Abstract
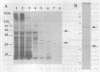
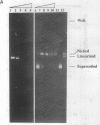
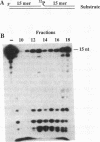
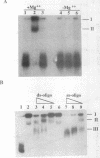
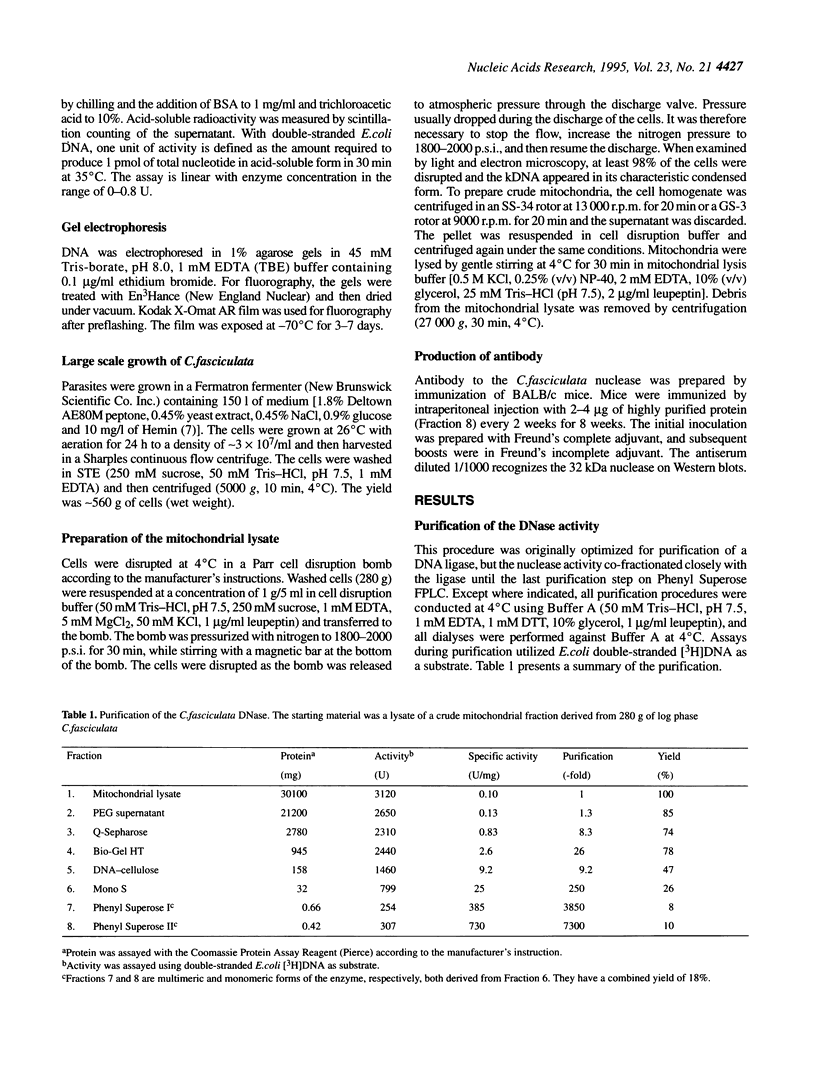
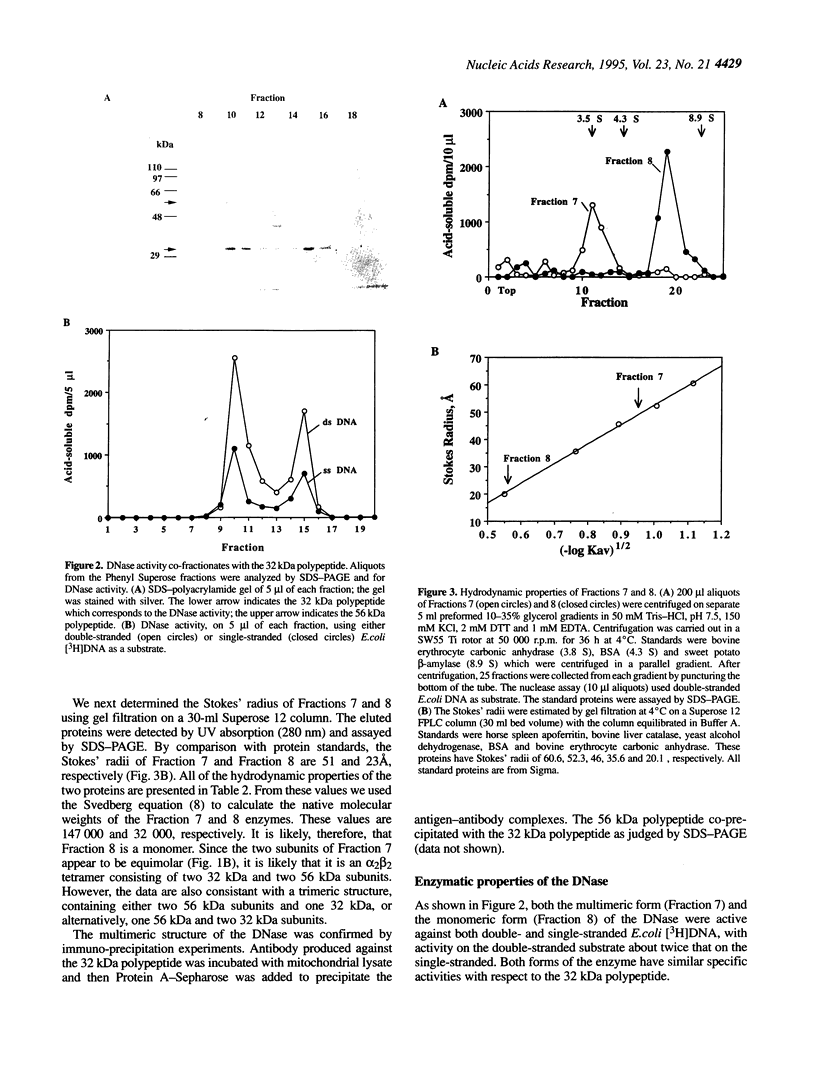
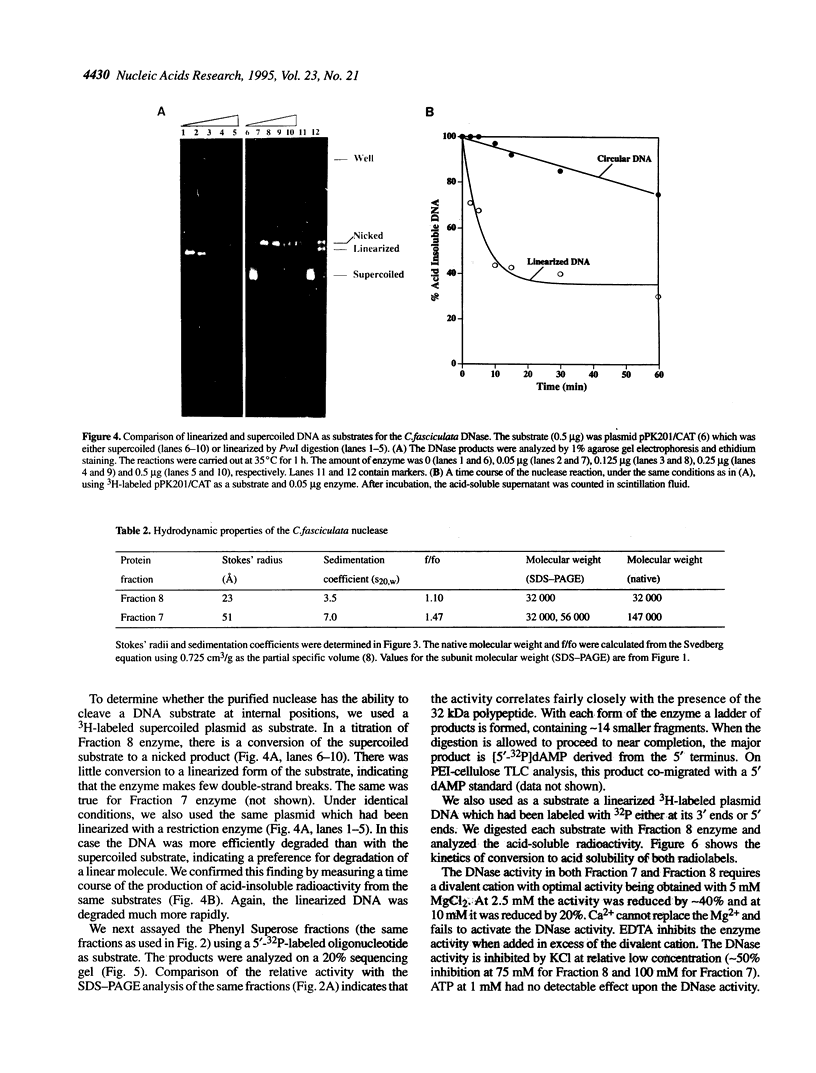
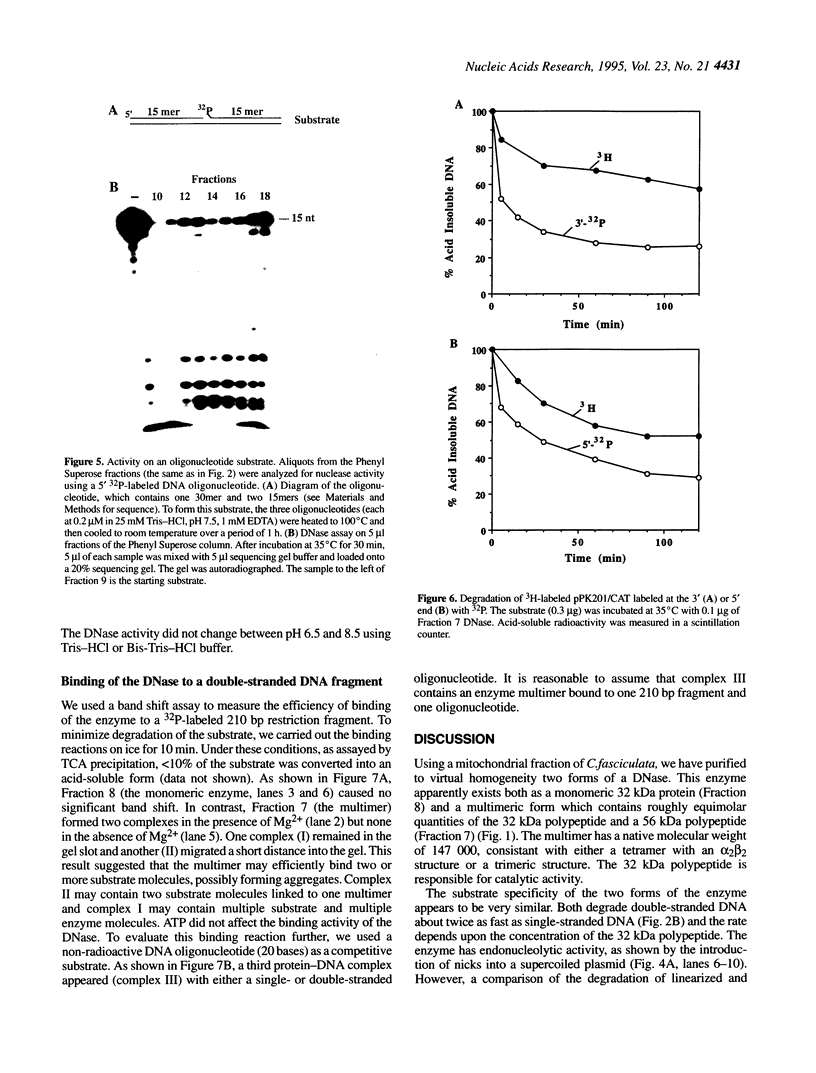
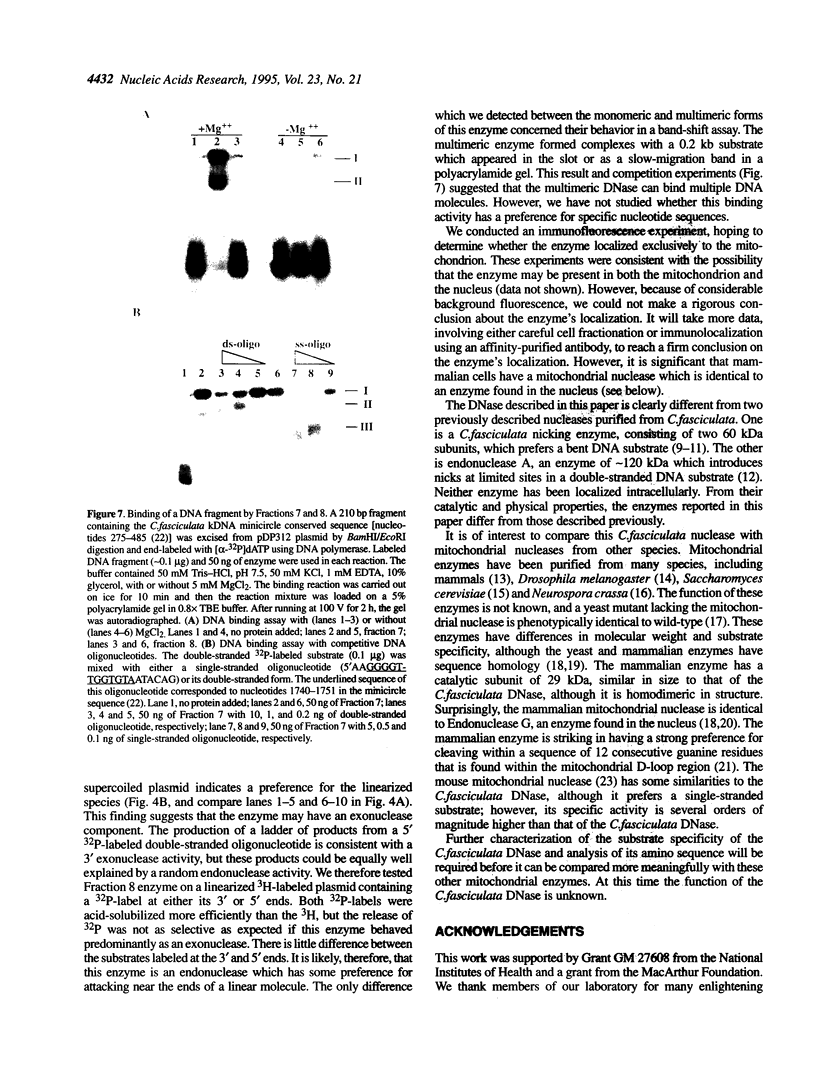
We have purified to homogeneity a DNase from a Crithidia fasciculata crude mitochondrial lysate. The enzyme is present in two forms, either as a 32 kDa polypeptide or as a multimer containing the 32 kDa polypeptide in association with a 56 kDa polypeptide. Native molecular weight measurements indicate that these forms are a monomer and possibly an alpha 2 beta 2 tetramer, respectively. The monomeric and multimeric forms of the enzyme are similar in their catalytic activities. Both digest double-stranded DNA about twice as efficiently as single-stranded DNA. They introduce single-strand breaks into a supercoiled plasmid but do not efficiently make double-strand breaks. They degrade a linearized plasmid more efficiently than a nickel plasmid. Both enzymes degrade a 5'-32P-labeled double-stranded oligonucleotide to completion, with the 5'-terminal nucleotide ultimately being released as a 5'-mononucleotide. One difference between the monomeric and multimeric forms of the enzyme, demonstrated by a band shift assay, is that the multimeric form binds tightly to double-stranded DNA, possibly aggregating it.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cummings O. W., King T. C., Holden J. A., Low R. L. Purification and characterization of the potent endonuclease in extracts of bovine heart mitochondria. J Biol Chem. 1987 Feb 15;262(5):2005–2015. [PubMed] [Google Scholar]
- Côté J., Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by endonuclease G. Science. 1993 Aug 6;261(5122):765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- Dake E., Hofmann T. J., McIntire S., Hudson A., Zassenhaus H. P. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1988 Jun 5;263(16):7691–7702. [PubMed] [Google Scholar]
- Gerschenson M., Houmiel K. L., Low R. L. Endonuclease G from mammalian nuclei is identical to the major endonuclease of mitochondria. Nucleic Acids Res. 1995 Jan 11;23(1):88–97. doi: 10.1093/nar/23.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harosh I., Mezzina M., Harris P. V., Boyd J. B. Purification and characterization of a mitochondrial endonuclease from Drosophila melanogaster embryos. Eur J Biochem. 1992 Dec 1;210(2):455–460. doi: 10.1111/j.1432-1033.1992.tb17442.x. [DOI] [PubMed] [Google Scholar]
- Holdsworth M. L., Hines J. C., Ray D. S. Characterization of a novel endonuclease from Crithidia fasciculata. Nucleic Acids Res. 1989 Jun 12;17(11):4047–4060. doi: 10.1093/nar/17.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchin P. A., Klein V. A., Ryan K. A., Gann K. L., Rauch C. A., Kang D. S., Wells R. D., Englund P. T. A highly bent fragment of Crithidia fasciculata kinetoplast DNA. J Biol Chem. 1986 Aug 25;261(24):11302–11309. [PubMed] [Google Scholar]
- Linial M., Shlomai J. A unique endonuclease from Crithidia fasciculata which recognizes a bend in the DNA helix. Specificity of the cleavage reaction. J Biol Chem. 1988 Jan 5;263(1):290–297. [PubMed] [Google Scholar]
- Linial M., Shlomai J. Sequence-directed bent DNA helix is the specific binding site for Crithidia fasciculata nicking enzyme. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8205–8209. doi: 10.1073/pnas.84.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M., Shlomai J. The sequence-directed bent structure in kinetoplast DNA is recognized by an enzyme from Crithidia fasciculata. J Biol Chem. 1987 Nov 5;262(31):15194–15201. [PubMed] [Google Scholar]
- Linn S., Lehman I. R. An endonuclease from mitochondria of Neurospora crassa. J Biol Chem. 1966 Jun 10;241(11):2694–2699. [PubMed] [Google Scholar]
- Low R. L., Cummings O. W., King T. C. The bovine mitochondrial endonuclease prefers a conserved sequence in the displacement loop region of mitochondrial DNA. J Biol Chem. 1987 Nov 25;262(33):16164–16170. [PubMed] [Google Scholar]
- Melendy T., Ray D. S. Purification and nuclear localization of a type I topoisomerase from Crithidia fasciculata. Mol Biochem Parasitol. 1987 Jun;24(2):215–225. doi: 10.1016/0166-6851(87)90108-3. [DOI] [PubMed] [Google Scholar]
- Ray D. S. Kinetoplast DNA minicircles: high-copy-number mitochondrial plasmids. Plasmid. 1987 May;17(3):177–190. doi: 10.1016/0147-619x(87)90026-6. [DOI] [PubMed] [Google Scholar]
- Shapiro T. A., Englund P. T. The structure and replication of kinetoplast DNA. Annu Rev Microbiol. 1995;49:117–143. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- Shlomai J. The assembly of kinetoplast DNA. Parasitol Today. 1994 Sep;10(9):341–346. doi: 10.1016/0169-4758(94)90244-5. [DOI] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966 Feb 7;112(2):346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Simpson L. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Annu Rev Microbiol. 1987;41:363–382. doi: 10.1146/annurev.mi.41.100187.002051. [DOI] [PubMed] [Google Scholar]
- Stuart K., Feagin J. E. Mitochondrial DNA of kinetoplastids. Int Rev Cytol. 1992;141:65–88. doi: 10.1016/s0074-7696(08)62063-x. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Ray D. S. DNA sequence of Crithidia fasciculata kinetoplast minicircles. Mol Biochem Parasitol. 1987 Apr;23(3):253–263. doi: 10.1016/0166-6851(87)90032-6. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Linn S. Purification and properties of a single strand-specific endonuclease from mouse cell mitochondria. Nucleic Acids Res. 1986 Dec 22;14(24):9579–9593. doi: 10.1093/nar/14.24.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent R. D., Hofmann T. J., Zassenhaus H. P. Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Apr 25;16(8):3297–3312. doi: 10.1093/nar/16.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zassenhaus H. P., Hofmann T. J., Uthayashanker R., Vincent R. D., Zona M. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 1988 Apr 25;16(8):3283–3296. doi: 10.1093/nar/16.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]