Abstract
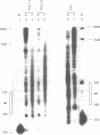
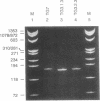
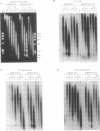
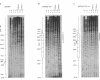
In competitive in vitro reconstitution experiments synthetic DNA composed of tandem repeats of the repetitive sequence (A/T)3NN(G/C)3NN, specifically the 20 bp 'TG sequence' (5'-TCGGTGTTAGAGCCTGTAAC-3'), was reported to associate with the histone octamer with an affinity higher than that of nucleosomally derived DNA. However, at least two groups have independently shown that tandem repeats of the TG sequence do not accommodate a stably positioned nucleosome in vivo. It was suggested that the anisotropic flexibility of the TG sequence, governed by a 10 bp sequence periodicity, is incompatible with the required underwinding of the DNA helix at the nucleosome pseudodyad while maintaining a bending preference that can be accommodated in the remainder of the nucleosome. Here we test this hypothesis directly by studying the in vivo nucleosomal structure of modified TG sequences designed to accommodate underwinding at the pseudodyad. We show that these modifications are not sufficient to allow stable incorporation of the TG sequence repeat into a nucleosome in vivo, but do note invasion from one end of the TG heptamer of a translationally random but rotationally constrained nucleosome. We discuss possible reasons for the absence of nucleosomes from the TG sequence in vivo.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arents G., Moudrianakis E. N. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan P. N., Wright E. B., Olins D. E. Core nucleosomes by digestion of reconstructed histone-DNA complexes. Nucleic Acids Res. 1979 Apr;6(4):1449–1465. doi: 10.1093/nar/6.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. Principles of sequence-dependent flexure of DNA. J Mol Biol. 1986 Dec 20;192(4):907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992 Jan 2;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo G., Di Mauro E., Salina G., Negri R. Attraction, phasing and neighbour effects of histone octamers on curved DNA. J Mol Biol. 1990 Nov 20;216(2):363–374. doi: 10.1016/S0022-2836(05)80327-4. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Calladine C. R. Sequence-specific positioning of core histones on an 860 base-pair DNA. Experiment and theory. J Mol Biol. 1987 May 5;195(1):143–173. doi: 10.1016/0022-2836(87)90333-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Can one measure the free energy of binding of the histone octamer to different DNA sequences by salt-dependent reconstitution? J Mol Biol. 1991 Jun 5;219(3):391–392. doi: 10.1016/0022-2836(91)90179-a. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Fascher K. D., Schmitz J., Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990 Aug;9(8):2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Gale J. M., Nissen K. A., Smerdon M. J. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D. A relational database of transcription factors. Nucleic Acids Res. 1990 Apr 11;18(7):1749–1756. doi: 10.1093/nar/18.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D., Wolffe A. P. The structure of DNA in a nucleosome. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J., Donald K. A., Griffiths D. E., Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991 Oct 25;19(20):5791–5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell. 1988 Feb 26;52(4):535–544. doi: 10.1016/0092-8674(88)90466-7. [DOI] [PubMed] [Google Scholar]
- Kefalas P., Gray F. C., Allan J. Precise nucleosome positioning in the promoter of the chicken beta A globin gene. Nucleic Acids Res. 1988 Jan 25;16(2):501–517. doi: 10.1093/nar/16.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Bird L. E., Mellor J. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 1993 Sep 25;21(19):4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Lutter L. C. The helical periodicity of DNA on the nucleosome. Nucleic Acids Res. 1981 Sep 11;9(17):4267–4283. doi: 10.1093/nar/9.17.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- Losa R., Omari S., Thoma F. Poly(dA).poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 1990 Jun 25;18(12):3495–3502. doi: 10.1093/nar/18.12.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. DNase II digestion of the nucleosome core: precise locations and relative exposures of sites. Nucleic Acids Res. 1981 Sep 11;9(17):4251–4265. doi: 10.1093/nar/9.17.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Precise location of DNase I cutting sites in the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 1979 Jan;6(1):41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer B., Linxweiler W., Hörz W. DNA engineering shows that nucleosome phasing on the African green monkey alpha-satellite is the result of multiple additive histone-DNA interactions. J Mol Biol. 1986 Aug 20;190(4):639–645. doi: 10.1016/0022-2836(86)90249-4. [DOI] [PubMed] [Google Scholar]
- Patterton H. G., Simpson R. T. Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol Cell Biol. 1994 Jun;14(6):4002–4010. doi: 10.1128/mcb.14.6.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterton H. G., von Holt C. Negative supercoiling and nucleosome cores. II. The effect of negative supercoiling on the positioning of nucleosome cores in vitro. J Mol Biol. 1993 Feb 5;229(3):637–655. doi: 10.1006/jmbi.1993.1069. [DOI] [PubMed] [Google Scholar]
- Pennings S., Muyldermans S., Meersseman G., Wyns L. Formation, stability and core histone positioning of nucleosomes reassembled on bent and other nucleosome-derived DNA. J Mol Biol. 1989 May 5;207(1):183–192. doi: 10.1016/0022-2836(89)90449-x. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Satchwell S. C., Travers A. A. Asymmetry and polarity of nucleosomes in chicken erythrocyte chromatin. EMBO J. 1989 Jan;8(1):229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J Mol Biol. 1990 Nov 5;216(1):69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning: occurrence, mechanisms, and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Zatchej M., Thoma F. Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J. 1992 Mar;11(3):1187–1193. doi: 10.1002/j.1460-2075.1992.tb05159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Thoma F., Zatchej M. Chromatin folding modulates nucleosome positioning in yeast minichromosomes. Cell. 1988 Dec 23;55(6):945–953. doi: 10.1016/0092-8674(88)90240-1. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Klug A. The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- Turnell W. G., Satchwell S. C., Travers A. A. A decapeptide motif for binding to the minor groove of DNA. A proposal. FEBS Lett. 1988 May 23;232(2):263–268. doi: 10.1016/0014-5793(88)80750-6. [DOI] [PubMed] [Google Scholar]
- Ulyanov N. B., Zhurkin V. B. Sequence-dependent anisotropic flexibility of B-DNA. A conformational study. J Biomol Struct Dyn. 1984 Oct;2(2):361–385. doi: 10.1080/07391102.1984.10507573. [DOI] [PubMed] [Google Scholar]
- Wallrath L. L., Lu Q., Granok H., Elgin S. C. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994 Mar;16(3):165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Drew H. R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]