84) animals at a single developmental stage. Each point represents a single gene, with an (x,y) coordinate defined by raw (non-normalized) counts of corresponding antisense small RNAs from the indicated libraries. A black line shows the ratio of total counts in each pair of samples giving the expected parity between samples. Two median lines are shown for each dataset: (i) median of the ratio of ego-1(om84)/ego-1(+) on a gene-by-gene basis (ii) median of the ratio of ego-1(om84)/ego-1(+) on a gene-by-gene basis using only those genes for which the sum of ego-1(om84) and ego-1(+) counts is greater than 200. Genes whose small RNA abundance increases [blue] or decreases [red] 3-fold (2-fold in L3) in ego-1(om84) (posterior probability ratio (PPR) < 0.005) in an independent replicate of the specified stage. (A) L3. (B) L4. (C) Adult.
Determination of putative EGO-1 targets
Comparing count numbers on a gene-by-gene basis for libraries from different stages, we found hundreds of genes that show a significant difference in small RNA abundance at each stage. These differences are highly reproducible in comparing distinct independent data sets (Figure 1). In addition, we observed a large number of genes that show a constant difference in multiple stages (Figure 2). Based on previous data showing that the majority of EGO-1 expression occurs in L4 and adult animals [14, 16], we initially focused our analysis on the genes affected in these two developmental stages.
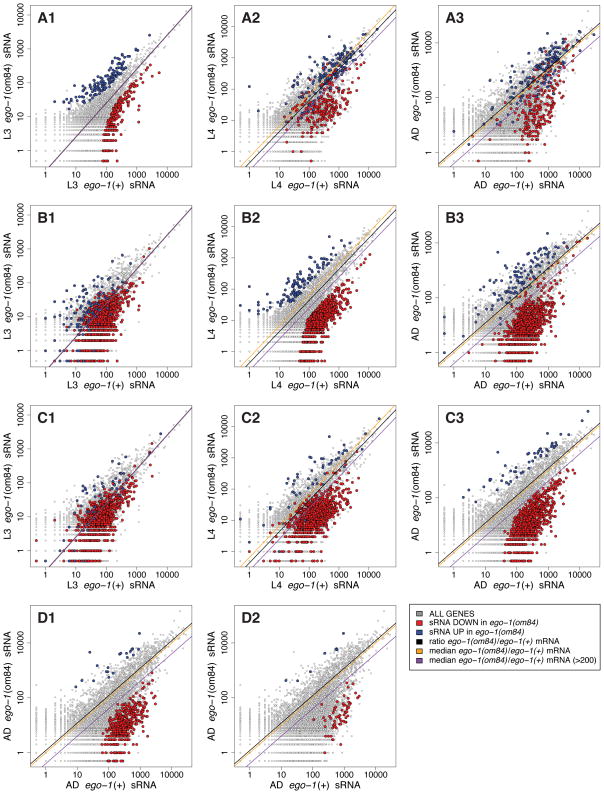
Figure 2. Stage-by-stage comparison of EGO-1 targets.
Scatter plots depict a gene-by-gene comparison of small RNA abundance. Plotted: Aggregated small RNA abundance for multiple replicates of L3, L4 or adult animals [gray]. Highlighted: Potential EGO-1 targets* in (A) L3. (B) L4. (C) Adult. (D1) L4/AD. (D2) L3/L4/AD. *Potential EGO-1 targets defined are as those genes whose antisense small RNA abundance changes at least 2-fold (L3) or 3-fold (L4 and adult) and who have a posterior probability ratio (PPR) < 0.005 in multiple replicates. Two median lines are shown for each dataset: (i) median of the ratio of ego-1(om84)/ego-1(+) on a gene-by-gene basis (ii) median of the ratio of ego-1(om84)/ego-1(+) on a gene-by-gene basis using only those genes for which the sum of ego-1(om84) and ego-1(+) counts is greater than 200. A black line shows the total ratio of total counts in each pair of samples giving the expected parity between samples.
We found that 437 gene loci had at least 3-fold fewer small RNAs (PPR < 0.005) in both L4 and adult ego-1(om84) samples. We also found that 20 genes showed at least 3-fold more small RNAs (PPR < 0.005) in both L4 and adult ego-1(om84) samples (Figure 2D-1, Supp Tables 1).
As a working hypothesis we consider small RNAs that are lost in ego-1(om84) animals to potentially represent direct products of EGO-1. RNAs that are enriched in ego-1(om84), would be unlikely to derive from synthesis by EGO-1 (these could show enrichment due to the loss of true EGO-1 products). We also note the alternative hypothesis that some or all of both sets of loci could be indirect targets or could be affected through downstream consequences of developmental defects in ego-1 mutant animals.
Despite the low levels of EGO-1 expression in L3 animals [16], we did find overlap between our candidate targets in L4 and adult samples with our L3 samples. We found that 60 genes that showed significantly fewer small RNAs in L4 and adult ego-1(om84) samples also showed at least 2-fold fewer small RNAs in L3 samples (p-value ~ 1.30 × 10−14). We found that 8 genes that showed significantly more small RNAs in L4 and adult ego-1(om84) samples also showed at least 2-fold more small RNAs in L3 samples (p-value ~ 1.64 × 10−19) (Supp Table 2, Figure 2D-2) [null hypothesis: no relationship between L3 targets and L4/adult targets].
To further characterize the temporal characteristics of EGO-1 action, we examined individual stage targets on each of the other stages. In Figure 3, we have plotted the aggregate sRNA counts from ego-1(om84) fem-1(hc17) and fem-1(hc17) for L3, L4, and adult small RNA libraries. For each stage (L3, L4, adult) we have highlighted putative EGO-1 targets from each of the three stages individually. For example, when we examine aggregate L3 small RNA abundance, we have highlighted those genes that were found to be significantly different in L3 (Figure 2A-1), in L4 (Figure 2B-1) and adult (Figure 2C-1). Similar comparisons were performed for aggregate L4 (Figure 2B) and adult (Figure 2C) samples. In analysis of these results, EGO-1 targets as a whole from any individual stage exhibit a strong parallel trend in their small RNA abundance through the other stages examined.
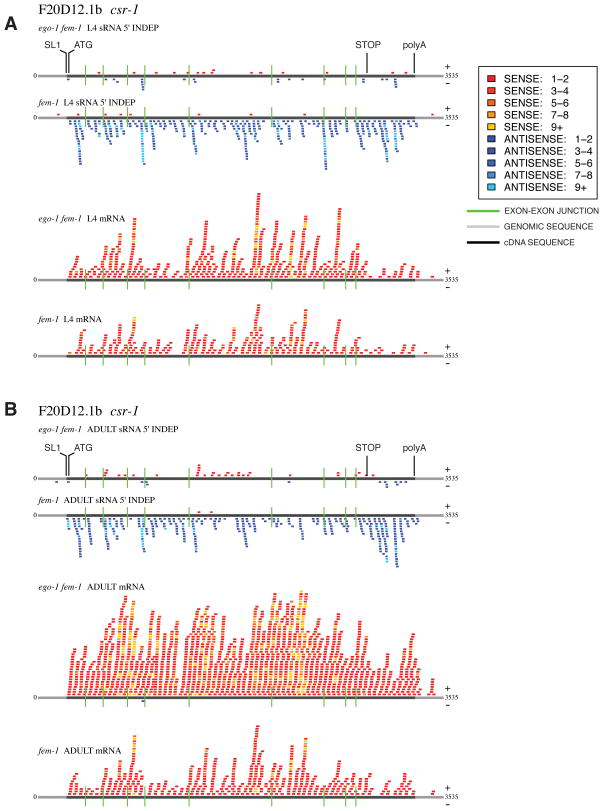
Figure 3. Small RNA and mRNA sequencing reads of csr-1.
Sense (shades of red) and antisense (shades of blue) reads of 5′-independent small RNAs and mRNA [experimental: ego-1(om84) fem-1(hc17) and control: fem-1(hc17)] mapped to csr-1 (spliced). Green lines represent exon-exon junctions. (A) L4-staged animals. (B) Adult-staged animals.
EGO-1-dependent small RNAs derive from germline-expressed mRNAs
In our analysis of EGO-1-dependent small RNAs we found that those specific RNAs that are missing in ego-1(−) animals are significantly more likely to be antisense to an mRNA molecule (p-value ~ 1.05 × 10−12). We also found that EGO-1 produces small RNAs that span the length of the target gene (Figure 3, Supp Figure 3). This suggests two features of the EGO-1-dependent sRNA populations. First, EGO-1 activity yields small RNAs antisense to a set of transcribed loci, and second, that these small RNAs generally span each targeted transcript.
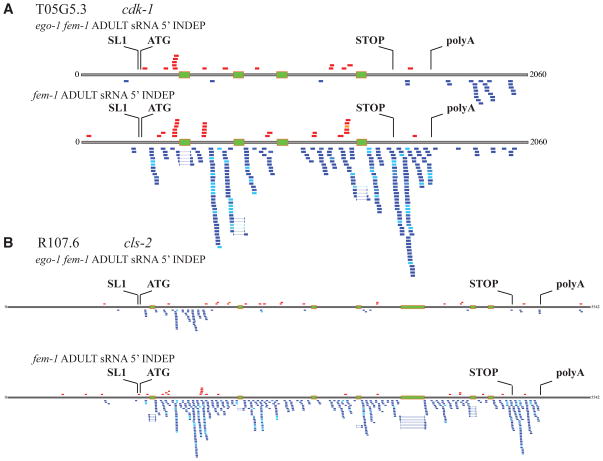
EGO-1 could conceivably copy genomic DNA, initial (unprocessed) transcripts, or processed mRNA. Our data are most consistent with the use of processed mRNA. First, we observe very few small RNAs that map to introns. Second, we found a substantial number of EGO-1-dependent small RNAs that span exon-exon junctions (Figure 4). Finally, we found small RNAs that span the 3′UTR/polyA junctions [29,30] for several L4/adult targets (Supp Figure 4); no such small RNAs were found in ego-1(−) mutant populations.
Figure 4. Small RNA and mRNA sequencing reads of cdk-1 and cls-2.
Sense (shades of red) and antisense (shades of blue) reads of 5′-independent small RNAs [experimental: adult ego-1(om84) fem-1(hc17) and control: adult fem-1(hc17)]. Green bars represent introns. (A) cdk-1. (B) cls-2.
Small RNA regulation can be used to extinguish residual expression of transcripts that are normally absent in a tissue [29] or to modulate expression of genes following their intended time of action [30]. Examination of putative EGO-1 targets inferred from our sequencing supports a focus for EGO-1 action on genes which have been active in the oogonial germline. First, examining a list of strongly oogonial-enriched mRNAs [31] shows a significant overrepresentation of these genes among putative EGO-1 targets. We found that both putative L3/L4/adult (p-value ~ 2.16 × 10−8) and putative L4/adult targets (p-value ~ 9.99 × 10−16) were significantly enriched for germline-expressed genes [31]. Second, we observe a relative underrepresentation of EGO-1 targets on the X chromosome (Figure 5, Supp Figure 5). The paucity of EGO-1 targets on the X chromosome would be consistent with a set of germline targets, given that the X chromosome is known to be substantially de-enriched for germline-expressed genes [31].
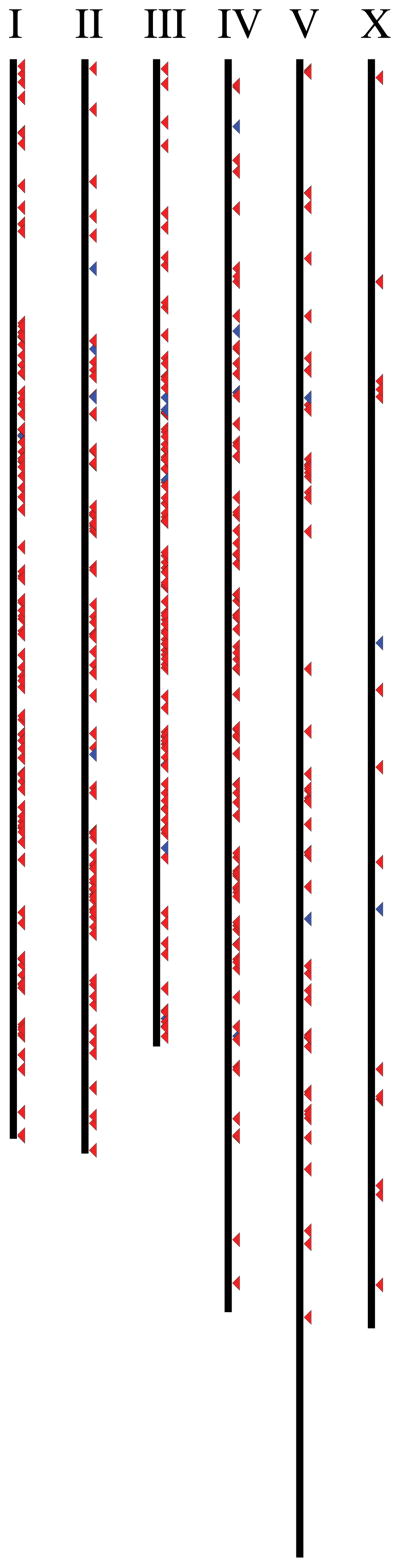
Figure 5. Chromosomal positions of EGO-1 targets.
L4 and adult. Genes whose small RNA abundance is decreased 3-fold [red, DOWN] or increased 3-fold [blue, UP] in ego-1[om84] relative to ego-1[+] with a posterior probability ratio [PPR] < 0.005. Chromosome size and gene positions are drawn to scale. [I] 104 genes DOWN, 1 gene UP. [II] 82 genes DOWN, 5 genes UP. [III] 101 genes DOWN, 6 genes UP. [IV] 79 genes DOWN, 4 genes UP. [V] 56 genes DOWN, 2 genes UP. [X] 15 genes DOWN, 2 genes UP. We found only 15 putative targets (437 total) to be on the X chromosome (p-value ~ 1.49×10−13). Correcting for multiple hypotheses, we found that EGO-1 targets are significantly underrepresented on the X chromosome.
Analysis of putative EGO-1 targets
We found several functionally related sets of genes to be prominent among EGO-1 targets, including genes involved in:
RNAi-related processes [24, 34–36] (L4/AD: p-value ~ 2.19 × 10−8)
Ras-ERK signaling [19] (L4/AD: p-value ~ 4.07 × 10−7)
Nuclear pore assembly/function [16] (L4/AD: p-value ~ 9.62 × 10−9)
Histone methyltransferases [17, 18] (L4/AD: p-value ~ 1.01 × 10−6)
Chromosome segregation [21] (L4/AD: p-value ~ 3.26 × 10−9)
These results are of particular interest given the phenotypic effects of EGO-1 on each of these cellular/biochemical processes.
Additionally, we found several unexpected groups of genes to be overrepresented amongst EGO-1 targets. Genes required for nonsense-mediated mRNA decay (L4/AD: p-value ~ 1.98 × 10−5), as well as a number of predicted ubiquitin ligases (L4/AD: p-value ~ 5.08 × 10−4) and serine/threonine phosphatases (L4/AD: p-value ~ 8.69 × 10−4) are highly enriched in our datasets. Groups of putative EGO-1 targets are summarized in Table 1.
Table 1. Putative EGO-1 target groups.
Gene groups and fold-enrichment (for the group) in L3/L4/AD and L4/AD ego-1 small RNA data sets. P-values for fold-enrichment: RNAi – (L3/L4/AD: 5.40 × 10−7; L4/AD: 2.19 × 10−8). Methyltransferases – (L3/L4/AD: 3.76 × 10−4; L4/AD: 1.01 × 10−6). Nuclear Pore Complex – L4/AD: 9.62 × 10−9). Centromere/Kinetochore – (L3/L4/AD: 8.64 × 10−7; L4/AD: 3.26 × 10−9). Kinesins – (L3/L4/AD: 1.24 × 10−4; L4/AD: 4.43 × 10−8). NMD – (L4/AD: 1.98 × 10−5). Ser/Thr Phosphatases – (L4/AD: 8.69 × 10−4). Ubiquitin Ligases – (L4/AD: 5.08 × 10−4). Ras-related – (L4/AD: 4.07 × 10−7). P granule – (L3/L4/AD: 0.020; L4/AD: 3.87 × 10−14). Transposon Silencing – (L3/L4/AD: 6.40 × 10−8; L4/AD: 4.10 × 10−9).
| Gene Group | L3/L4/AD | L4/AD | Examples |
|---|---|---|---|
| RNAi | 33.4X | 10.1X | CSR-1, TSN-1, C16C10.3, F58G1.1, T23D8.7 |
| Methyltransferases | 21.9X | 9.0X | SET-21(H3K9), SET-25(H3K9), SET-33(H3K9), SET-2(H3K4), SET-4(H3K20) |
| Nuclear Pore Complex | - | 15.2X | LMN-1, NPP-7(Nup98), NPP-11(Nup98), NPP-5(Nup107), NPP-8(Nup155), NPP-3(Nup205) |
| Centromere/Kinetochore | 61.1X | 18.9X | HCP-1(CENP-F), HCP-3(CENP-A), HCP-4(CENP-C) HCP-6, KNL-1, KNL-3, ROD-1 |
| Kinesins | 32.1X | 13.2X | KLC-1, KLP-7, KLP-15, KLP-16, KLP-18, ZEN-4 |
| Nonsense-Mediated mRNA Decay (NMD) | - | 15.7X | SMG-2, SMG-3, PAA-1, RUVB-1, RUVB-2 |
| Ser/Thr Phosphatases | - | 5.4X | GSP-1, PPH-6, SUR-6, PPTR-1 |
| Ubiquitin Ligases | - | 3.1X | BRC-1, CUL-3, NCL-1, UBC-9, UBC-13, WWP-1 |
| RAS-related | - | 4.8X | CCR-4, LET-60, LIN-15b, RAB-8, RHO-1, TRR-1 |
| P granule | 5.0X | 7.3X | CAR-1, GLD-3, GLH-4, PRG-1, SNR-1, XYG-9 |
| Transposon Silencing | 51.7X | 14.2X | MUT-14, MUT-16, PRG-1, K07C5.4 |
Genetic requirements for EGO-1-dependent small RNA production
To determine what other RNAi factors might be required for the production of EGO-1 dependent small RNAs, we analyzed a series of other small RNA libraries from several sources. We analyzed 5′-phosphate independent libraries from four RNAi-related mutants (rde-1(ne300), rde-4(ne299), rrf-1(pk1417) glp-4(bn2), and MAGO (WM126)) (Supp Figure 2A–H). In addition, we also analyzed data from CSR-1 protein complexes and several other RNAi-related mutants (csr-1(tm892), ego-1(om97), drh-3(ne4253), ekl-1(tm1599), rrf-3(pk1426), rrf-1(pk1417), eri-1(mg366), dcr-1(mg375), ergo-1(gg098)) (Supp Figure 2I-AB). Some libraries were from previously published work (including ego-1(om97) adults) [9, 21, 37], while others were from ongoing studies of the exogenous RNAi response in this lab (Julia Pak, personal communication). Although different conditions and staging were used in some of these cases (L4 versus adult and RNAi versus standard growth media), we have been able in each case to compare each mutant to an isogenic control under identical conditions.
As validation of the analysis, we found that our putative direct EGO-1 targets are also lost in a single ego-1(om97) library reported by Claycomb, et al., when matched to a comparable ego-1(+) library from the same investigators [21] (p-value ~ 5.13×10−28, Supp Figure 2K). ego-1(om84) and ego-1(om97) both contain stop codons early in the coding region and EGO-1 protein is not detected in extracts from either mutant [16].
The critical RNAi Argonaute factor RDE-1 has been shown to function in the response to foreign double-stranded RNA and may recruit RdRPs to small RNA/mRNA complexes [2, 38, 39]. We saw no requirement for RDE-1 in the production of small RNAs derived from putative EGO-1 target loci (Supp Figure 2A, 2E).
Interestingly, we did see a requirement for the double-stranded RNA binding protein RDE-4 [38, 37] for maximal accumulation of putative EGO-1 products from both L4/adult and L3/L4/adult targets (p-values of ~0.044 and ~ 6.22 × 10−3 respectively, Supp Figures 2B, 2F).
Additionally, we found putative products from EGO-1 targets to be decreased in rrf-1(pk1417) glp-4(bn2) animals (Supp Figures 2C, 2G). Although putative products from EGO-1 targets are decreased in rrf-1(pk1417) glp-4(bn2) they do not appear to be decreased in rrf-1(pk1417) (Supp Figure 2U, 2Z). This result is not surprising given that RRF-1 expression is highest in somatic tissues [12, 14] while EGO-1 expression is highest in germline tissues [14] we might predict that these two proteins would have non-overlapping targets (with limited effects in rrf-1 mutants). The loss of EGO-1 target small RNAs in rrf-1(pk1417) glp-4(bn2) animals would be consistent with the loss of germline tissue in these animals.
We did not find a significant shift in the multiple Argonaute mutant WM126 (Supp Figures 2D, 2H) or in mutants lacking the RNAi components ERI-1, ERGO-1, or RRF-3 (Supp Figure 2S, 2T, 2W, 2X, 2Y, 2AB). We did however find that EGO-1 targets became a significantly larger part of the small RNA pool in the the helicase domain-specific allele dcr-1(mg375) (Supp Figures 2V, 2AA) The latter (modest) effect may represent the loss of helicase-dependent small RNAs from other pathways.
As we described previously, EGO-1 has been functionally linked to CSR-1, DRH-3, and EKL-1 in a number of cellular processes. CSR-1 is a C. elegans Argonaute that physically interacts with small RNAs and is required for proper chromosome segregation [20, 21, 24]. DRH-3 interacts with DCR-1 and is required for germline development and efficient germline RNAi [25]. EKL-1 is also required for efficient RNAi as well as transgene silencing and cosuppression in the germline [19]. Using our statistical criteria, it appears that none of these three factors is required in EGO-1-dependent small RNA accumulation (Supp Figure 2L, 2M, 2Q, 2R). However, in examining the data, the number of antisense small RNA counts in these drh-3(ne4253) and ekl-1(tm1599) datasets is quite low. These low counts create more extreme median values and may cause a skewed perspective of the data.
While CSR-1 may not be involved in EGO-1 target small RNA production, we see a dramatic shift in EGO-1 targets in RNAs selected by CSR-1 immunoprecipitation [21]. We found that 295 of 437 L4/adult EGO-1 targets are enriched in CSR-1 complexes (p-value ~ 2.05×10−13, Supp Figure 2I). Included among these 295 genes are several RNAi-related genes (csr-1, tsn-1, mut-14, mut-16), centromere and kinetochore components (hcp-1, hcp-3, hcp-4, klp-19), and Ras-related genes (cdc-42, lin-9, rho-1, trr-1). The strong overlap between CSR-1-bound small RNAs and EGO-1 targets suggests that while CSR-1 may not be involved in the production of EGO-1-dependent small RNAs, it may be important in the function of these small RNAs.
Analysis of 5′-monophosphate-enriched small RNA populations
To further probe the physical structure of EGO-1-dependent small RNAs we sequenced a pool of small RNAs using an alternative procedure that relies on a 5′ monophosphate (thus capturing primarily 5′-P small RNAs). This analysis was performed on two biological replicates of adult experimental (ego-1(om84) fem-1(hc17)) and adult control (fem-1(hc17)) animals. Each library was amplified in two independent reactions and sequenced (for a total of four sequenced groups each from experimental and control animals). These libraries, as well as adult 5′-P independent libraries, were aligned to the C. elegans genome and transcriptome and small RNAs that matched perfectly antisense to genes were counted. In addition to alignment of these small RNAs, all libraries were also aligned to C. elegans miRNAs and 21U RNAs.
When we compared 5′-P-dependent and 5′-P-independent libraries, we found that small RNAs antisense to genes are greater than 100-fold enriched relative to miRNAs in 5′-P-independent sequencing. We observed a similar fold enrichment in small RNAs antisense to genes in 21U RNA datasets. We found that small RNAs antisense to genes are almost 74-fold enriched relative to 21U RNAs in 5′-P-independent sequencing. EGO-1 targets show a similar level of fold-enrichment.
This dramatic fold enrichment strongly suggests that the vast majority of small RNAs captured in our 5′-P independent capture procedure are not 5′-monophosphorylated. Based upon the two 5′-end capture procedures and several pieces of published data [9, 13, 41], it seems likely that the majority of these non-5′-monophosphorylated RNAs are in fact triphosphorylated and are products of RdRPs. We found that small RNAs antisense to L4/adult EGO-1 target loci are approximately 64-fold enriched to miRNAs and 43-fold enriched to 21U RNAs in 5′-P-independent sequencing and are therefore likely triphosphorylated products of EGO-1 RdRP function.
Analysis of messenger RNA levels
To determine if changes in sRNA abundance correlate with mRNA levels, we performed mRNA tag sequencing on L3, L4, and adult animals. We isolated poly(A)+ RNA and fragmented this RNA to the 100–200 nucleotide range. Using sense reads we calculated fold changes and posterior probability ratios for these mRNA libraries.
Using a 2-fold change with posterior probability ratio (PPR) < 0.005, we found that 132 of 19231 distinct gene models tested differ in abundance between experimental (ego-1(om84) fem-1(hc17)) and control (fem-1(hc17)) in the L3 stage (47 up in ego-1(om84), 85 down in ego-1(om84)). We found 119 genes differ in abundance in L4 (25 up, 94 down) and 113 genes differ in abundance in adult (56 up, 57 down).
Comparison of EGO-1 small RNA targets with mRNA abundance
We performed a sensitive evaluation of reciprocity in mRNA and sRNA changes in ego-1 mutant animals through a quantitative comparison over all genes. A sensitive comparison is important in that it might be expected that some targets would show only modest differences in mRNA levels. Of the 437 genes whose 5′-triphosphorylated small RNA levels decrease in an ego-1(om84) background in L4 and adult, we found 288 show increased mRNA levels in L4 (p-value ~ 1.40×10−11) (Supp Figure 6D) and 330 show increased mRNA levels in adult samples (p-value ~ 7.98×10−28) (Figure 6D). Of the 60 genes whose 5′-triphosphorylated small RNA levels decrease in an ego-1(om84) background in L3, L4 and adult, we found 49 show increased mRNA levels in L4 (p-value ~ 3.78×10−7) (Supp Figure 6E) and 55 in adult (p-value ~ 5.19×10−12) (Figure 6E). Of the 20 genes whose 5′-triphosphorylated small RNA levels increase in an ego-1(om84) background in L4 and adult animals we saw no significant shift in mRNA levels at any stage (Figure 6, Supp Figure 6).
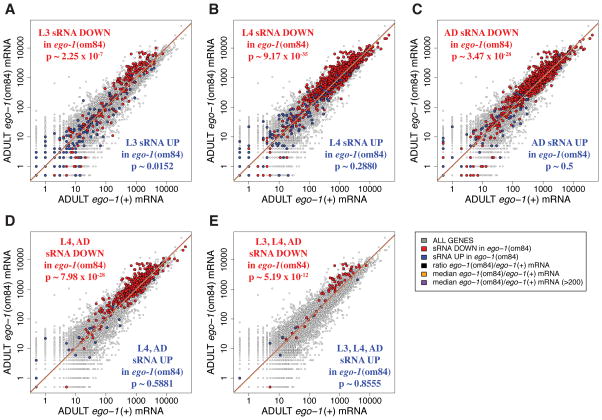
Figure 6. Summary of adult mRNA abundance.
Scatter plots depict a gene-by-gene comparison of mRNA abundance in staged adult animals (gray), and highlighted are genes whose sRNA abundance is down in ego-1(om84) (red) and genes whose sRNA abundance is up in ego-1(om84) (blue). As the key question for these data was the existence of an inverse relationship between small RNA (sRNA) and mRNA abundance, a central aspect of the data is the median values of the ratio of mRNA levels in ego-1(om84)/mRNA levels in ego-1(+). Two median lines are shown for each dataset: (i) median of the ratio of ego-1(om84)/ego-1(+) on a gene-by-gene basis (ii) median of the ratio of ego-1(om84)/ego-1(+) on a gene-by-gene basis using only those genes for which the sum of ego-1(om84) and ego-1(+) counts is greater than 200. A black line shows the total ratio of total counts in each pair of samples giving the expected parity between samples. Gene counts summary: (A) Changes in L3 sRNA abundance: 243 genes down 2-fold (red, p-value ~ 2.25×10−7) and 145 genes up 2-fold (blue, p ~ 0.0152). (B) Changes in L4 sRNA abundance: 1066 genes down 3-fold (red, p-value ~ 9.17×10−35) and 115 genes up 3-fold (blue, p ~ 0.2880). (C) Changes in adult sRNA abundance: 880 genes down 3-fold (red, p-value ~ 3.47×10−28) and 51 genes up 3-fold (blue, p ~ 0.5). (D) Changes in L4 and adult sRNA abundance: 437 genes down 3-fold (red, p-value ~ 7.98×10−28) and 20 genes up 3-fold (blue, p ~ 0.5881). (E) Changes in L3, L4, and adult sRNA abundance: 60 genes down 2-fold in L3 and 3-fold in L4 and adult (red, p-value ~ 5.19×10−12) and 115 genes up (blue, p ~ 0.8555).
Further focusing on mRNA abundance in L4 and adult stages we found 16 genes (10 up, 6 down) that have significant abundance differences in both stages. Of these 16 genes, 6 also show significant change in 5′-P-independent small RNA levels at both the L4 and adult stage (Table 2). For each of these 6 genes, we see an inverse relationship between 5′-P-independent small RNAs (up in ego-1(om84)) and mRNA abundance (down in ego-1(om84)). These 6 genes represent several classes that may be important in understanding the ego-1(om84) phenotype, including chromosome segregation and RNAi (csr-1, Figure 4), cell division (klp-7, Supp Figure 2A, 2B), germline development (mes-6, Supp Figure 2C, 2D), and heterochromatin formation (T12E12.2, Supp Figure 2E, 2F).
Table 2. Summary of genes with significant change in small RNA and mRNA abundance.
Fold-change (FC) and posterior probability ratios (P) for six genes whose small RNA abundance differs by at least 3-fold and mRNA abundance differs by at least 2-fold in L4 and adult animals [ego-1(om84) fem-1(hc17) vs fem-1(hc17)]. Calculations for fold-change and P-ratios are found in Methods.
| L4 | ADULT | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| sRNA | mRNA | sRNA | mRNA | ||||||
| FC | P | FC | P | FC | P | FC | P | ||
| mes-6 | Polycomb-like | 0.15–0.17 | <10−15 | 2.18–2.46 | <10−15–10−3 | 0.01–0.04 | <10−15–10−6 | 2.14–4.16 | <10−15 |
| F01G4.4 | Unknown function | 0.03–0.14 | <10−15 | 3.27–5.31 | <10−15 | 0.01–0.06 | <10−15 | 2.81–3.15 | <10−15 |
| csr-1 | Argonaute | 0.02–0.04 | <10−15 | 2.09–2.73 | <10−15 | 0.002–0.02 | <10−15 | 2.56–5.61 | <10−15 |
| klp-7 | Kinesin | 0.03–0.14 | <10−15 | 3.11–4.35 | <10−15 | 0.01–0.09 | <10−15 | 6.12–8.27 | <10−15 |
| T12E12.2 | Heterchromatin-assoc. protein HP1 | 0.07–0.08 | <10−15 | 2.26–4.33 | <10−15–10−3 | 0.02–0.17 | <10−15 | 2.64–2.97 | <10−15 |
| T12E12.3 | Unknown function | 0.12–0.15 | <10−15 | 2.94–6.29 | <10−15 | 0.01–0.10 | <10−15 | 5.38–9.90 | <10−15 |
These data support a reciprocal relationship between small RNA populations and consequences at the mRNA level. In particular, loci which yield large numbers of EGO-1-dependent small RNAs show substantially greater expression (relief of inhibition) in ego-1 mutant animals.
DISCUSSION
We have described a large class of genes that behave as targets of EGO-1 in both L4 and adult animals, with our results showing that small RNAs antisense to these genes are substantially reduced in ego-1(−) animals. These EGO-1 targets include histone methyltransferases, kinetochore and centromeric components, P granule components, RNAi-related factors and Ras-related genes. Additional classes of EGO-1 targets include a number of regulatory factors including Ser/Thr phosphatases and ubiquitin ligases that are not currently associated with specific aspects of the observed ego-1 mutant phenotype.
In addition to these 437 targets, it appears very likely that there are also additional EGO-1 targets. We found a number of interesting genes that fall just outside of our rather strict fold-change and/or statistical criteria. Included amongst this group are ego-1 itself, drh-3 and ekl-1, as well as critical RNAi components, RRF-3, ERI-1, DCR-1 (Supp Table 3). The number and function of putative EGO-1 targets points to EGO-1 having a central role in the modulation of a number of cellular processes including endogenous RNAi.
One possible mode of action of EGO-1 and EGO-1 products would be in the downregulation of target mRNAs. Although the mechanisms by which triphosphorylated small RNAs lead to mRNA turnover remain unclear, these RNAs are associated with downregulation of target mRNAs during RNAi and in the endogenous RRF-3 pathway [9–11, 13] We found that EGO-1 target mRNAs as a whole show a significant shift in their expression in ego-1(om84) animals. The regulatory shifts that are observed in mRNAseq data are not “all or nothing.” Rather, it appears that the RdRP system is used to achieve a globally-modest regulation of a large class of mRNA targets.
Does the EGO-1 target family comprise the genes where expression is valuable in specific stages of germline development but which are not needed or damaging at subsequent stages? This idea would closely correspond to another C. elegans RdRP, RRF-3, which downregulates a set of target mRNAs during spermatogenesis [8, 9]. In both spermatogenesis and oogenesis, a defined and intricate developmental pathway requires a carefully choreographed engagement of specific protein factors at specific times [40–42].
Many mRNAs expressed in oogenesis may encode factors that are only needed in transient stages with much of the regulation occurring post-transcriptionally [43]. One mechanism to effect a temporal “bump” in expression is to have a specific negative regulator turn on precisely at the time that expression of the target needs to be extinguished. As making new specific regulatory machinery for each stage of oogenesis may be evolutionary expensive, RdRPs may provide a modular mechanism that allows any mRNA to acquire a signal that will serve as a negative regulator that only activates after the mRNA has accumulated.
The identification of large numbers of target loci highlights the questions of both identity and number of physiologically-critical targets. The dramatic germline defects and unconditional sterility of ego-1 mutants could be caused by a specific misregulated target locus or a combined misregulation of many such loci, with the complexity of both the target pool and the observed phenotype certainly consistent with a contribution of many targets to aspects of the phenotype. Critical analysis of individual functional contributions of specific EGO-1 targets to organized germline development should emerge as biochemical/genetic analysis of individual target loci proceeds, as a more detailed view of the subcellular ego-1(−) mutant phenotype emerges, and as tools for examining multigenic influences on the critical subcellular phenotypes are developed.
RdRP-based modulation of gene expression has been characterized in fungi, plants, and animals [8, 9, 14, 46–48]. While not present in vertebrates, the eukaryotic RdRP protein superfamily that includes EGO-1 is present in vertebrate ancestors [47]. Observed RdRP activities in higher systems reserve the possibility of similar mechanisms modulating development in these systems [48, 49].
Supplementary Material
Acknowledgments
We are grateful to Julia Pak, Weng-Onn Lui, Jonathan Gent, Ayelet Lamm, Sam Gu, Poornima Parameswaran, Chaya Krishna, Karen Artiles, Michael Stadler, Cheryl Smith, Ziming Weng, Phil Lacroute, Arend Sidow, Virginia Walbot, Daniela Witten and Robert Tibshirani for their advice and assistance with experimental protocols and analysis; to Anne Villeneuve and John Pringle for comments on the manuscript; the Caenorhabditis Genetics Center for strains; and for financial support from NIH (R01GM377 06 [A.Z.F], T32HG00044 [J.M.M.]) and the NSF Graduate Research Fellowship Program [J.M.M.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 2.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 3.Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhettes S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- 4.Ratcliff F, Harrison BD, Baulcombe DC. A similarity between viral defense and gene silencing in plants. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics. 2009;183:1297–1314. doi: 10.1534/genetics.109.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci USA. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 13.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 14.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 15.Qiao L, Lissemore JL, Shu P, Smardon A, Gelber MB, Maine EM. Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics. 1995;141:551–569. doi: 10.1093/genetics/141.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vought VE, Ohmachi M, Lee M, Maine EM. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics. 2005;170:1121–1132. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maine EM, Hauth J, Ratliff T, Vought VE, She X, Kelly WG. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocheleau CE, Cullison K, Huang K, Bernstein Y, Spilker AC, Sundaram MV. The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics. 2008;178:1431–1443. doi: 10.1534/genetics.107.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 23.Robert VJP, Sijen T, van Wolfswinkel J, Plasterk RHA. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 25.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 26.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JK, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 28.Robert VJP, Sijen T, van Wolfswinkel J, Plasterk RHA. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 30.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 31.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 32.Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RHA. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- 33.Sijen T, Plasterk RHA. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- 37.Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- 38.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 39.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 40.Kimble J, Crittenden SL. WormBook. 2005. Germline proliferation and its control; pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenstein D. WormBook. 2005. Control of oocyte meiotic maturation and fertilization; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strome S. WormBook. 2005. Specification of the germ line; pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Racher H, Hansen D. Translational control in the C. elegans hermaphrodite germ line. Genome. 2010;53:83–102. doi: 10.1139/g09-090. [DOI] [PubMed] [Google Scholar]
- 44.Dougherty WG, Parks TD. Transgenes and gene suppression: telling us something new? Curr Opin Cell Biol. 1995;7:399–405. doi: 10.1016/0955-0674(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 45.Schiebel W, Pélissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sänger HL, Wassenegger M. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell. 1998;10:2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiu PKT, Metzenberg RL. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics. 2002;161:1483–1495. doi: 10.1093/genetics/161.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zong J, Yao X, Yin J, Zhang D, Ma H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene. 2009;447:29–39. doi: 10.1016/j.gene.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Chao M. RNA recombination in hepatitis delta virus: implications regarding the abilities of mammalian RNA polymerases. Virus Res. 2007;127:208–215. doi: 10.1016/j.virusres.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.