Abstract
Many environmental risk factors for common, complex human diseases have been revealed by epidemiologic studies, but how genotypes at specific loci modulate individual responses to environmental risk factors is largely unknown. Gene-environment interactions will be missed in genome-wide association studies and may account for some of the ‘missing heritability’ for these diseases. In this review, we focus on asthma as a model disease for studying gene-environment interactions because of relatively large numbers of candidate gene-environment interactions with asthma risk in the literature. Identifying these interactions using genome-wide approaches poses formidable methodological problems and elucidating molecular mechanisms for these interactions has been challenging. We suggest that studying gene-environment interactions in animal models, while more tractable, is not likely to shed light on the genetic architecture of human diseases. Lastly, we propose avenues for future studies to find gene-environment interactions.
The Problem: Accounting for the Heritability of Common Human Diseases
Dissecting the genetics of common human diseases with complex etiologies continues to be challenging and the genetic architectures of these diseases remain elusive. Despite the many successes of genome-wide association studies (GWAS) during the past four years (refs.1, 2 as examples), there is a growing consensus that the common variants with modest effects on disease risk discovered through GWAS do not account for the majority of the estimated heritabilities of these diseases. This observation was initially termed ‘missing heritability’3–5, to suggest that additional genetic variation (such as rare variants or copy number variants) or interactions that are not explicitly modeled (such as epistasis or gene-environment interactions [GEIs]) have been missed by GWAS and may account for a significant proportion of the heritability. More recently, the term ‘hidden heritability’6 was suggested in response to the argument that the joint or simultaneous analysis of individual common risk alleles may account for more of the heritability than the sum of each allele7, 8. Of course, it is likely that the genetic architecture of most, if not all, common human diseases will include all of the above components, and it is timely in the post-GWAS era to begin to directly assess the contribution of each to the overall heritability of complex diseases. Here, we will focus on the role of GEIs on risk for human disease and propose asthma as a model disease for understanding the importance of genetic modifiers of environmental risk factors. We will first review the role of environmental exposures on asthma risk, using the term ‘environment’ to reflect any endogenous or exogenous non-genetic factor that influences risk; and then provide examples of specific GEIs that represent different patterns of associations. Finally, we will discuss potential mechanisms for GEIs and speculate on future directions for this field.
Asthma as a Model for Studying GEIs
Asthma is a heterogeneous disease that is characterized by reversible airway obstruction and airway inflammation. It is among the most common chronic diseases, affecting more than 300 million people worldwide9. Similar to other immune-mediated diseases, the prevalence of asthma is highest among developed countries and has risen significantly over the past few decades10, especially in countries transitioning to a western lifestyle9, attesting to the importance of environmental modifiers of disease risk.
Epidemiologic studies of asthma have revealed numerous associations between exposures and subsequent risk for asthma. The sheer number of validated risk factors for asthma, many of which are often known and measurable, have placed asthma at the forefront of studies on GEIs and some of the best and most replicated examples of such interactions have come from this field (see, for examples, recent reviews on this topic11–14). Moreover, these studies have established that timing of exposure in the lifecycle is a critical variable in determining risk (Figure 1), and that risks differ for childhood-onset and adult-onset asthma.
Figure 1.
Risk and Protection Factors Influence Asthma Risk Throughout the Lifecycle. Epidemiologic studies of asthma have established numerous risk and protection factors that exert their effects at specific stages of the lifecycle. The relationship of some investigated exposures that have equivocal associations with risk or protection are not shown. These include breastfeeding effects and cat in the home at birth on subsequent risk for asthma in the child, as examples. A. Environmental exposures that have been associated with increased risk for asthma. B. Stages of the lifecycle that are “sensitive” to the epidemiologic risk and protective factors. C. Environmental exposures associated with protection from asthma.
The prenatal environment is the first exposure that establishes lifelong risks for asthma in the fetus. During this period, maternal smoking is a significant risk for subsequent asthma15–17. Maternal asthma is among the most significant and consistent risk factors, and a greater risk factor than paternal asthma, for childhood asthma18–20, thereby suggesting that the in utero environment differs between asthmatic and non-asthmatic mothers and contributes to subsequent risk of asthma in the fetus. Sex, which is established at conception, has opposite effects on asthma risk in the pre- and post-puberty periods, whereby many more boys receive a diagnosis of asthma by the age of 6 but diagnoses in girls predominate after puberty (reviewed in reference21). Moreover, asthma resolves in many boys with early-onset asthma so that by adulthood there are significantly more women than men with asthma. Sex may exert its effects on risk prenatally or throughout life by modifying responses to other environmental risk factors, or it may directly affect risk by differentially modifying gene expression (discussed in detail in reference 21).
During the first year of life, and in some cases prenatally, a number of exposures have been associated with protection from asthma in childhood. For example, exposure to large animals among European farmers22–25, having a dog in the home26–28, attending daycare29–31, or drinking unprocessed cow’s milk32, 33 has been associated with protection against asthma in childhood, and gives support to the ‘Hygiene Hypothesis’ (Box 1) as an explanation for the rising prevalence of asthma risk in westernized countries34. In contrast, some ‘exposures’ during the first three years of life are associated with higher risk for asthma. These include allergic sensitization and the presence of wheezing illnesses with respiratory viral infections, particularly those due to rhinovirus or respiratory syncytial virus35–38. Whether these conditions themselves alter the risk profile of the child or are merely early manifestations of asthma in genetically susceptible children is not known, but their associations with the subsequent development of asthma by age 6 is undisputed.
The Hygiene Hypothesis
The Hygiene Hypothesis is an evolving concept. The hypothesis was initially formulated to explain the protective effect of having older siblings on risk of hay fever and eczema34 and later extended to explain the marked increase in the prevalence of allergies observed in western societies over the last few decades. Reduced exposure to infections in early childhood not only due to smaller family sizes, but also to improved living standards and higher personal hygiene, has been proposed to result in increased risk of developing allergic diseases. This theory was then integrated with the then dominant Th1/Th2 paradigm and further suggested that the hygienic environments associated with western lifestyles deprive the immune system of stimuli required to boost Th1 responses in early childhood, leading to a surge in the prevalence of Th2-mediated allergic disease105. When it became clear that both Th2-mediated allergic disorders and Th1-mediated autoimmune diseases are on the rise in the western world10, the hygiene hypothesis was further updated with a regulatory twist. According to its most recent version, immuno-regulatory mechanisms are activated by interactions between the innate immune system and the microbial environment, particularly when these interactions occur in utero and/or in early life106, 107. These mechanisms balance and fine tune both Th1 and Th2 responses, but are currently compromised by a decrease in, and possibly by qualitative alterations of, the microbial burden the immune system encounters in western societies108. This hypothesis awaits further integration with our still incomplete understanding of the role that genetic variation plays in shaping immune responses to environmental stimuli109.
Beyond infancy, additional exposures are important in establishing risk for asthma, including obesity or high body mass index (BMI)39–41, occupational exposures42, 43, and air pollution44–46. These risks are likely increasing the penetrance of asthma in genetically susceptible individuals, but it is noteworthy that the risk alleles associated with occupational asthma47, 48, for example, are often at loci that have not been implicated in asthma in non-exposed individuals. Moreover, it is likely that childhood- and adult-onset asthma (i.e., age of onset of asthma) have only partially overlapping genetic etiologies2, 49, 50.
Demonstrating that a particular exposure is associated with risk for or protection from a disease does not necessarily imply that GEIs are at play, but the fact that not all exposed individuals develop the disease suggests that genetic variation between individuals may be important. To establish a GEI one must show that specific genotypes have different responses to the same ‘exposure’ by demonstrating that the effects of ‘exposure’ (for example, the development of or protection from a disease) vary between individuals with different genotypes. GEIs can show different patterns of associations, which are described below for asthma and in Table 1 for other phenotypes and diseases. The different patterns of GEIs are illustrated in Figure 2.
Table 1.
Examples of recent (2009–2010) reports of GEIs for behavioral, gene expression, and disease phenotypes.
| Disease or Phenotype(Sample) | Environmental Exposure(s) | Genetic Variant(s) | Results | Reference |
|---|---|---|---|---|
| Cleft lip with/without cleft palate (N=326 cases-parent trios) | Multivitamin supplementation, ETS, parent-of-origin | 22 SNPs in IRF1 | GEI with multiple SNPs and vitamin supplement; risk reduced for specific genotypes in ‘exposed’ group | 110 |
| Delinquency (1825 high school students) | Maltreatment (self-reported) and gender | Functional VNTR in MAOA | GEI with ‘flip-flop’ association by gender; opposite alleles interact with maltreatment in males and females | 111 |
| Mortality (184 adults age 70–80 yrs at initial assessment) | Depression | Functional IL6 promoter SNP | GEI with −174C/G genotype; increased mortality among depressed individuals with GG genotype | 112 |
| Preterm birth (743 women) | Bacterial vaginosis | 1536 SNPs in candidate genes in inflammatory pathways | GEI with SNPs in PRKCA, FLT1, IL6; risk increased in or confined to exposed women | 113 |
| Global gene expression in human aortic endothelial cells (96 heart transplant donors) | Oxidized phospholipid treatment | ~500k genome-wide SNPs | GEI at multiple SNPs (cis and trans); fold-changes in expression in treated cells confined to specific genotypes | 103 |
| Coronary artery disease (735 cases and 519 controls undergoing coronary angiography) | Sex, presence of hypertension (HTN) | 6 SNPs in AGT | GEI with ‘flip-flop’ association of a haplotype in men and women with HTN (risk increased in women and decreased in men); haplotype effect not significant in men or women without HTN | 114 |
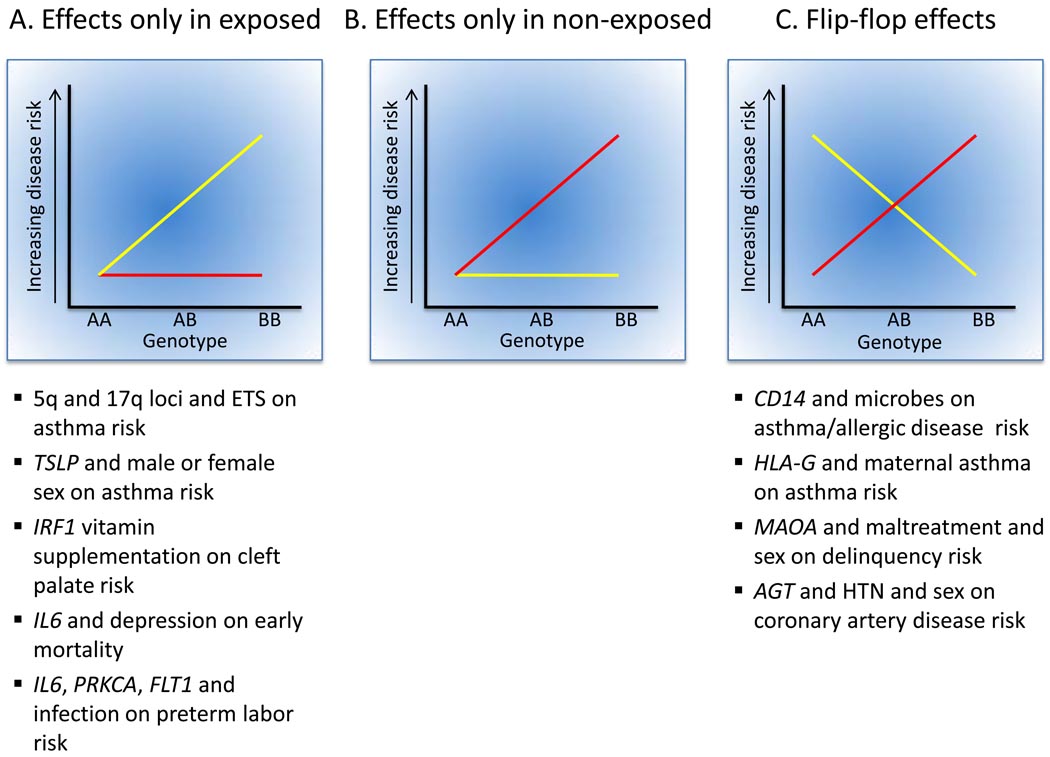
Figure 2.
Types of GEIs. The yellow line in each panel corresponds to risk in exposed individuals; the red line corresponds to risk in non-exposed individuals. Examples from this review that illustrate each type of interaction are shown below the panels. It is notable that no clear examples of specific SNPs or genes involved in GEIs limited to non-exposed individuals were reported in the studies reviewed here, including the recent GWIS of farming exposures and asthma99. Not all interactions shown have been replicated or substantiated by other reports in the literature.
Examples of GEIs in Asthma
Microbial Exposure, Genotype, and Protection from Asthma
One of the most established and best replicated GEI on risk for asthma or allergic disease is that resulting from interactions between genotype for a promoter polymorphism (−159C/T, a.k.a. −260C/T, rs2569190) at the locus encoding a subunit of the endotoxin receptor on mononuclear cells, CD14, and exposure to microbes, as assessed by house dust endotoxin levels51, 52, having a pet in the house at birth53, 54, working with laboratory animals55, contact with farm animals56, and country living during childhood57. In these studies, the −159T allele is associated with asthma or allergic disease among the highly “exposed” subjects, whereas the −159C allele is associated with asthma or allergic disease among “non-exposed” subjects. No associations with this polymorphism is seen among subjects with intermediate levels of exposure. This “flip-flop” pattern of association results in overall poor replicability of associations with the CD14 −159C/T polymorphism when environmental exposures are not considered, as has been reviewed elsewhere58–60.
Genetic modifiers of environmental tobacco smoke (ETS) exposure on asthma risk
ETS exposure in early life is a well established risk factor for reduced lung function, increased numbers of lower respiratory tract infections, and asthma15–17. However, not all exposed children show these effects. Interactions between in utero or neonatal exposures to ETS and a number of genetic loci were first suggested by family-based linkage studies61–63, with some chromosomal regions showing linkage to asthma or bronchial hyperresponsiveness (BHR) only in exposed children (chromosomes 1p61, 1q63, 3p62, 4q63, 5q61, 62, 17p61) or only in non-exposed children (chromosomes 1q61, 5p63, 6p61, 9q61, 17p63, 19p62). The 5q region, which harbors many asthma candidate genes, was linked to asthma in the exposed children in two studies; one other region on 1q43 showed interaction effects in two studies but in opposite groups (in exposed children in one study and in non-exposed children in the other). In the past three years alone, interactions between in utero or neonatal ETS exposure and genotypes at candidate loci on risk for asthma or wheezing have been reported, including but not limited to single nucleotide polymorphisms (SNPs) in ADAM metallopeptidase domain 33 (ADAM33)64, glutathione S-transferase M1 (GSTM1)65, interleukin 13 (IL13)66, interleukin 1 receptor antagonist (IL1RN)67, tumor necrosis factor (TNF)68, 69, and the 17q21 asthma locus49, 70. In all but one of these studies69, associations with asthma were significant in exposed children only69. It is notable that the IL13 gene resides at chromosome 5q31 within the locus showing interactions in two linkage studies. In all three studies, the linkage61, 62 or association66 with asthma was only among children exposed to ETS in utero or in the first year of life, providing consistent evidence for an ‘IL13-ETS exposure’ association with asthma risk. Accounting for this easily measurable exposure in genetic studies may enhance our ability to identify important risk loci.
Maternal asthma as a prenatal ‘environmental’ exposure
Maternal asthma remains among the most significant risk factors for the development of childhood asthma in her offspring18–20. Because asthma risk alleles are inherited from both parents, this suggests that the in utero environment differs between mothers with and without asthma. Such differences could result in differential ‘prenatal programming’71 of immune cells in the fetus depending on the asthma status of the mother, or from exposure to asthma medications (e.g. corticosteroids) taken by asthmatic mothers during pregnancy. Evidence that fetal genotype interacts with ‘maternal asthma’ to determine risk for asthma in the child was first provided by a positional cloning study that identified human leukocyte antigen (HLA)-G as an asthma susceptibility gene72. In that study, the −964G allele was associated with asthma only in families with an affected mother, and the −964A allele was associated with asthma in families with an unaffected mother. Paternal asthma status had no effect. Moreover, the parental origin of the fetal alleles had no effect on risk, indicating that paternal imprinting is not involved. Subsequent studies73 implicated a SNP (+3142C/G; rs1063320) that resides in the 3’ untranslated region (UTR) within a target site for three microRNAs (miRNAs)73. In the presence of any of these miRNAs, expression of the G allele is suppressed whereas expression levels are unchanged for the C allele73. The GG genotype, which is suppressed in the presence of the miRNAs, was associated with significant protection from asthma among children of mothers with asthma but with modest risk for asthma among children of mothers without asthma. The mechanism through which the ‘maternal asthma-fetal HLA-G genotype’ interaction influences subsequent risk for asthma in the child is still unknown, but modulation of expression via miRNA targeting could be involved. Because asthma in the mother is such a strong predictor of asthma in her children, it is likely that other, as yet undiscovered, genes also play a role in modulating this effect.
Genotype-by-sex interactions and asthma risk
Although sex itself is genetically determined, the in utero environment with respect to sex hormones and all downstream targets differs between male and female fetuses beginning in the first trimester of pregnancy. Sex-specific differences in hormonal milieu and gene expression persist throughout life, and likely account for observed sex differences in disease prevalences21. Thus, it is possible that risk alleles for common diseases may have different effects in males and females, or affect males and females differently at specific stages of the lifecycle. In fact, sex-specific genetic effects are ubiquitous in the genomes of model organisms74–76, and there are likely to be significant sex-specific genetic architectures for human quantitative traits in general21, 77, and for asthma-related quantitative traits in particular (reviewed in references 21, 78).
For example, sex-specific linkages78–and associations79, 81, 82 with asthma traits have been reported. One recent example is quite intriguing. The thymic stromal lymphopoietin (TSLP) gene encodes thymic stromal lymphopoietin, an IL-7-like cytokine that regulates allergic asthma in mouse models and shows increased expression in human airway epithelial cells from patients with asthma compared to controls (reference 81 and citations therein). Expression of Tslp in mice leads to a more severe phenotype in females compared to males83. In a follow-up of a linkage study in Costa Rican families, a SNP upstream of TSLP showed a female-specific association with elevated IgE levels84. A subsequent study of TSLP in more than 13,000 subjects, revealed a complex pattern of interaction at this locus emerged. The T allele at a SNP upstream of the gene (rs1837253) was inversely associated with asthma in boys but not in girls; however, the T allele at a second SNP in exon 1 (rs2289276) showed a stronger inverse association with asthma in girls than in boys, although the effect of this SNP on asthma risk was more variable between study samples. The investigators proposed that variation in TSLP may account for some of the curious epidemiological observations of age-sex interactions on asthma risk, as discussed above21.
GEIs in the nucleus
The nuclear milieu is a powerful determinant of GEIs, particularly for SNPs in regulatory regions. For example, the relative transcriptional activities of the −159C and −159T alleles of CD14 (rs2569190), discussed above, differ between monocytes and hepatocytes, depending on the relative expression of SP1 and SP3, the transcription factors that bind the polymorphic promoter region and control its activity85. An IL13 reporter construct carrying the minor −1112T allele (rs1800925) is significantly more active than the −1112C allele in primary human polarized T helper 2 cells (Th2 cells), but is less active than the −1112C allele in CD4+ Jurkat T cells. The two IL13−1112 alleles had comparable activity in non-polarized, freshly isolated CD4+ T cells. The allele-specific patterns of transcription factor binding in these different cellular models determine, at least in part, the differential activity of the IL13 promoter86, an observation that has recently been validated in humanized mice87. Remarkably, this “flip-flop” pattern of gene expression was detected at distinct stages of differentiation of the same cell, rather than the more commonly reported differences between distinct cell types (as described above for the CD14 promoter variants). The implication of these data is that specific genetic variants may remain functionally silent until the carriers of that variant are exposed to relevant environmental stimuli (allergens and/or parasites, in the case of IL13) that promote the differentiation of the cell. These findings illustrate the very essence of GEIs: both the functional variant and the environmental stimulus that modifies the cellular environment are required for the interaction to be manifested.
Epigenetics: The Link Between Genes and Environment?
Understanding the mechanisms that underlie GEIs is a formidable challenge. It is difficult enough to characterize the impact of polymorphisms on gene expression and function under static conditions (i.e., within a constant environment); it is considerably more challenging to elucidate the dynamic intertwining of polymorphic variation and environmental stimuli. This is why very little has been published to date about the mechanisms of GEIs, and no major finding has yet been reported for asthma. Nonetheless, there is consensus within the community that epigenetic mechanisms could explain the interplay between genes and the environment. The dynamic nature of this interplay is a quasi-perfect fit for the plasticity of epigenetic processes, which can be seen as the functional transducers of environmental cues88. Stable epigenetic alterations can arise during cell development and proliferation, making it possible for cells of multicellular organisms to be genetically identical but structurally and functionally heterogeneous89. Modifications in gene expression in response to environmental and/or developmental cues can result from modifications of DNA (e.g., by methylation) or from proteins that intimately associate with DNA (e.g., acetylation, methylation or phosphorylation of histones) (reviewed in references 90, 91).
Precisely how epigenetic processes impact gene regulation, and to a lesser extent how environmental exposures impact epigenetic marks (such as methylation), has been extensively studied in recent years, and many striking examples of the latter have emerged (see reference 92 for a review of asthma genes). For example, diet can modulate gene expression by impacting DNA methylation in mice93. The fidelity of methylation in the newly synthesized DNA strand depends on the availability of dietary methyl donors and cofactors required for S-adenosylmethionine synthesis, and the concentration of S-adenosylmethionine affects DNA methyl transferase activity94. In a mouse model, a maternal diet supplemented with methyl donors enhanced the severity of allergic airway disease that was inherited transgenerationally by litters exposed prenatally, but not among litters exposed during lactation or adulthood95. Genome-wide methylation revealed 82 genes that were differentially methylated after prenatal supplementation with a methyl-rich diet; and methylation was associated with decreased transcriptional activity and increased disease severity. In particular, the runt-related transcription factor 3 gene (Runx3), which negatively regulates allergic airway disease, was excessively methylated and Runx3 mRNA and protein levels were suppressed in progeny exposed in utero to a high-methylation diet. These findings indicated that dietary factors can modify the heritable risk of allergic airway disease through epigenetic mechanisms during a vulnerable period of fetal development. Consistent with this observation, maternal folic acid supplementation in the Norwegian Mother and Child Cohort Study (>32,000 children) resulted in a modest increase in wheezing and lower respiratory tract infections in children up to 18 months of age96, although neither gene expression nor methylation studies were performed in these children. Therefore, the mechanism for the observed association can not necessarily be attributed to epigenetic modifications.
In summary, the evidence in favor of a strong relationship between environmental exposure, epigenetic changes, and asthma-related phenotypes is plentiful. What is still missing is an explicit link to specific genetic variants; that is, demonstration that an observed GEI results from an allele-specific epigenetic change at the implicated SNP.
GWAS, GWIS, and GEIs
The question as to how much of the ‘missing’ heritability in GWAS will be accounted for by GEIs remains unanswered at this time, although genome-wide interaction studies (GWIS) are beginning to emerge. One of the greatest challenges for this unbiased approach to gene (or interaction) discovery is that of power (see references 97, 98 for further discussion). The first GWIS for childhood asthma and farming exposures has recently been completed in approximately 1700 children from four rural regions in central Europe who participated in the larger GABRIEL study99. This carefully designed investigation of interactions between genome-wide SNP genotypes and farming exposures on asthma risk highlights some of the challenges inherent to taking studies of GEIs to the genome-wide level. This study was well-powered to detect interactions with common alleles in the frequency range of 40–70%. Nevertheless, no interactions in the GWIS were significant, not even those involving SNPs in genes previously identified in an asthma GWAS2 or SNPs in genes that showed interactions with farming exposures in previous candidate gene studies (including the CD14 polymorphism discussed above). That no interactions were found with the SNPs identified in a previous GWAS is not surprising because those variants were selected based on their very significant main effects on asthma risk and because the earlier GWAS examined largely non-farming populations. However, the lack of interactions with the SNPs in genes previously showing interactions was surprising, particularly because many of the original studies were conducted in central European farming populations. Only two of the latter genes showed interactions, and both were quite modest in effect: SNPs at the toll-like receptor 4 (TLR4) and nucleotide oligomerization domain containing 1 (NOD1) genes interacted with ‘contact with cows’ to modify asthma risk. The investigators attributed the lack of interactions with SNPs in these genes in part to the fact that they did not directly measure endotoxin levels, an important environmental variable in many of the earlier studies. In contrast, using a two-step strategy that allows for less stringent thresholds of significance in the second step97, 97 SNPs showed interactions with at least one environmental exposure on asthma risk, providing at least eight intriguing candidate genes for further studies. Of the 97 SNPs, 38% showed protection only in the exposed group, none showed protection only in the non-exposed group, and 62% showed a ‘flip-flop’ pattern of association (Figure 2).
Overall, the authors argue that the absence of significant GEIs in the GWIS suggests that interacting alleles may be in the frequency range <40%, for which their study was not well powered. Alternatively, differences in the microenvironments between the four population samples included in the study may have influenced the ability to detect GEIs. It is even possible that subtle genetic substructure, particularly at key loci, is not accounted for by the global genomic control parameter typically used in such studies (for example, as in reference 2). Because very large samples of human subjects are required for studies of interactions, these studies will nearly always be composed of individuals who differ with respect to relevant environmental exposures and to some degree by ethnicity or genetic backgrounds, which may mask true interactions. That is, the genetic and environmental heterogeneity that is inevitably present in large samples may make this approach less amenable to revealing GEIs than more focused studies. While some exposures, such as prenatal exposure to ETS or sex, may be more straightforward to assess and sufficiently robust in their effects to withstand subtle sample heterogeneity, others may not be amenable to very large studies of human subjects. The fact that the GWIS did not detect interactions between a SNP in nearly perfect linkage disequilibrium (LD) with the CD14 −159C/T variant and exposure to animals, despite the high replicability of this association in previous smaller studies, as discussed above, suggests to us cryptic heterogeneity between study samples.
In fact, both background genes and gene-gene interactions (epistasis) can mask or enhance GEIs, as elegantly demonstrated in yeast100, and at least one example of a gene-by-gene-by-environment interaction in humans has been put forth101. Using yeast as a model system, pairwise epistatic interactions between four variants accounted for a majority of the heritability of sporulation efficiency in strains grown in different media (environments), but the nature of the pairwise interactions differed between strains grown in different environments so that when environment and strain are ignored, none of the pairwise gene-gene interactions are significant. In other words, the relative importance of alleles and their interactions varied with respect to both genetic background and environment, and were not constant between individuals100. We can expect that the genetic architecture of common diseases and quantitative phenotypes in humans will be at least this complex and it may therefore be unrealistic to assume that the same constellation of SNPs will explain the same proportion of the heritability in all populations and in all environments. Thus, the ‘missing’ heritability may not just be ‘hidden’ due to failure to consider the combined effects of associated SNPs but rather because we are averaging over environments and genetic backgrounds, i.e., ignoring ‘context’.
Conclusions and Future Perspectives
It is for this and related reasons that animal models are often proposed as powerful tools for studies of complex interactions102. While we agree that studies in model systems can provide valuable insights into the architecture of gene-gene, gene-environment, and gene-gene-environment interactions100, it is unclear that animal models will offer insights into specific interactions contributing to disease risk in humans because of the context-dependency of these effects. In contrast, cellular human models may provide one approach and a reasonable compromise for studies of GEIs. Although these studies will sacrifice organismal context, cellular systems can reveal a finite number of genotype-dependent interactions occurring in response to specific ‘exposures’ under relatively controlled conditions and, in addition, allow direct studies of mechanism. For example, using a systems genetics approach to identify GEIs, transcript abundance was measured in primary endothelial cells from 96 individuals in the basal state and after exposure to proinflammatory oxidized phospholipids103. In parallel, the same cultures were genotyped for genome-wide SNPs. Roughly one-third of the 59 transcripts that were most regulated (i.e., more than two-fold change in transcript levels) and heritable across cell passages showed evidence of interactions. More than one-quarter of individual SNPs showing GEIs explained 24–32% of the fold-change in transcript levels and six gene-environment ‘hotspots’ were identified as co-localized SNPs that regulated the response of 10 or more transcripts to proinflammatory oxidized phospholipids. Causal candidates were identified and subjected to further functional and validation studies. The investigators suggested that GEIs affecting gene expression might be an important attribute of common, complex human diseases. We further suggest that this approach, applied to additional cell types and exposures, is a powerful tool both for identifying candidates that can be subjected to more targeted GEI studies in human populations and for elucidating the mechanism(s) that underlie GEIs.
A complementary, and equally important, avenue of investigation is to characterize GEIs in human populations. The important question of how to maximize our ability to detect GEIs given their subtle effects and therefore elusive nature remains. A critical consideration in this context is that most of the well-validated GEIs have not been found by chance. Rather, they were discovered using models based on our knowledge of biological processes and/or pathways. How, then, do these prior successes inform our ability to discover additional GEIs and where should we look for them? We propose that GEIs are most likely to occur in, and impact on, signaling pathways regulated by threshold effects – that is, whenever quantitative differences in the intensity of the signal delivered by exogenous or endogenous stimuli result in qualitative differences in the outcome of that signal. In the immune system, for instance, weak versus strong signals can result in distinct and often opposite effects, particularly on cell fate determination. The choice between a Th2 and a Th1 cell fate in response to weak or strong T cell receptor cross-linking is a paradigmatic case104. Such environmental stimuli and gain- or loss-of function variants in the receptors and/or adaptors that transduce these signals are excellent candidates for GEIs. Indeed, the reason why candidate gene studies have been so successful in discovering and characterizing GEIs may be that these studies have explicitly, or more often implicitly, focused on processes, such as innate immunity, in which the strength of the environmental signal plays a pivotal role.
In conclusion, the evidence for GEIs in the literature is compelling, as is the argument that the failure to model GEIs in genetic studies will result in missing potentially important loci that show interactions, particularly those with ‘flip-flop’ patterns of association. Thus, although the ‘environment’ may be considered a ‘nuisance’ to genetic studies, we prefer to think of it as an outstanding ‘opportunity’ to understand disease heterogeneity, to provide clues to the causative pathways in asthma pathogenesis, and to inform us on the complex genetic architecture of common diseases and quantitative phenotypes.
Acknowledgments
This work was supported by National Institutes of Health grants HL085197, HL7831, HL101651 to C.O. and HL100800, HL66391, AI076715 to D.V. The authors acknowledge S.A. Willis-Owen and W. Valdar for first using the expression “nuisance or opportunity” in the context of GEIs in their 2009 review and F. D. Martinez, J. E. Gern, R. J. Lemanske, E. von Mutius, and M. Ege for helpful discussions. The authors apologize to those investigators whose work was not cited due to space constraints.
Glossary
- Atopic dermatitis
also called eczema, is an inflammatory skin disease; it is particularly common in young children
- Bronchial hyperresponsiveness (BHR)
a clinical measure of airway reactivity, usually in response to inhaled methacholine, which many studies use as a criterion for asthma diagnosis
- CD4+ T cells
the population of T cells (typically, with helper function) that express the CD4 surface marker and, depending on the cytokine milieu, will differentiate along the Th1 or Th2 pathway
- Endotoxin
a structural component of the gram-negative bacterial cell that is recognized by the innate immune system and has been associated with complex effects on human diseases
- Epigenetic
heritable changes in gene expression that are not encoded in the DNA sequence, but attributed to mechanisms such as methylation of CpG dinucleotides or post-translational histone modifications
- Epistasis
interaction between genes that are not additive; or when the expression of one gene is dependent on the expression of a second gene
- Flip-flop
when opposite alleles are associated with risk for a disease or a phenotype in different groups (e.g., ethnic groups, groups stratified by an environmental exposures)
- Heritability
the proportion of the total phenotypic variance that is attributed to genetic variance
- Penetrance
the proportion of individuals with the risk genotype that express the phenotype (or disease)
- Th2 cells
polarized CD4+ T cells that differentiate in response to allergen exposure or parasitic infections and provide signals (cytokines) responsible for production of IgE antibodies, eosinophilia, mucus metaplasia, alternative macrophage activation and fibrosis. In the lung, these immune responses are associated with bronchial hyperresponsiveness and ultimately allergic inflammation and asthma.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 6.Gibson G. Hints of hidden heritability in GWAS. Nat Genet. 2010;42:558–560. doi: 10.1038/ng0710-558. [DOI] [PubMed] [Google Scholar]
- 7.Park CC, et al. Fine mapping of regulatory loci for mammalian gene expression using radiation hybrids. Nat Genet. 2008;40:421–429. doi: 10.1038/ng.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masoli M, et al. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 10.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 11.Garantziotis S, Schwartz DA. Ecogenomics of respiratory diseases of public health significance. Annu Rev Public Health. 2010;31:37–51. doi: 10.1146/annurev.publhealth.012809.103633. 31 p following 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vercelli D. Gene-environment interactions in asthma and allergy: the end of the beginning? Curr Opin Allergy Clin Immunol. 2010;10:145–148. doi: 10.1097/ACI.0b013e32833653d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Mutius E. Gene-environment interactions in asthma. J Allergy Clin Immunol. 2009;123:3–11. doi: 10.1016/j.jaci.2008.10.046. quiz 12–13. [DOI] [PubMed] [Google Scholar]
- 14.London SJ, Romieu I. Gene by environment interaction in asthma. Annu Rev Public Health. 2009;30:55, 80. doi: 10.1146/annurev.publhealth.031308.100151. [DOI] [PubMed] [Google Scholar]
- 15.Gergen PJ, et al. The burden of environmental tobacco smoke exposure on the respiratory health of children 2 months through 5 years of age in the United States: Third National Health and Nutrition Examination Survey, 1988 to 1994. Pediatrics. 1998;101:E8. doi: 10.1542/peds.101.2.e8. [DOI] [PubMed] [Google Scholar]
- 16.Gilliland FD, et al. Effects of maternal smoking during pregnancy and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med. 2001;163:429–436. doi: 10.1164/ajrccm.163.2.2006009. [DOI] [PubMed] [Google Scholar]
- 17.Gold DR. Environmental tobacco smoke, indoor allergens, and childhood asthma. Environ Health Perspect. 2000;108 Suppl 4:643–651. doi: 10.1289/ehp.00108s4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdulrazzaq YM, et al. Association of allergic symptoms in children with those in their parents. Allergy. 1994;49:737–743. doi: 10.1111/j.1398-9995.1994.tb02096.x. [DOI] [PubMed] [Google Scholar]
- 19.Holberg CJ, et al. Differences in familial segregation of FEV1 between asthmatic and nonasthmatic families. Role of a maternal component. Am J Respir Crit Care Med. 1998;158:162–169. doi: 10.1164/ajrccm.158.1.9706117. [DOI] [PubMed] [Google Scholar]
- 20.Litonjua AA, et al. Parental history and the risk for childhood asthma. Am J Respir Crit Care Med. 1998;158:176–181. doi: 10.1164/ajrccm.158.1.9710014. [DOI] [PubMed] [Google Scholar]
- 21.Ober C, et al. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douwes J, et al. Farm exposure in utero may protect against asthma, hay fever and eczema. Eur Respir J. 2008;32:603–611. doi: 10.1183/09031936.00033707. [DOI] [PubMed] [Google Scholar]
- 23.Riedler J, et al. Austrian children living on a farm have less hay fever, asthma and allergic sensitization. Clin Exp Allergy. 2000;30:194–200. doi: 10.1046/j.1365-2222.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- 24.Von Ehrenstein OS, et al. Reduced risk of hay fever and asthma among children of farmers. Clin Exp Allergy. 2000;30:187–193. doi: 10.1046/j.1365-2222.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 25.Von Mutius E, Vercelli D. Farm living: Effects on childhood asthma and hay fever. Nature Rev Immunol. 2010 doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 26.Bufford JD, et al. Effects of dog ownership in early childhood on immune development and atopic diseases. Clin Exp Allergy. 2008;38:1635–1643. doi: 10.1111/j.1365-2222.2008.03018.x. [DOI] [PubMed] [Google Scholar]
- 27.Ownby DR, et al. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. Jama. 2002;288:963–972. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- 28.Campo P, et al. Influence of dog ownership and high endotoxin on wheezing and atopy during infancy. J Allergy Clin Immunol. 2006;118:1271–1278. doi: 10.1016/j.jaci.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodner C, et al. Childhood exposure to infection and risk of adult onset wheeze and atopy. Thorax. 2000;55:383–387. doi: 10.1136/thorax.55.5.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nafstad P, et al. Early respiratory infections and childhood asthma. Pediatrics. 2000;106:E38. doi: 10.1542/peds.106.3.e38. [DOI] [PubMed] [Google Scholar]
- 31.Ponsonby AL, et al. Relationship between early life respiratory illness, family size over time, and the development of asthma and hay fever: a seven year follow up study. Thorax. 1999;54:664–669. doi: 10.1136/thx.54.8.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riedler J, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 33.Perkin MR, Strachan DP. Which aspects of the farming lifestyle explain the inverse association with childhood allergy? J Allergy Clin Immunol. 2006;117:1374–1381. doi: 10.1016/j.jaci.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Strachan DP. Hay fever, hygiene, and household size. Bmj. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gern JE. Rhinovirus respiratory infections and asthma. Am J Med. 2002;112 Suppl 6A:19S–27S. doi: 10.1016/s0002-9343(01)01060-9. [DOI] [PubMed] [Google Scholar]
- 36.Johnston SL. Influence of viral and bacterial respiratory infections on exacerbations and symptom severity in childhood asthma. Pediatr Pulmonol Suppl. 1997;16:88–89. doi: 10.1002/ppul.1950230851. [DOI] [PubMed] [Google Scholar]
- 37.Pattemore PK, et al. Viruses as precipitants of asthma symptoms. I. Epidemiology. Clin Exp Allergy. 1992;22:325–336. doi: 10.1111/j.1365-2222.1992.tb03094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein RT, et al. Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. Lancet. 1999;354:541–545. doi: 10.1016/S0140-6736(98)10321-5. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, et al. Obesity may increase the incidence of asthma in women but not in men: longitudinal observations from the Canadian National Population Health Surveys. Am J Epidemiol. 2002;155:191–197. doi: 10.1093/aje/155.3.191. [DOI] [PubMed] [Google Scholar]
- 40.Gilliland FD, et al. Obesity and the risk of newly diagnosed asthma in school-age children. Am J Epidemiol. 2003;158:406–415. doi: 10.1093/aje/kwg175. [DOI] [PubMed] [Google Scholar]
- 41.Kubzansky LD, et al. Angry breathing: A prospective study of hostility and lung function in the Normative Aging Study. Thorax. 2006;61:863–868. doi: 10.1136/thx.2005.050971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bardana EJ., Jr 10. Occupational asthma. J Allergy Clin Immunol. 2008;121:S408–S411. doi: 10.1016/j.jaci.2007.08.005. quiz S421. [DOI] [PubMed] [Google Scholar]
- 43.Dykewicz MS. Occupational asthma: current concepts in pathogenesis, diagnosis, and management. J Allergy Clin Immunol. 2009;123:519–528. doi: 10.1016/j.jaci.2009.01.061. quiz 529–530. [DOI] [PubMed] [Google Scholar]
- 44.Islam T, et al. Relationship between air pollution, lung function and asthma in adolescents. Thorax. 2007;62:957–963. doi: 10.1136/thx.2007.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz J. Air pollution and children's health. Pediatrics. 2004;113:1037–1043. [PubMed] [Google Scholar]
- 46.Trasande L, Thurston GD. The role of air pollution in asthma and other pediatric morbidities. J Allergy Clin Immunol. 2005;115:689–699. doi: 10.1016/j.jaci.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 47.Choi JH, et al. The HLA DRB1*1501-DQB1*0602-DPB1*0501 haplotype is a risk factor for toluene diisocyanate-induced occupational asthma. Int Arch Allergy Immunol. 2009;150:156–163. doi: 10.1159/000218118. [DOI] [PubMed] [Google Scholar]
- 48.Kim SH, et al. Pharmacogenetics of aspirin-intolerant asthma. Pharmacogenomics. 2008;9:85–91. doi: 10.2217/14622416.9.1.85. [DOI] [PubMed] [Google Scholar]
- 49.Bouzigon E, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 50.Thomsen SF, et al. Genetic influence on the age at onset of asthma: A twin study. J Allergy Clin Immunol. 2010 doi: 10.1016/j.jaci.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 51.Zambelli-Weiner A, et al. Evaluation of the CD14/−260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics Study. J Allergy Clin Immunol. 2005;115:1203–1209. doi: 10.1016/j.jaci.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Simpson A, et al. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med. 2006;174:386–392. doi: 10.1164/rccm.200509-1380OC. [DOI] [PubMed] [Google Scholar]
- 53.Gern JE, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol. 2004;113:307–314. doi: 10.1016/j.jaci.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 54.Bottema RW, et al. Interleukin 13, CD14, pet and tobacco smoke influence atopy in three Dutch cohorts: the allergenic study. Eur Respir J. 2008;32:593–602. doi: 10.1183/09031936.00162407. [DOI] [PubMed] [Google Scholar]
- 55.Pacheco KA, et al. Gene-environment interactions influence airways function in laboratory animal workers. J Allergy Clin Immunol. 2010;126:232–240. doi: 10.1016/j.jaci.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eder W, et al. Opposite effects of CD 14/−260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol. 2005;116:601, 607. doi: 10.1016/j.jaci.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Smit LA, et al. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med. 2009;179:363–368. doi: 10.1164/rccm.200810-1533OC. [DOI] [PubMed] [Google Scholar]
- 58.Koppelman GH. Gene by environment interaction in asthma. Curr Allergy Asthma Rep. 2006;6:103–111. doi: 10.1007/s11882-006-0047-y. [DOI] [PubMed] [Google Scholar]
- 59.Martinez FD. CD14, endotoxin, and asthma risk: actions and interactions. Proc Am Thorac Soc. 2007;4:221–225. doi: 10.1513/pats.200702-035AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vercelli D. Learning from discrepancies: CD14 polymorphisms, atopy and the endotoxin switch. Clin Exp Allergy. 2003;33:153–155. doi: 10.1046/j.1365-2222.2003.01606.x. [DOI] [PubMed] [Google Scholar]
- 61.Colilla S, et al. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol. 2003;111:840–846. doi: 10.1067/mai.2003.170. [DOI] [PubMed] [Google Scholar]
- 62.Meyers DA, et al. Genome screen for asthma and bronchial hyperresponsiveness: interactions with passive smoke exposure. J Allergy Clin Immunol. 2005;115:1169–1175. doi: 10.1016/j.jaci.2005.01.070. [DOI] [PubMed] [Google Scholar]
- 63.Dizier MH, et al. Evidence for gene × smoking exposure interactions in a genome-wide linkage screen of asthma and bronchial hyper-responsiveness in EGEA families. Eur J Hum Genet. 2007;15:810–815. doi: 10.1038/sj.ejhg.5201830. [DOI] [PubMed] [Google Scholar]
- 64.Reijmerink NE, et al. Smoke exposure interacts with ADAM33 polymorphisms in the development of lung function and hyperresponsiveness. Allergy. 2009;64:898–904. doi: 10.1111/j.1398-9995.2009.01939.x. [DOI] [PubMed] [Google Scholar]
- 65.Rogers AJ, et al. The interaction of glutathione S-transferase M1-null variants with tobacco smoke exposure and the development of childhood asthma. Clin Exp Allergy. 2009;39:1721–1729. doi: 10.1111/j.1365-2222.2009.03372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sadeghnejad A, et al. IL13 gene polymorphisms modify the effect of exposure to tobacco smoke on persistent wheeze and asthma in childhood, a longitudinal study. Respir Res. 2008;9:2. doi: 10.1186/1465-9921-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramadas RA, et al. Interleukin-1R antagonist gene and pre-natal smoke exposure are associated with childhood asthma. Eur Respir J. 2007;29:502–508. doi: 10.1183/09031936.00029506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panasevich S, et al. Interaction between early maternal smoking and variants in TNF and GSTP1 in childhood wheezing. Clin Exp Allergy. 2010;40:458–467. doi: 10.1111/j.1365-2222.2010.03452.x. [DOI] [PubMed] [Google Scholar]
- 69.Wu H, et al. Parental smoking modifies the relation between genetic variation in tumor necrosis factor-alpha (TNF) and childhood asthma. Environ Health Perspect. 2007;115:616–622. doi: 10.1289/ehp.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smit LA, et al. 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 71.Pfefferle PI, et al. Cord blood cytokines are modulated by maternal farming activities and consumption of farm dairy products during pregnancy: the PASTURE Study. J Allergy Clin Immunol. 2010;125:108–115. doi: 10.1016/j.jaci.2009.09.019. e101-103. [DOI] [PubMed] [Google Scholar]
- 72.Nicolae D, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76:349–357. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan Z, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mackay TF. Mutations and quantitative genetic variation: lessons from Drosophila. Philos Trans R Soc Lond B Biol Sci. 2010;365:1229–1239. doi: 10.1098/rstb.2009.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korstanje R, et al. Influence of sex and diet on quantitative trait loci for HDL cholesterol levels in an SM/J by NZB/BlNJ intercross population. J Lipid Res. 2004;45:881–888. doi: 10.1194/jlr.M300460-JLR200. [DOI] [PubMed] [Google Scholar]
- 76.Ueno T, et al. Rat model of familial combined hyperlipidemia as a result of comparative mapping. Physiol Genomics. 2004;17:38–47. doi: 10.1152/physiolgenomics.00043.2003. [DOI] [PubMed] [Google Scholar]
- 77.Weiss LA, et al. The sex-specific genetic architecture of quantitative traits in humans. Nat Genet. 2006;38:218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- 78.Ober C, et al. Sex-specific genetic architecture of asthma-associated quantitative ttrait loci in a founder population. Curr Allergy Asthma Rep. 2006;6:241–246. doi: 10.1007/s11882-006-0041-4. [DOI] [PubMed] [Google Scholar]
- 79.Aschard H, et al. Sex-specific effect of IL9 polymorphisms on lung function and polysensitization. Genes Immun. 2009;10:559–565. doi: 10.1038/gene.2009.46. [DOI] [PubMed] [Google Scholar]
- 80.Raby BA, et al. Sex-specific linkage to total serum immunoglobulin E in families of children with asthma in Costa Rica. Hum Mol Genet. 2007;16:243–253. doi: 10.1093/hmg/ddl447. [DOI] [PubMed] [Google Scholar]
- 81.Hunninghake GM, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy. 2010 doi: 10.1111/j.1398-9995.2010.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seibold MA, et al. An african-specific functional polymorphism in KCNMB1 shows sex-specific association with asthma severity. Hum Mol Genet. 2008;17:2681–2690. doi: 10.1093/hmg/ddn168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taneda S, et al. Cryoglobulinemic glomerulonephritis in thymic stromal lymphopoietin transgenic mice. Am J Pathol. 2001;159:2355–2369. doi: 10.1016/S0002-9440(10)63085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hunninghake GM, et al. Sex-stratified linkage analysis identifies a female-specific locus for IgE to cockroach in Costa Ricans. Am J Respir Crit Care Med. 2008;177:830–836. doi: 10.1164/rccm.200711-1697OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.LeVan TD, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J. Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 86.Cameron L, et al. Th2-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J. Immunol. 2006;177:8633–8642. doi: 10.4049/jimmunol.177.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pivniouk V, et al. Faithful epigenetic and transcriptional regulation of a transgenic human Th2 cytokine gene locus in the murine nuclear environment. American Association of Immunologists. 2010 [Google Scholar]
- 88.Vercelli D. Genetics, epigenetics, and the environment: switching, buffering, releasing. J Allergy Clin Immunol. 2004;113:381–386. doi: 10.1016/j.jaci.2004.01.752. quiz 387. [DOI] [PubMed] [Google Scholar]
- 89.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 90.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 91.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 92.Ho SM. Environmental epigenetics of asthma: An update. J Allergy Clin Immunol. 2010;126:453–465. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gonzalgo M, Jones P. Mutagenic and epigenetic effects of DNA methylation. Mutat Res. 1997;386:107–118. doi: 10.1016/s1383-5742(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 95.Hollingsworth JW, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–3469. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Haberg SE, et al. Folic acid supplements in pregnancy and early childhood respiratory health. Arch Dis Child. 2009;94:180–184. doi: 10.1136/adc.2008.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Murcray CE, et al. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009;169:219–226. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lanktree MB, Hegele RA. Gene-gene and gene-environment interactions: new insights into the prevention, detection and management of coronary artery disease. Genome Med. 2009;1:28. doi: 10.1186/gm28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ege MJ, et al. Gene-environment interaction for childhood asthma and exposure to farm animals. J Allergy Clin Immunol. 2010 doi: 10.1016/j.jaci.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 100.Gerke J, et al. Gene-environment interactions at nucleotide resolution. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wenten M, et al. Functional variants in the catalase and myeloperoxidase genes, ambient air pollution, and respiratory-related school absences: an example of epistasis in gene-environment interactions. Am J Epidemiol. 2009;170:1494–1501. doi: 10.1093/aje/kwp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Willis-Owen SA, Valdar W. Deciphering gene-environment interactions through mouse models of allergic asthma. J Allergy Clin Immunol. 2009;123:14–23. doi: 10.1016/j.jaci.2008.09.016. quiz 24-15. [DOI] [PubMed] [Google Scholar]
- 103.Romanoski CE, et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sheikh A, Strachan DP. The hygiene theory: fact or fiction? Curr Opin Otolaryngol Head Neck Surg. 2004;12:232–236. doi: 10.1097/01.moo.0000122311.13359.30. [DOI] [PubMed] [Google Scholar]
- 106.Wills-Karp M, et al. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 107.Vuillermin PJ, et al. Microbial exposure, interferon gamma gene demethylation in naive T-cells, and the risk of allergic disease. Allergy. 2009;64:348–353. doi: 10.1111/j.1398-9995.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 108.Schaub B, et al. The many faces of the hygiene hypothesis. J Allergy Clin Immunol. 2006;117:969–977. doi: 10.1016/j.jaci.2006.03.003. quiz 978. [DOI] [PubMed] [Google Scholar]
- 109.Vercelli D. Mechanisms of the hygiene hypothesis--molecular and otherwise. Curr Opin Immunol. 2006;18:733–737. doi: 10.1016/j.coi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 110.Wu T, et al. Evidence of gene-environment interaction for the IRF6 gene and maternal multivitamin supplementation in controlling the risk of cleft lip with/without cleft palate. Hum Genet. 2010;128:401–410. doi: 10.1007/s00439-010-0863-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aslund C, et al. Maltreatment, MAOA, and Delinquency: Sex Differences in Gene-Environment Interaction in a Large Population-Based Cohort of Adolescents. Behav Genet. 2010 doi: 10.1007/s10519-010-9356-y. [DOI] [PubMed] [Google Scholar]
- 112.Cole SW, et al. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci U S A. 2010;107:5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gomez LM, et al. Evidence of a gene-environment interaction that predisposes to spontaneous preterm birth: a role for asymptomatic bacterial vaginosis and DNA variants in genes that control the inflammatory response. Am J Obstet Gynecol. 2010;202(386):e381–e386. doi: 10.1016/j.ajog.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 114.Tsai CT, et al. Interaction of gender, hypertension, and the angiotensinogen gene haplotypes on the risk of coronary artery disease in a large angiographic cohort. Atherosclerosis. 2009;203:249–256. doi: 10.1016/j.atherosclerosis.2008.06.004. [DOI] [PubMed] [Google Scholar]