Abstract
During cell migration, the movement of the nucleus must be coordinated with the cytoskeletal dynamics at the leading edge and trailing end, and, as a result, undergoes complex changes in position and shape, which in turn affects cell polarity, shape, and migration efficiency. We here describe the steps of nuclear positioning and deformation during cell polarization and migration, focusing on migration through 3-dimensional matrices. We discuss molecular components that govern nuclear shape and stiffness, and review how nuclear dynamics are connected to and controlled by the actin, tubulin and intermediate cytoskeleton-based migration machinery and how this regulation is altered in pathological conditions. Understanding the regulation of nuclear biomechanics has important implications for cell migration during tissue regeneration, immune defence and cancer.
Introduction
Cell migration is a complex physicochemical process which leads to translocation of the cell body across two-dimensional (2D) surfaces, through basement membranes, or through three-dimensional (3D) interstitial tissues [1]. During migration through 3D tissues, the stiffness and density of the surrounding extracellular matrix (ECM) presents an additional physical challenge to the moving cell body. Two principal mechanisms are known by which migrating cells can overcome these constraints: (i) proteolytic ECM degradation leading to gap widening and cell-generated trail formation and (ii) elastic and plastic deformations of the cell body to fit through the available space [2]. If a cell is unable to “squeeze” through a particularly narrow region, it employs a third mechanism to maintain migration, i.e., retraction of already established protrusions and repolarization to explore the adjacent environment for an alternative route, thereby bypassing the obstacle [3]. Consequently, translocation through 3D tissues is dependent on both, deformability of the environment and the cell body.
The cell body consists of the plasma membrane surrounding the gel-like cytoplasm including small organelles (dimensions below the 1-2 micron range) and cytoskeletal filaments. These structures are morphologically adaptable enough to vigorously change and adjust to virtually any shape via a combination of (i) fast (passive) mechanical deformation and (ii) slower cytoskeletal remodelling [4-6]. Ultimately, cytoplasmic protrusions pass through gaps in the submicron range; this adaptability can be experimentally exploited to isolate protrusive cell regions including pseudopodia, filopodia and invadopodia using filters with pores sized of 1 micrometer and below [7,8]. In contrast, the nucleus, the largest cellular organelle, is approximately 5 to 10-times stiffer than the surrounding cytoskeleton as it is mechanically stabilized by a constitutive network of structural proteins (see below); therefore it commonly resists large changes in shape [5,9-12]. Consequently, for migration through small pores or 3D scaffolds, the nucleus can become the rate-limiting organelle [7,13,14].
Nuclear shapes and sizes can vary widely between different cell types and even within cell lines; nonetheless, most cells imaged in situ or cultured in 3D substrates have ovoid or spherical shaped nuclei with diameters of 5 to 15 μm [15]. In 2D culture, cells often spread out significantly, resulting in more disk-shaped nuclei 10 to 20 μm in diameter and only a few micrometers in height [16]. In a few often highly mobile cell types, however, including myeloid and cancer cells, the nuclei are bean-shaped, lobulated or segmented (“polymorphic”) (Table 1) and thus may develop greater morphological flexibility [14,17] (Fig. 1).
Table 1. Overview of factors influencing nuclear shape and stiffness.
| Factor | Effect on nuclear structure and mechanics |
|---|---|
| Lamins A and C | Loss of lamins A/C causes irregular shape and reduced nuclear stiffness, as well as loss of heterochromatin; increased expression can cause nuclear invaginations. [37,39,79,92] |
| Lamin B1 and B2 | Loss of lamin B1 causes nuclear blebs; overexpression can result in nuclear lobulation; lamins B1 and B2 have little or no effect on nuclear stiffness. [39,92,93] |
| Lamin B receptor (LBR) | (Over-)expression of LBR causes nuclear lobulation and formation of excess nuclear envelope. [52,94] |
| Chromatin configuration | Stem cell-like chromatin organization causes increased nuclear plasticity and reduced stiffness; condensed chromatin increased nuclear stiffness. [53,95] |
Figure 1.
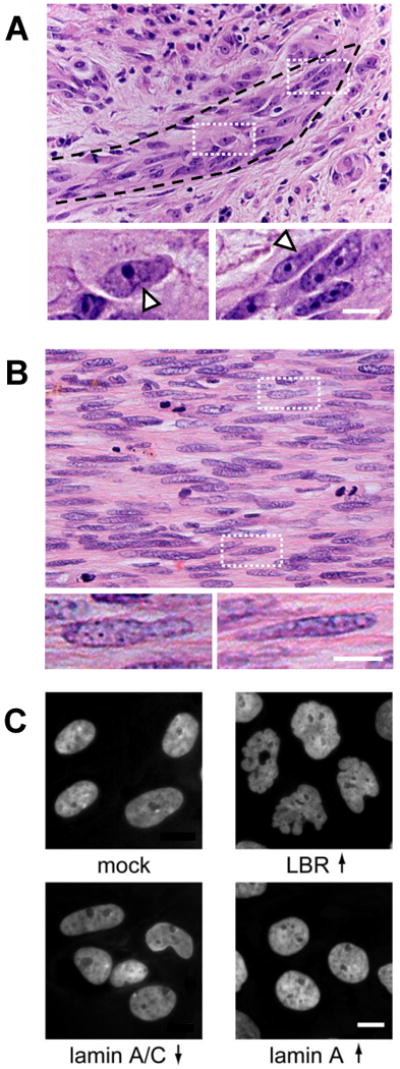
Nuclear diversity and deformation in cancer cells.
(A) Tip of an invasion zone of a primary human melanoma, showing heterogeneously shaped, elongated and partly deformed nuclei. Arrowheads in details, local deformation. (B) Central region of an invasion zone in human fibrosarcoma exhibiting elongated and partly deformed cigar-shaped nuclei of variable size. (C) Effect of altered nuclear envelope composition on mammary epithelial cells. Genetically modified MCF10A breast epithelial cells that stably express lamin B receptor (LBR↑), lamin A (Lamin A↑) or shRNA directed against lamins A and C (Lamin A/C ↓) or control vector (mock). Cells overexpressing LBR or having reduced expression of lamins A/C show striking similarities to the nuclear shape observed in many cancer cells. Both modifications result in decreased nuclear stiffness (J Lammerding, unpublished). Overexpression of lamin A results in rounder (and stiffer) nuclei. Bars, 10μm.
In vivo, the dimensions of structural pores available for migrating cells in basement membranes and interstitial connective tissue vary considerably, ranging from large gaps in the range of 30 μm and larger in loose connective tissue and lymph nodes [18,19] down to submicron pores in basement membranes [20]. For cells passing through pores which are smaller than the cell diameter, the extent of required cell deformation depends on whether the cell degrades the ECM proteolytically in regions of local compression, or not [21]. Surface-localized proteolytic cleavage of ECM fibers enlarges the available space and leads to the de novo formation of small tracks; the diameter of these tracks approximates the cross section of the cell and thereby reduces required cell deformation [13,22]. In both proteolytic and non-proteolytic migration through 3D tissues, the shapes of both cytoplasm und nucleus thus adopt their morphology and thereby minimize resistance towards tissue structures [3]. We here aim to integrate nuclear dynamics into the multistep model of cell migration through interstitial tissue and discuss the implications of nuclear mechanics for physiological and neoplastic cell migration and invasion.
Nuclear dynamics during cell migration
Steps of cell migration
Dependent on whether proteases are utilized or not, cell migration in 3D environments consists of four or five respective steps which are executed in a concurrent and cyclic manner [1,23] (Fig. 2). First the cell polarizes by actin assembly into filaments which push the plasma membrane outward and form protrusions (step 1), followed by the interaction of cell protrusions to the extracellular tissue matrix (step 2). In proteolytic migration through 3D tissues, the proteolytic degradation and realignment of ECM fibers results in the generation or widening of tracks (optional step 3) [23]. Myosin II mediated contraction of actin filament networks leads to tension between the leading and trailing edge (step 4) which facilitates the gradual release of adhesive bonds at the cell rear and rear-end sliding along the substrate (step 5).
Figure 2.

Nuclear dynamics and deformation during cell migration.
(A) HT-1080 cells expressing DsRed2 in the cytoplasm and H2B/eGFP in the nucleus migrating in 3D collagen lattice. Confocal time-lapse sequence in mid-density (3.3 mg/ml) collagen shows phases of shape change. Bar, 10 μm. (B) Initial cell polarization leads to rotational dynamics (I) followed by initial translocation of the nucleus (II). When physical barriers are encountered, the nucleus deforms during forward sliding (III). In the absence of ECM degradation capability, the nuclear deformation is more pronounced (IIIa). Alternatively, the cell degrades ECM structures proteolysically (green color) and generates a small trail-like matrix defect (IIIb, asterisk), thereby minimizing nuclear deformation.
Nuclear positioning during cell movement
With the exception of initial cell protrusion formation, all other steps of the migration cycle involve dynamic interactions between the cytoskeleton and the nucleus, resulting in changes in nuclear shape, orientation, and position within the cell [24,25]. First, cytoskeletal cell elongation is followed by nuclear rotation along the length axis of the cell [26]. Next, depending on the cell type, the nucleus first moves towards the cell rear or the leading edge, whereas the cell rear still remains in a stable position. In polarizing epithelial, neuronal and mesenchymal cells, the nucleus moves rearward of the centrosome and other cell organelles, including the ER and Golgi [27].
Conversely, in amoeboid-moving leukocytes, the nucleus moves towards the leading edge, anterior to the centrosome [28]; the reason for the difference between both migration types is unclear. In cells that retain their cell-cell junctions during migration and move as multicellular groups (collective cell migration), cadherin-based cell-cell junctions control the nucleus in rearward position to the ER and Golgi [29]. With the onset of rear-end sliding, the cell moves in a persistent manner, and the nucleus with it [30].
Mechanically, translocation of the nucleus is dependent on myosin-II mediated contraction of actin filaments and shortening of the cell rear while the leading edge remains anchored to the substrate, resulting in forward pushing of the nucleus [31]. Consequently, inhibition of myosin II, or its upstream regulators ROCK and the small GTPase Rho, leads to defects in rear retraction and nuclear forward movement [14,27,30,32]. These findings indicate that pushing from the cell rear is rate-limiting for nuclear translocation [31].
Nuclear deformation in regions of cell confinement
During cell passage through the basement membrane or 3D interstitial tissue, the nucleus further undergoes remarkable deformation, dependent on outside cues. Thereby, nuclear deformation results from several concurrent parameters, including tissue porosity, the shape of tissue discontinuities, traction force generated by moving cells, and the presence or absence of pericellular proteolysis (Fig. 2) [13]. Whereas in loose tissues, the nucleus retains its original ellipsoid shape during translocation (K. Wolf, unpublished), dense tissues with narrow regions in the path result in local compression of the cell, generating large intracellular forces that are transmitted across the cytoskeleton to the nucleus, leading to deformation of the nucleus (Fig. 2A) [13,14].
During this process (Box 1), the changes in nuclear shape are transient and reflect forces imposed by the geometry of outside tissue structure and the intracellular counterforces. The mechanics of cell constriction are poorly understood and likely comprise both, the passive physical pushing of extracellular scaffold into the cell body and nucleus, and active actomyosin-mediated constriction, similar to the contractile ring generating two daughter cells during cytokinesis [33]. In addition to these transient, external effectors on nuclear shape, high end-to-end tension of the cell leads to spindle-shaped cell elongation and to nuclear deformation into a cigar-like shape in fibroblasts or mesenchymal tumor cells migrating through 3D tissues (compare Fig. 1B). In difference to single-cell migration, collective cell migration, which commonly occurs along smooth ECM interfaces, such as a basement membrane or trails of degraded ECM, nuclei are not exposed to rigorous tissue discontinuities and usually remain in non-compressed, ellipsoid configuration [13,34,35]. Thus, both location and extent of deformation of the nucleus during cell migration depends upon the size of the nucleus, tissue type and migration mode.
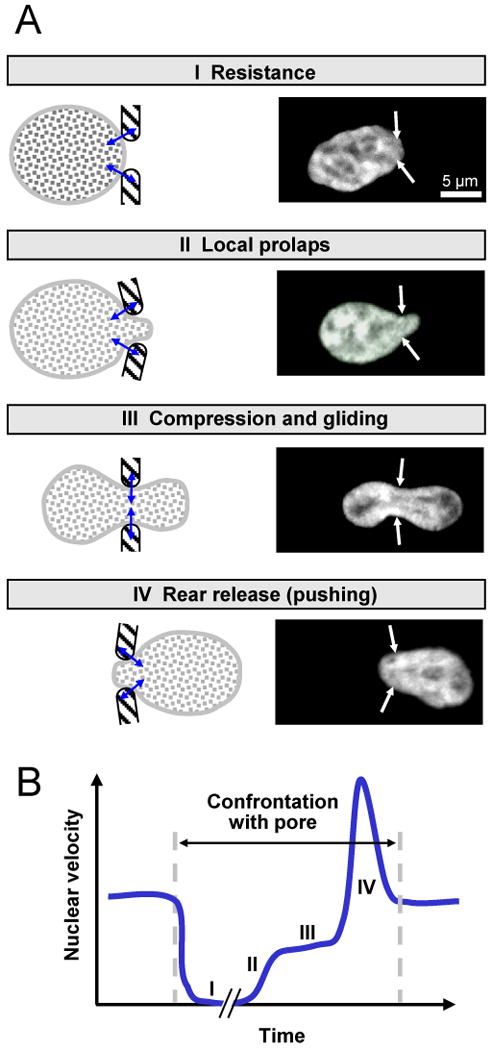
Box 1. Phases of nuclear deformation during single-cell migration.
When cells migrate through a narrow tissue pore, the nucleus undergoes temporal shape change in 4 consecutive steps. First, punctual pressure acts upon the nuclear membrane (A, step I; pressure indicated by small double-arrows) followed by the reciprocal local deformation of both, the slightly elastic ECM and the nuclear envelope and contents (A, step II). If the nuclear deformability is sufficient to accommodate the pore width, the initially small prolaps progresses into a circumferentially confined zone through which the nucleus glides while adopting a transient hour-glass shape (A, step III). As last step, the rear of the nucleus slips rapidly forward (outward pushing) and readapts its original shape (A, step IV). (B) The engagement of both forward-pushing forces and confinement by the ECM are reflected by an oscillarory speed profile (B). Whereas the initial pressure until early proplaps prompts nuclear arrest of variable duration in Phase I (up to an hour or longer; K. Wolf, unpublished data), forward pushing during the last phase is fast and leads to an propulsive acceleration lasting only few minutes (compare Fig. 1; 150 to 160 min). Thus, the shape of the nucleus in migrating cells represents a steady-state “negative image” of the physical space and discontinuities of the extracellular matrix.
Molecular regulation of nuclear shape and rigidity
In interphase nuclei, the size, shape, and stiffness are determined by the nuclear envelope and the nuclear interior. The nuclear envelope consists of the inner and outer nuclear membrane, interconnected at the sites of nuclear pores, and the underlying nuclear lamina, a dense protein network mostly made of lamin proteins that are part of the intermediate filament family (Fig. 3) [5].
Figure 3.
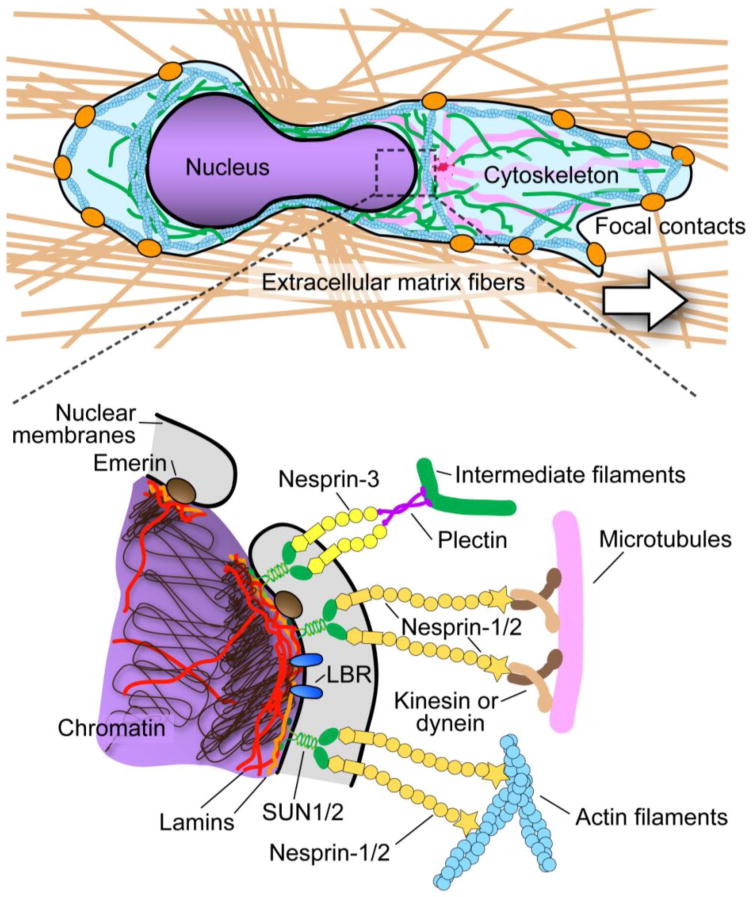
Structural proteins involved in nuclear morphology, stability and interaction with the cytoskeleton.
Top panel: Cell migrating through 3D matrix of extracellular matrix fibers (brown) and encountering a narrow constriction (not drawn to scale). Orange dots denote focal contacts; pink lines microtubules, green lines intermediate filaments, and blue structures actin filaments. (Inset). The lower panel shows molecules contributing to nuclear stiffness and nuclear-cytoskeletal coupling. Lamins underly the inner nuclear membrane and also form stable structures within the nuclear interior. Membrane proteins such as emerin, LBR, and SUN1/2 are retained at the inner nuclear membrane through their interactions with lamins, chromatin, or other nuclear proteins. Nesprins bind to SUN proteins across the perinuclear space and can directly interact with actin filaments (nesprins-1 and -2) or with intermediate filaments via plectin (nesprin-3). Nesprins-1 and -2 have been proposed to bind to microtubules via kinesin or dynein.
Lamins
Nuclear lamins are important structural determinants of nuclear shape and stiffness. The nuclear lamina acts as a nuclear “shock-absorber”, particularly for small deformations and when subjected to tensile forces [5,36]. Mammalian cells express two types of lamins, A-type lamins (mainly lamins A and C) and B-type lamins (lamins B1 and B2 in somatic cells). They form distinct but overlapping networks at the nuclear envelope with unique effects on nuclear shape and stiffness [37-39]. Fibroblasts deficient for lamin B1 develop nuclei with chromatin protrusions (nuclear blebs) but have otherwise normal nuclear stiffness [39]. Cells lacking lamin A and/or lamin C have irregularly shaped nuclei (Fig. 1C) with severely reduced nuclear stiffness [39,40]. Consistent with the role of lamin A as key regulator of nuclear stiffness, cells ectopically expressing lamin A have stiffer nuclei that resist experimental deformation, for example when subjecting cells to substrate strain (J.Lammerding, unpublished). Importantly, in addition to their structural function, lamins bind to several transcription factors, including retinoblastoma protein (Rb), SREBP-1, and c-Fos, making it often difficult to discern mechanical from gene-regulatory effects.
Other determinants of nuclear shape and stiffness
Besides intranuclear lamins, structural proteins traditionally associated with the cytoskeleton can be present within the nuclear interior, including actin, myosin, spectrins and titin [41-43]. There, αII-spectrin [44] and nuclear 4.1R [45] could potentially contribute to nuclear shape and stability by forming stable networks similar to the cortical network in erythrocytes [46-48]. Similarly, the inner nuclear envelope protein emerin can, at least in vitro, promote actin polymerization [46] and also connect to nuclear myosin I [44]. Consistent with a role of emerin on nuclear stiffness, emerin-deficient fibroblasts have altered nuclear deformability in micropipette aspiration assays [49].
The nuclear membranes further contain more than 80 nuclear envelope transmembrane proteins that determine the nuclear shape and anchoring to the surrounding cytoskeleton. As one example, overexpression of the inner nuclear membrane protein, lamin B receptor (LBR), results in increased nuclear surface area (Fig. 1C); this mechanism is utilized during granulopoiesis, resulting in neutrophils with highly lobulated nuclei [50-52].
Besides these structural proteins, chromatin structure and assembly impact nuclear deformability. The nuclei of embryonic stem cells which contain an open, less condensed chromatin configuration are relatively soft upon experimental deformation; during differentiation and development of a more closed chromatin structure, representing a transcriptionally more restrictive state, their nuclear stiffness increases by 6-fold [53].
However, as stem cells after initiating differentiation also increase the expression of lamins A and C, it remains unclear which factor is responsible for the observed increase in nuclear stiffness. Nonetheless, the nuclear interior, consisting of chromatin and nuclear matrix, significantly contributes to the stiffness of the nucleus during large, compressive deformations [36]. For example, cells aspirated into narrow micropipettes show first a strong reduction in nuclear size but then reach a plateau in which the chromatin can be no longer compressed [36,49]. Unlike the nuclear interior and the nuclear lamina, the lipid nuclear membranes offer little resistance to nuclear deformation [36,49]. Thus, multiple structural proteins control nuclear shape and rigidity.
Mechanisms that integrate the nucleus to the actin and intermediate filament cytoskeleton
The intracellular positioning and shape of the nucleus are controlled by structural anchoring to the cytoskeleton. Lamins, chromatin, and nuclear pore complexes interact with SUN proteins located at the inner nuclear membrane [54,55] (Figure 3). SUN1 and SUN2 bind to nesprins located in the outer nuclear membrane via their conserved KASH domain. The giant isoforms of nesprins1 and -2 contain an N-terminal actin binding domain facing the cytoplasm [56-59] and thereby forma physical link between the nuclear interior and the actin cytoskeleton that is often referred to as the LINC (Linker of the Nucleus and Cytoskeleton) complex [60].
The interaction of retrograde moving dorsal actin filaments with the SUN2/nesprin-2 complex at the nuclear envelope, termed transmembrane actin-associated nuclear (TAN) lines, mechanically contributes to the rearward nuclear positioning in fibroblasts before the onset of migration, and depletion or dominant negative mutants of SUN2 or nesprin-2 results in slower migrating cells [61]. Whereas nesprins-1 and -2 bind to actin filaments, nesprin-3 can connect the nucleus to the intermediate filament network via its interaction with plectin [62,63].
Nesprin-4, the most recently discovered member of the nesprin family, binds to kinesin-1 and can modulate localization of the centrosome and Golgi complex; however, its expression is limited to secretory epithelial cells, where nesprin-4 may contribute to establish apicobasal polarization [64]. Supporting the role of lamins in nuclear-cytoskeletal coupling, cells lacking lamins A and C or expressing specific lamin A/C mutations have disturbed perinuclear cytoskeletal organization and reduced cytoskeletal stiffness, resulting in impaired cell migration [40,65,66]. Likewise, cells lacking functional lamin B1 display a nuclear anchoring defect characterized by spontaneous and sustained nuclear rotation [67], and lamin B2-deficient mice have severe brain abnormalities due to defective neuronal migration [68]. Thus, both lamins A/C and B1 and B2 contribute to nuclear-cytoskeletal coupling and mechanotransduction.
Studies in neurons, fibroblasts and epithelial cells further implicate cytoplasmic dynein, cell polarity genes, and microtubule-associated proteins in coordinating the rotation, intracellular position and dynamics of the nucleus [25,26]. MTOC positioning ahead of the nucleus requires dynein and signalling through the Rho GTPase Cdc42 [27] as well as the dynein motor protein components Lis1 and Ndel1 [25]. Likewise, in Caenorhabditis elegans, interaction of kinesin-1 and dynein with the UNC-83/KASH domain protein, which forms a functional complex with sun, control nuclear positioning and movement along microtubules in forward and backward direction, respectively [69].
Implications for cell dynamics in immune cell function and cancer
Alterations of the expression of proteins involved in nuclear morphology and plasticity occur during physiological cell differentiation and activation, as well as in deregulated form in cancer.
Leukocyte differentiation ad function
Neutrophils are the fastest-migrating cells in the mammalian body to be rapidly recruited to sites of inflammation. The segmented shape and deformability of the nucleus likely contributes to the capability of neutrophils to penetrate through submicron gaps [17,70]. Accordingly, during granulopoiesis the differentiation of promyelocytes into mature neutrophils results in an increased expression of LBR and downregulation of A-type lamins, leading to increasingly lobulated nuclei [50-52]. Neutrophil deformability is further enhanced by the downregulation of cytoskeletal proteins such as vimentin and proteins involved in nuclear-cytoskeletal coupling, e.g., nesprins [71] and by changes in chromatin structure [72].
In contrast to neutrophils, T lymphocytes contain a uniformly-shaped ellipsoid nucleus. T cell activation which ultimately leads to T cell recirculation to peripheral tissues, causes the upregulation of A-type lamins [73] and the phosphorylation of type B-type lamins [74]. Likewise, during thymocyte development, levels of A-type lamins increase [75], which is indicative of increased nuclear stability with differentiation. Consistently, T cells from lamin A deficient mice develop defects in development, homing to lymphoid organs, and survival [76].
The cause for the differential regulation of nuclear protein composition in neutrophils and T cells which both generate rapid amoeboid migration is unknown but may reflect their different function and life-spans. Mature neutrophils shut down their DNA replication and execute antibacterial effector function until they undergo apoptosis within a few hours; thus these cells are not dependent on protecting their chromosomal DNA. Conversely, activated T cells that undergo differentiation towards memory cells are long-lived and may therefore require preserving mechanic nuclear and genomic DNA integrity during recirculation. Increasing nuclear stiffness with T cell activation might impede migration efficiency through dense interstitial tissues and intact basement membranes, however may sufficiently comply with more porous tissue regions, including the lymph node stroma [18], partially disintegrated basemement membranes [20] and edematous perivascular tissue tracks during inflammatory processes [77], all which often offer pore dimensions from 5 to 40 micrometers [19] that match the small diameter of lymphocyte nuclei (5 to 7 μm).
Cancer
Neoplastic cells acquire significant aberrations in nuclear morphology, including size, shape, and loss of nuclear domains [78]. This nuclear plasticity closely resembles alterations observed in diseases caused by mutations or deletions in nuclear envelope proteins such as lamin A/C or LBR [39,78-80] (Fig. 1), suggesting that changes in the expression of these proteins could also be responsible for the abnormal nuclear structure in cancer.
Deregulation of lamin expression has been reported in several cancers types. Lamins A and C are over-expressed in ovarian cancers [81], colorectal cancer [82] and cancers of the skin and prostate [83,84]. Rapid growth in basal cell carcinoma in skin cancer correlates with reduced levels of lamin A [85], and silencing of A-type lamins was reported in leukemias and lymphomas [86], gastrointestinal cancer [87] and small cell lung cancer [88]. Many tumor cells further show strong downregulation [89] or mutations [90] of nesprins. The reason for neoplastic regulation of lamins and nesprin in different cancer types and its contribution to cancer progression are unclear. Reduced levels of A-type lamins could increase the deformability of the nucleus [39] and thereby enhance cell squeezing through narrow tissue spaces or basement membranes, similar to increased nuclear plasticity in terminally differentiated neutrophils [50-52]. In addition to altered lamin and/or nesprin expression, changes in chromatin configuration often associated with cancer cells could also result in increased nuclear plasticity, as a more open chromatin configuration typically associated with non-differentiated cells results in more deformable nuclei [53]. Whether neoplastic nuclear abnormalities contribute to cancer cell invasion and metastatic dissemination is unclear; Consequently, future experiments that systematically study the effects of modulation of lamins and other nuclear envelope proteins expression on migration, invasion, and metastatic potential of cancer cells in vitro and in vivo are required to to delineate their contribution to cancer progression.
Conclusions
Through its connection to the actin, tubulin and intermediate filament cytoskeleton, the nucleus is a major mechanosensory integrator. Because of its size and high rigidity, compared to all other cell compartments, it further imposes a major physical challenge for cells moving in 3D environments. Consequently, studies on nuclear mechanics and its implications for cell migration should preferentially focus on 3D in vitro and in vivo models. Whereas increased nuclear deformability offers an advantage for efficient migration in cells with large nuclei, other cells with small but more rigid nuclei such as in highly motile T cells may confine their tissue distribution towards default migration routes with sufficient preexisting tissue porosity.
Besides merely indicating cell deformation in dense tissue, nuclear deformation may further exert secondary effects, such as altered DNA transcription and replication [91], albeit migration-associated changes in location and function of chromosmal DNA and nucleoli remain to be tested directly. Thereby, besides testing cells with different natural nuclear shapes and lamin composition, targeting defined nuclear structural proteins is required to elucidate their dual function in facilitating (or preventing) nuclear deformation alongside with protecting structural DNA integrity and other cellular functions. In conclusion, altered nuclear morphological and mechanical plasticity during cell differentiation and neoplastic transformation may represent a central determinant of cell mobility during physiologic and pathologic invasion and recirculation.
Acknowledgments
We gratefully acknowledge Uta Flucke for human soft tissue sarcoma and Willeke Blokx for melanoma samples, and Monika Zwerger for generating the lamin-modified MCF-10A cells. This work was supported by the European Community (T3Net, European Training Network) to P.F., grant 917.10.364 by the Dutch Science Foundation (NWO) to K.W. and the National Institutes of Health grants HL082792 and NS059348 and the Cardiovascular Leadership Group award to J.L..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Wolf K, Muller R, Borgmann S, Brocker EB, Friedl P. Amoeboid shape change and contact guidance: T-lymphocyte crawling through fibrillar collagen is independent of matrix remodeling by MMPs and other proteases. Blood. 2003;102:3262–3269. doi: 10.1182/blood-2002-12-3791. [DOI] [PubMed] [Google Scholar]
- 4.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 5.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri O, Parekh SH, Fletcher DA. Reversible stress softening of actin networks. Nature. 2007;445:295–298. doi: 10.1038/nature05459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia Z, Barbier L, Stuart H, Amraei M, Pelech S, Dennis JW, Metalnikov P, O'Donnell P, Nabi IR. Tumor cell pseudopodial protrusions. Localized signaling domains coordinating cytoskeleton remodeling, cell adhesion, glycolysis, RNA translocation, and protein translation. J Biol Chem. 2005;280:30564–30573. doi: 10.1074/jbc.M501754200. [DOI] [PubMed] [Google Scholar]
- 8.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caille N, Tardy Y, Meister JJ. Assessment of strain field in endothelial cells subjected to uniaxial deformation of their substrate. Ann Biomed Eng. 1998;26:409–416. doi: 10.1114/1.132. [DOI] [PubMed] [Google Scholar]
- 10.Caille N, Thoumine O, Tardy Y, Meister JJ. Contribution of the nucleus to the mechanical properties of endothelial cells. J Biomech. 2002;35:177–187. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 11.Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J Biomech. 1995;28:1529–1541. doi: 10.1016/0021-9290(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 12.Guilak F, Tedrow JR, Burgkart R. Viscoelastic properties of the cell nucleus. Biochem Biophys Res Commun. 2000;269:781–786. doi: 10.1006/bbrc.2000.2360. [DOI] [PubMed] [Google Scholar]
- 13.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 14.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng D, Nagy JA, Pyne K, Dvorak HF, Dvorak AM. Neutrophils emigrate from venules by a transendothelial cell pathway in response to FMLP. J Exp Med. 1998;187:903–915. doi: 10.1084/jem.187.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajenoff M, Egen JG, Qi H, Huang AY, Castellino F, Germain RN. Highways, byways and breadcrumbs: directing lymphocyte traffic in the lymph node. Trends Immunol. 2007;28:346–352. doi: 10.1016/j.it.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Voisin MB, Larbi KY, Dangerfield J, Scheiermann C, Tran M, Maxwell PH, Sorokin L, Nourshargh S. Venular basement membranes contain specific matrix protein low expression regions that act as exit points for emigrating neutrophils. J Exp Med. 2006;203:1519–1532. doi: 10.1084/jem.20051210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf K, Friedl P. Mapping proteolytic cancer cell-extracellular matrix interfaces. Clin Exp Metastasis. 2009;26:289–298. doi: 10.1007/s10585-008-9190-2. [DOI] [PubMed] [Google Scholar]
- 22.Friedl P, Maaser K, Klein CE, Niggemann B, Krohne G, Zanker KS. Migration of highly aggressive MV3 melanoma cells in 3-dimensional collagen lattices results in local matrix reorganization and shedding of alpha2 and beta1 integrins and CD44. Cancer Res. 1997;57:2061–2070. [PubMed] [Google Scholar]
- 23.Friedl P, Wolf K. Proteolytic interstitial cell migration: a five-step process. Cancer Metastasis Rev. 2009;28:129–135. doi: 10.1007/s10555-008-9174-3. [DOI] [PubMed] [Google Scholar]
- 24.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J Cell Sci. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 28.Friedl P, Weigelin B. Interstitial leukocyte migration and immune function. Nat Immunol. 2008;9:960–969. doi: 10.1038/ni.f.212. [DOI] [PubMed] [Google Scholar]
- 29.Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cramer LP. Forming the cell rear first: breaking cell symmetry to trigger directed cell migration. Nat Cell Biol. 12:628–632. doi: 10.1038/ncb0710-628. [DOI] [PubMed] [Google Scholar]
- 32.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 34.Andresen V, Alexander S, Heupel WM, Hirschberg M, Hoffman RM, Friedl P. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr Opin Biotechnol. 2009;20:54–62. doi: 10.1016/j.copbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowat AC, Lammerding J, Herrmann H, Aebi U. Towards an integrated understanding of the structure and mechanics of the cell nucleus. Bioessays. 2008;30:226–236. doi: 10.1002/bies.20720. [DOI] [PubMed] [Google Scholar]
- 37.Schape J, Prausse S, Radmacher M, Stick R. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophys J. 2009;96:4319–4325. doi: 10.1016/j.bpj.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 40.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young KG, Kothary R. Spectrin repeat proteins in the nucleus. Bioessays. 2005;27:144–152. doi: 10.1002/bies.20177. [DOI] [PubMed] [Google Scholar]
- 42.Zastrow MS, Flaherty DB, Benian GM, Wilson KL. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci. 2006;119:239–249. doi: 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann WA, Johnson T, Klapczynski M, Fan JL, de Lanerolle P. From transcription to transport: emerging roles for nuclear myosin I. Biochem Cell Biol. 2006;84:418–426. doi: 10.1139/o06-069. [DOI] [PubMed] [Google Scholar]
- 44.Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krauss SW, Chen C, Penman S, Heald R. Nuclear actin and protein 4.1: essential interactions during nuclear assembly in vitro. Proc Natl Acad Sci U S A. 2003;100:10752–10757. doi: 10.1073/pnas.1934680100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:E231. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holaska JM. Emerin and the nuclear lamina in muscle and cardiac disease. Circ Res. 2008;103:16–23. doi: 10.1161/CIRCRESAHA.108.172197. [DOI] [PubMed] [Google Scholar]
- 48.Zhong Z, Wilson KL, Dahl KN. Beyond lamins other structural components of the nucleoskeleton. Methods Cell Biol. 2010;98:97–119. doi: 10.1016/S0091-679X(10)98005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J. 2006;91:4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma. 2007;116:227–235. doi: 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]
- 51.Yabuki M, Miyake T, Doi Y, Fujiwara T, Hamazaki K, Yoshioka T, Horton AA, Utsumi K. Role of nuclear lamins in nuclear segmentation of human neutrophils. Physiol Chem Phys Med NMR. 1999;31:77–84. [PubMed] [Google Scholar]
- 52.Olins AL, Zwerger M, Herrmann H, Zentgraf H, Simon AJ, Monestier M, Olins DE. The human granulocyte nucleus: Unusual nuclear envelope and heterochromatin composition. Eur J Cell Biol. 2008;87:279–290. doi: 10.1016/j.ejcb.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hasan S, Guttinger S, Muhlhausser P, Anderegg F, Burgler S, Kutay U. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 55.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 57.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 59.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 60.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiche G. Role of plectin in cytoskeleton organization and dynamics. J Cell Sci. 1998;111(Pt 17):2477–2486. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- 63.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Houben F, Willems CH, Declercq IL, Hochstenbach K, Kamps MA, Snoeckx LH, Ramaekers FC, Broers JL. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim Biophys Acta. 2009;1793:312–324. doi: 10.1016/j.bbamcr.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ji JY, Lee RT, Vergnes L, Fong LG, Stewart CL, Reue K, Young SG, Zhang Q, Shanahan CM, Lammerding J. Cell nuclei spin in the absence of lamin b1. J Biol Chem. 2007;282:20015–20026. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- 68.Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci U S A. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mackarel AJ, Cottell DC, Russell KJ, FitzGerald MX, O'Connor CM. Migration of neutrophils across human pulmonary endothelial cells is not blocked by matrix metalloproteinase or serine protease inhibitors. Am J Respir Cell Mol Biol. 1999;20:1209–1219. doi: 10.1165/ajrcmb.20.6.3539. [DOI] [PubMed] [Google Scholar]
- 71.Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, Karakesisoglou I, Hodzic D, Olins DE. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88:203–214. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- 73.Andrade R, Alonso R, Pena R, Arlucea J, Arechaga J. Localization of importin alpha (Rch1) at the plasma membrane and subcellular redistribution during lymphocyte activation. Chromosoma. 2003;112:87–95. doi: 10.1007/s00412-003-0247-3. [DOI] [PubMed] [Google Scholar]
- 74.Hornbeck P, Huang KP, Paul WE. Lamin B is rapidly phosphorylated in lymphocytes after activation of protein kinase C. Proc Natl Acad Sci U S A. 1988;85:2279–2283. doi: 10.1073/pnas.85.7.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guilly MN, Kolb JP, Gosti F, Godeau F, Courvalin JC. Lamins A and C are not expressed at early stages of human lymphocyte differentiation. Exp Cell Res. 1990;189:145–147. doi: 10.1016/0014-4827(90)90267-e. [DOI] [PubMed] [Google Scholar]
- 76.Hale JS, Frock RL, Mamman SA, Fink PJ, Kennedy BK. Cell-extrinsic defective lymphocyte development in Lmna(-/-) mice. PLoS One. 2010;5:e10127. doi: 10.1371/journal.pone.0010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friedl P, Brocker EB. T cell migration in three-dimensional extracellular matrix: guidance by polarity and sensations. Dev Immunol. 2000;7:249–266. doi: 10.1155/2000/56473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 79.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muchir A, Medioni J, Laluc M, Massart C, Arimura T, van der Kooi AJ, Desguerre I, Mayer M, Ferrer X, Briault S, et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 81.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci U S A. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willis ND, Cox TR, Rahman-Casans SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruine A, et al. Lamin A/C is a risk biomarker in colorectal cancer. PLoS One. 2008;3:e2988. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tilli CM, Ramaekers FC, Broers JL, Hutchison CJ, Neumann HA. Lamin expression in normal human skin, actinic keratosis, squamous cell carcinoma and basal cell carcinoma. Br J Dermatol. 2003;148:102–109. doi: 10.1046/j.1365-2133.2003.05026.x. [DOI] [PubMed] [Google Scholar]
- 84.Coradeghini R, Barboro P, Rubagotti A, Boccardo F, Parodi S, Carmignani G, D'Arrigo C, Patrone E, Balbi C. Differential expression of nuclear lamins in normal and cancerous prostate tissues. Oncol Rep. 2006;15:609–613. [PubMed] [Google Scholar]
- 85.Venables RS, McLean S, Luny D, Moteleb E, Morley S, Quinlan RA, Lane EB, Hutchison CJ. Expression of individual lamins in basal cell carcinomas of the skin. Br J Cancer. 2001;84:512–519. doi: 10.1054/bjoc.2000.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agrelo R, Setien F, Espada J, Artiga MJ, Rodriguez M, Perez-Rosado A, Sanchez-Aguilera A, Fraga MF, Piris MA, Esteller M. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. J Clin Oncol. 2005;23:3940–3947. doi: 10.1200/JCO.2005.11.650. [DOI] [PubMed] [Google Scholar]
- 87.Moss SF, Krivosheyev V, de Souza A, Chin K, Gaetz HP, Chaudhary N, Worman HJ, Holt PR. Decreased and aberrant nuclear lamin expression in gastrointestinal tract neoplasms. Gut. 1999;45:723–729. doi: 10.1136/gut.45.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Broers JL, Raymond Y, Rot MK, Kuijpers H, Wagenaar SS, Ramaekers FC. Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am J Pathol. 1993;143:211–220. [PMC free article] [PubMed] [Google Scholar]
- 89.Marme A, Zimmermann HP, Moldenhauer G, Schorpp-Kistner M, Muller C, Keberlein O, Giersch A, Kretschmer J, Seib B, Spiess E, et al. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int J Cancer. 2008;123:2048–2056. doi: 10.1002/ijc.23763. [DOI] [PubMed] [Google Scholar]
- 90.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 91.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prufert K, Vogel A, Krohne G. The lamin CxxM motif promotes nuclear membrane growth. J Cell Sci. 2004;117:6105–6116. doi: 10.1242/jcs.01532. [DOI] [PubMed] [Google Scholar]
- 93.Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci U S A. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen, Zhai Z, Zhang C. Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci. 2007;120:520–530. doi: 10.1242/jcs.03355. [DOI] [PubMed] [Google Scholar]
- 95.Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Highlighted references
- *.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]; First detailed analysis of the contribution of nuclear lamina und chromatin to the mechanical stiffness of nuclei using osmotic swelling and micropipette aspiration of xenopus oocytes.
- *.Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]; A systematic approach to identify proteins specific to the nuclear envelope found more than 80 proteins, many with still unknown function.
- **.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]; Demonstrates that A-type and B-type lamins have distinct effects on nuclear shape and stiffness, with lamin A identified as the major contributor to nuclear stiffness.
- *.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstrates the importance of chromatin configuration on nuclear mechanics and illustrating that non-differentiated cells have significantly softer nuclei.
- **.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed characterization of the nesprin/SUN protein complex connecting the nucleus with the cytoskeleton. This manuscript introduced the term “LINC complex” for Linking Nucleus and Cytoskeleton.
- **.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]; First functional demonstration of the interaction between SUN and nesprin and actin cables in nuclear movement during migration.
- **.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]; First description of impaired nuclear stiffness, increased nuclear fragility, altered cytoskeletal stiffness, and impaired activation of mechanosensitive genes in fibroblasts lacking A-type lamins.
- *.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]; Shows that proteolytic ECM degradation reduces nuclear deformation during cell migration through 3D collagen lattices.
- *.Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma. 2007;116:227–235. doi: 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]; Comprehensive overview on altered structure of nucleus in differentiating granulocytes.
- *.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sythematic mapping of the pore sizes present in collagen-rich in vitro and in vivo substrates used for cell migration studies.


