Abstract
Plant microRNAs (miRNAs) have been shown to play critical roles in regulating gene expression at the post-transcriptional level. In this study, we employed high throughput sequencing combined with computational analysis to survey miRNAomes from the seedlings of rice under normal conditions and treatments of H2O2 that result in oxidative stress. Comparison of the miRNAomes and subsequent northern blot analysis identified seven miRNA families differentially expressed under H2O2 stress. Predicted and experimentally validated targets of these H2O2-responsive miRNAs are involved in different cellular responses and metabolic processes including transcriptional regulation, nutrient transport, auxin homeostasis, cell proliferation and programmed cell death. This indicates that diverse miRNAs form a complex regulatory network to coordinate plants’ responses under oxidative stress. In addition, we also discovered 32 new miRNAs in the seedlings of rice. Interestingly, of these new miRNAs, miR3981 was originally found to be a putative exonic miRNA located in the exon of AK106348, suggesting that plants may also use some exons as an miRNA source. This study is the first genome-wide investigation of H2O2-regulated miRNAs in plants and broadens our perspectives on the important regulatory roles of miRNAs in plant oxidative stress and physiological adaption.
INTRODUCTION
In plants, hydrogen peroxide (H2O2) and other reactive oxygen species (ROS) are unavoidable by-products of aerobic metabolic pathways, including photosynthesis and respiration (1). H2O2 is highly reactive and toxic and thus capable of injuring cells, but it is also known to act as a signaling molecule in regulating basic biological processes, such as growth and development, hormone signaling, biotic and abiotic stress responses and programmed cell death (PCD) (1,2). In particular, most forms of biotic or abiotic stress lead to an increase in H2O2 production, suggesting that H2O2 can be regarded as a cellular indicator of various stresses and as a secondary messenger involved in the stress-response signal transduction pathways. To date, global expression profiling studies of plants at transcriptomic or proteomic levels have revealed that changes in gene expression in response to H2O2 stress include the induction of expression and synthesis of proteins involved in redox homeostasis and cell rescue or defense, as well as the inhibition of expression of genes related to normal growth and development (3,4). It was reported that 713 expressed sequence tags (ESTs) were regulated by high light-induced H2O2 in catalase-deficient tobacco plants, and their transcriptional responses mimicked those that have been reported during other biotic and abiotic stresses (3). The expression profiling corroborated by physiological experiments showed that a short term H2O2 exposure of the catalase-deficient plants triggered an increased tolerance against subsequent severe oxidative stress, indicating that cross-tolerance is mediated by H2O2 (3). Comparative proteomics revealed that 144 proteins were differentially expressed in the leaves of rice seedlings under H2O2 stress. This indicates that there is a H2O2 stress-responsive protein network and provides clues to help understand the adaptation strategies of rice seedlings to oxidative challenges at the protein level (4). Large-scale transcriptomic and proteomic analyses have already documented a large number of genes or proteins that respond to H2O2 stress; however, the mechanisms by which the dual functions of this molecule are regulated, particularly in plants, are far from being completely understood.
Small RNA molecules including microRNAs (miRNAs) and short interfering RNAs (siRNAs) are important modulators of gene expression in eukaryotic cells (5,6). Among these small RNA molecules, plant miRNAs are a class characterized by endogenous non-coding small RNAs (20–24 nt) derived from primary transcripts that are capable of forming characteristic stem-loop structures (7). Plant miRNAs function as important regulators of gene expression at the post-transcriptional level by guiding mRNA degradation or translational repression (7–9). It is well-known that mature miRNAs typically direct cleavage of their mRNA targets based on extensive complementarity, and play crucial roles in basic developmental processes such as leaf development, flowering time, organ polarity and auxin signaling (8–11). Recently, increasing evidence has indicated that these miRNAs have important functions in the adaptive responses to diverse abiotic and biotic stresses, including antibacterial resistance (12), sulfate or phosphate starvation (13–15), cold, drought, salinity (16,17), UV-B radiation (18), mechanical strain (19) and heavy metals (20). Interestingly, an Arabidopsis miRNA (Ath-miR398) was downregulated under oxidative stress treatments including high light, heavy metal and methyl viologen, while the expression of its targets, two copper superoxide dismutase enzyme (CSD) genes, was induced under the same conditions (21). Transgenic plants overexpressing the Ath-miR398-resistant CSD gene were more resistant to oxidative stresses than plants overexpressing a regular CSD gene (21). Therefore, it is reasonable to believe that there is a diverse set of miRNAs involved in the regulation of redox homeostasis in plants.
Advanced high-throughput sequencing technologies have allowed the discovery of a large and diverse class of non-conserved miRNAs, particularly those that are not abundant and of a recent evolutionary origin (22). As a result, hundreds of new miRNAs have been identified in various plant species including Arabidopsis (22,23) and rice (24–27), rapidly enlarging the total number of identified plant miRNAs. Due to its good reproducibility, deep sequencing can also be used as a powerful tool for small RNA expression profiling (22), in which miRNAs with tissue or developmental stage-specific expression patterns can be identified by comparing the read frequencies of miRNAs from different tissues or developmental stages. The recent application of this method has resulted in the identification of some miRNAs that are preferentially expressed at different developmental stages in the developing rice grains (26) or in the developing inflorescence of rice (28), indicating that there is still room to discover new and/or specific miRNAs in rice.
In the present study, we aimed to identify putative H2O2-regulated miRNAs expressed in rice seedlings under oxidative stress by using a deep sequencing approach developed by Solexa (now Illumina Inc.). Two small RNA libraries were constructed from H2O2-treated and control rice seedlings, and more than five million small RNA sequence reads were generated for each library. We identified seven H2O2-responsive miRNA families (miR169, miR397, miR528, miR1425, miR827, miR319a.2 and miR408-5p) via expression profiling of the miRNAs based on a comparative miRNAomic analysis in combination with experimental validation. The predicted targets were experimentally validated and found to be involved in different cellular responses and metabolic processes, indicating the importance of diverse miRNAs in response to oxidative stress. Furthermore, 32 miRNAs, 18 from known miRNA loci and 14 from novel miRNA loci, were also newly identified from the sequencing data. To the best of our knowledge, this is the first report of a systematic investigation of H2O2-regulated miRNAs and their targets in plants.
MATERIALS AND METHODS
Plant growth
Rice (Oryza sativa L. ssp indica cv. 93-11) seedlings were grown in a growth chamber with 28/21°C (16 h day/8 h night) and relative humidity of 70% as reported earlier (4). For H2O2 treatment, 12-day-old seedlings were treated with three H2O2 concentrations (0.6, 3.0 and 15.0 mM) for 6 h in plastic containers, respectively (4). Seedlings immersed in dd H2O were used as a control. The rice seedlings were snap-frozen in liquid nitrogen, and then stored at −80°C for RNA extraction.
Small RNA library construction
Total RNA was isolated from the frozen seedlings by using the Trizol Reagent (Invitrogen) according to the manufacturer's instructions. The RNA concentration and purity were determined photometrically by measuring absorbance at 260 nm and A260/A280 ratio using the Ultrospec 3000 pro spectrophotometer (Amersham Biosciences). The RNA integrity was evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies). For small RNA library construction and deep sequencing, RNA samples were prepared as follows: equal quantities (10 μg) of total RNA isolated from rice seedlings treated with three H2O2 concentrations were mixed together to construct the H2O2-treated small RNA library, and total RNA prepared from the control sample was used to construct the control small RNA library.
In brief, total RNA was separated by 15% polyacrylamide gel electrophoresis (PAGE), and RNA molecules in the range of 18–30 nt were enriched and ligated with proprietary adapters to the 5′ and 3′ termini. The samples were used as templates for cDNA synthesis and the resulting cDNA was amplified to produce sequencing libraries. The Solexa sequencing was performed by Beijing Genomics Institute (BGI).
Analysis of sequencing data
The overall procedure for analyzing Solexa small RNA libraries is shown in Supplementary Figure S1. After trimming the 3′ adaptor sequence, sequences shorter than 18 nt were excluded from further analysis. Next, sequences were perfectly mapped onto the BGI 93-11 genome [http://rice.genomics.org.cn/rice/; (29)] using the program SOAP (30). According to rice (Oryza sativa L. ssp japanica) defined mature miRNAs and stem-loop miRNA precursors from miRBase [version 14; http://www.mirbase.org; (31)], their homologs of indica (93-11) were found and regarded as 93-11 mature miRNAs and miRNA precursors. We use the following criteria to define a known miRNA in rice indica (93-11): (i) sequences can be perfectly mapped onto 93-11 miRNA precursors; (ii) the start position of the alignment must be between +2 and −2 nt away from the 5′ end of the mature miRNA on the precursor. To identify sequences originating from protein-coding genes, repeats, rRNA, tRNA, snRNA, and snoRNA, we used 93-11 full length cDNA (FLcDNA) and FGeneSH (29), TIGR Oryza Repeat Database [ftp://ftp.tigr.org/pub/data/TIGR_Plant_Repeats/; (32)], and Sanger Rfam data [ftp://ftp.sanger.ac.uk/pub/databases/Rfam/; (33)].
For novel miRNA prediction, unique sequences that have more than 10 hits to the genome or have match to known non-coding RNAs were removed. After this, 150 nt upstream and 150 nt downstream genome sequences surrounding each unique sequence were extracted for secondary structure analysis. As described earlier (13), inverted repeats were found in the 93-11 genome and their secondary structures were predicted by RNAfold (34). Inverted repeats with good hairpin-like structures were identified as putative miRNA precursors and unique reads in the precursors were evaluated by MirCheck (13). The miRNA candidates that passed MirCheck were deemed as high probability if their corresponding miRNA*s were also found in the small RNA libraries. After prediction, the resulting potential miRNA loci were examined carefully based on the distribution and numbers of small RNAs on the entire precursor regions, and those loci without precise excision of an ∼21 nt miRNA/miRNA* duplex were filtered (35).
Target predictions
A search for miRNA target genes was performed using an approach described earlier (36). All newly identified miRNA sequences were used to query 93-11 FgeneSH and FLcDNA for potential target genes using PatScan (37). Only no more than three mismatches were allowed between the miRNA and its target in our predictions. A G–U bond counts as a 0.5 mismatch and no mismatch was allowed at position 10 or 11.
Northern blot analysis
Small RNA northern blot analysis was carried out as described earlier (25). In brief, total RNA was extracted from 12-day-old rice seedlings using Trizol Reagent, and low-molecular-weight (LMV) RNA was isolated from total RNA by PEG8000/NaCl precipitation. About 20 μg LMV RNA per lane was separated on a denaturing 15% polyacrylamide gel, transferred electrophoretically to Hybond-N+ membranes (Amersham Biosciences), and fixed on the membranes by 5 min UV crosslinking and 2 h baking at 80°C. DNA oligonucleotide probes complementary to miRNA sequences were end labeled with γ-32P-ATP by T4 polynucleotide kinase (New England Biolabs) and purified by Microspin G-25 columns (GE Healthcare). The membranes were prehybridized for at least 2 h and hybridized overnight using Ultrahyb-Oligo hybridization buffer (Ambion) at 42°C. After the hybridization, the membranes were washed three times with washing buffer (2 × SSC + 0.5% SDS) at 42°C, and then exposed to X films for a week.
RNA ligase-mediated 5′ RACE
A modified RNA ligase-mediated rapid amplification of 5′ cDNA ends (5′ RACE) was used to map the cleavage sites in target mRNAs (24). Poly(A)+ mRNA was enriched from total RNA using the Oligotex-dT30 mRNA purification kit (Takara), and then ligated directly to the 5′ RACE adapter using FirstChoice RLM-RACE kit (Ambion) according to the manufacturer’s instruction. Two gene-specific primers were used for each RACE (Supplementary Table S1). The PCR products were gel-purified, cloned (pMD19-T vector, Takara) and sequenced.
Quantitative real-time PCR
About 2 μg of total RNA were reverse-transcribed using Takara RNA PCR Kit to generate cDNA. Quantitative real-time PCR was performed on iCycler iQ5 Multicolor real-time PCR detection system (Bio-Rad) by using Power SYBR Green PCR Master Mix (Applied Biosystems). Gene expression levels were presented as fold-change calculated using the comparative threshold cycle (CT) methods as described (38) with β-tubulin as the internal reference (39). All the primers used were listed in Supplementary Table S1.
RESULTS AND DISCUSSION
An overview of our high-throughput sequencing datasets
In our previous studies, the exposure of 12-day-old rice (Oryza sativa L. cv. 93-11) seedlings to three different concentrations (0.6, 3.0 and 15.0 mM) of exogenous H2O2 resulted in dramatic morphological and physiological changes (4). Furthermore, we identified 144 differentially expressed proteins through a comparative proteomic analysis, which revealed a dynamic protein network responsive to H2O2 stress (4). In this study, to examine the H2O2 stress-responsive miRNAs, two small RNA libraries were constructed from a mixture of 12-day-old 93-11 rice seedlings treated with three H2O2 concentrations and a control sample, and were then subjected to Solexa deep sequencing. The resulting raw sequence reads (more than five million for each library) were processed computationally to remove the 3′ adapter and this yielded a total of 5 123 001 genome-matching reads (≥18 nt) from the two libraries (2 397132 and 2 725 869 reads from the H2O2-treated and control libraries, respectively). The remaining sequences that could not be mapped onto the rice genome were possibly derived from unsequenced 93-11 genomic regions, sequence errors or contaminants, and thus were excluded from subsequent analysis. More than 60% of these genome-mapped small RNAs were 20–24 nt in length with 24 and 21 nt as the major size groups (Supplementary Figure S2), consistent with the size of Dicer-like (DCL) cleavage products.
The genome-matched small RNA sequences were clustered into several RNA classes such as known miRNAs, trans-acting siRNAs (ta-siRNAs), repeats, rRNA, tRNA, snRNA/snoRNA and others (Table 1). Similar to a previous report in rice (24), known rice miRNAs account for 20.5% of all sequence reads for the control library and 17.2% for the H2O2-treated library, suggesting that mature miRNAs were highly enriched in our small RNA libraries. However, after analyzing the number of unique sequences, the proportion of small RNA sequences derived from known miRNAs represented only a very small fraction (∼0.1%) of the total number (Table 1). The highest fraction of unique sequences (∼84%) was unclassified small RNA sequences, which probably included novel miRNA candidates and other classes of regulatory RNAs.
Table 1.
Distribution of the genome-mapped sequence reads in small RNA libraries
| RNA class | Control library |
H2O2-treated library |
||
|---|---|---|---|---|
| Reads | Unique sequences | Reads | Unique sequences | |
| Total | 23 971 32 | 920 697 | 27 258 69 | 998 349 |
| Known miRNAsa | 491 660 (20.5%) | 753 (0.08%) | 470 462 (17.3%) | 781 (0.08%) |
| ta-siRNAsb | 9129 (0.4%) | 173 (0.02%) | 9187 (0.3%) | 188 (0.02%) |
| Repeats | 411 055 (17.1%) | 104 975 (11.4%) | 549 151 (20.1%) | 112 492 (11.3%) |
| rRNA | 238 086 (9.9%) | 19 441 (2.1%) | 346 657 (12.7%) | 22 222 (2.2%) |
| tRNA | 99 667 (4.2%) | 5083 (0.6%) | 129 425 (4.8%) | 6177 (0.6%) |
| snRNA/snoRNA | 4696 (0.2%) | 2658 (0.3%) | 6798 (0.3%) | 4459 (0.4%) |
| Protein-coding genes | 93 841 (3.9%) | 15 008 (1.6%) | 137 753 (5.0%) | 17 634 (1.8%) |
| Othersc | 10 489 98 (43.8%) | 772 606 (83.9%) | 10 764 36 (39.5%) | 834 396 (83.6%) |
aThe number of reads includes the defined mature miRNAs and their ±2 nt variants.
bMatching known rice ta-siRNA loci including TAS3a1, TAS3a2, TAS3b1 and TAS3b2.
cContains all of the unclassified sequences that possibly include new miRNAs.
Known rice miRNAs expressed in seedlings
Currently, miRBase (release 14) lists over 400 mature miRNA sequences cloned or predicted in rice. Out of these, 264 rice miRNAs were detected in our sequencing datasets (Supplementary Table S2) and all of the 22 miRNA families (including the recently identified miR827) conserved in Arabidopsis and rice were included. This further demonstrates that the deep sequencing approach is very sensitive. As reported earlier (24–26), conserved miRNAs are far more abundant than non-conserved miRNAs in our libraries. The most abundant miRNA families are miR156 and miR168, accounting for about 49% and 34% of the total sequence reads from the known miRNAs datasets, respectively. On the other hand, the reasons for those undetected miRNAs in our sequencing data might be explained by the following facts: (i) some miRNAs (miR413 to miR426) were only predicted by computational methods and have not been found by deep sequencing thus far (24–27,40); (ii) homologs in 93-11 for some miRNAs such as miR810, miR1423 and miR1437 and their pre-miRNAs cannot be found due to the genome difference between japonica and indica and (iii) some miRNAs might be tissue- or cell-specific or of extremely low abundance.
Similar to other deep sequencing studies (24,26), many variants were observed within a ±2 nt range away from the 5′ or 3′ ends of the annotated miRNA sequences. Generally, the sequence reads of variants are less abundant than those of annotated miRNA sequences. However, in some cases, the variants are more abundant, indicating that they are probably dominant and might substitute for the reported miRNA sequences in the miRBase. For example, the annotated miR156l sequence is 21-nt long and had only five reads in our libraries, whereas its most abundant 20-nt variant had 449 reads, and the detected mi156l* is just of the variant (Supplementary Figure S3A). Similarly, miR156k, miR167d-j, miR319a,b, miR820a-c and miR1318/1432 were predominantly found at lengths of 20, 22, 21, 24 and 22 nt, respectively (Supplementary Figure S3A), and this is partly supported by a previous deep-sequencing study in rice (26). Intriguingly, for three miRNAs (miR171i and miR397a,b), the position of the most abundant sequence was shifted from the annotated miRNA by 3 nt (Supplementary Figure S3A). Because these 3-nt variants are predicted to target identical mRNAs with the annotated miRNAs, they might be regarded as the authentic miRNA sequences in place of the annotated sequences.
The high sensitivity of the deep sequencing approach makes it feasible to investigate the distributions and numbers of the unique sequences along the miRNA precursors. We identified 150 miRNA* sequences out of 264 known rice miRNAs expressed in our libraries (Supplementary Table S2). As expected, most miRNA*s had far less sequence reads (<10 reads) than the corresponding miRNAs. However, several miRNA*s showed an even higher sequencing frequency than did the corresponding miRNAs (Supplementary Figure S3B). Similar to a previous report (26), we found that miR408* and miR529* were much more abundant than miR408 and miR529 (Supplementary Figure S3B). In this case, the miRNA* might be the authentic miRNA, or alternatively, both the miRNA and miRNA* can function simultaneously, indicating that there is a complex mechanism of miRNA biogenesis. Additionally, probably due to the long stems of plant miRNA precursors, two or even more distinct miRNAs can be produced in a single precursor (24,41). For example, pre-miR159a generated two totally different miRNAs, miR159a.1 and miR159a.2, and both were found to function in target cleavage (41). Other examples were also observed including pre-miR159b, pre-miR169a, pre-miR319a,b and pre-miR2871a (Supplementary Figure S3C). For these precursors, newly identified miRNAs were expressed at a level similar to the annotated miRNAs, and the corresponding miRNA*s were also found for most of them, indicating they are unlikely to be the byproducts of DCL1 processing. Therefore, with the exception of the annotated miRNAs, the small RNA sequences from the same precursor and with a similar expression level can also be annotated as a new miRNA member (Supplementary Table S3). Furthermore, nearly half of the non-conserved miRNAs could not be well-supported by our sequencing results, indicating that they were either borderline miRNA candidates or originated from siRNA-like clusters (24,26). For these loci with weak miRNA-like characteristics, numerous small RNAs were generated from both strands, just from the antisense strand of the precursor, or showed a smeared distribution pattern over the entire precursor region (Supplementary Figure S3D).
Newly identified miRNAs in both libraries
We identified 18 new miRNAs derived from known miRNA loci by investigating the distribution of small RNAs on known miRNA precursors. However, there are still abundant unclassified small RNAs in our libraries, some of which might be originated from novel miRNA loci. According to the criteria for annotation of plant miRNAs (35), a reliable miRNA locus should generate the majority of small RNAs (>75%) precisely from the positions of miRNA and miRNA*. Although almost 1000 loci with miRNA precursor-like stem-loop structures were found to generate small RNAs in our sequencing datasets, only 14 novel miRNA loci were identified based on the criteria that miRNA and/or miRNA* were the major small RNA species derived from the precursor (Supplementary Table S3, Supplementary Figure S4). For most of new miRNAs, miRNA* species were detected in our libraries, which is the strong evidence for the cleavage by DCL1 during the miRNA biogenesis (23). Out of these new miRNAs, some (miR811d, miR812k-o, miR1862f,g and miR2873b) belong to known miRNA families for the homology with known miRNAs or miRNA precursors. According to the sequencing reads, miR812k-m is more abundant than the previously reported miR812 family members (Supplementary Table S3). None of the novel miRNAs has been found in Arabidopsis or Populus trichocarpa, indicating that they are rice-specific. Interestingly, we identified ppt-miR894 like sequences with abundant sequence reads in both libraries (Supplementary Table S4), but these sequences cannot map to the rice genome, and neither their origin nor their functions are known.
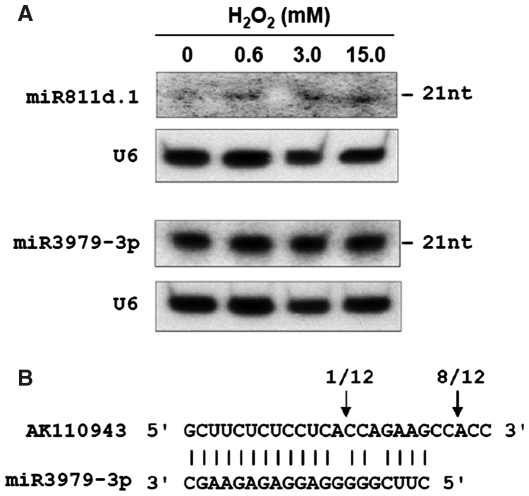
Although several new miRNAs such as miR812k-m was found to be of a relatively high abundance, most new miRNAs have sequence reads less than 100 and might be expressed at low levels. Despite this, two representative miRNAs (miR811d.1 and miR3979-3p) were detected using northern blots (Figure 1A). Cleavage of the miR3979-3p target gene has been validated by RNA ligase-mediated 5′ RACE. miR3979-3p targets the AK110943 mRNA, which encodes a bHLH (basic helix–loop–helix) transcription factor. The main cleavage site of AK110943 is 2 nt downstream from the miR3979-3p complementary region (Figure 1B), which might result from an unstable cleavage product or other potential mechanisms such as secondary siRNAs triggered by miRNA-mediated cleavage of target mRNAs (42).
Figure 1.
(A) Northern blot analysis of the expression of novel miRNAs in rice seedlings treated with aqueous solutions containing 0 (control), 0.6, 3.0 or 15.0 mM H2O2 for 6 h. U6 was used as a loading control. (B) Mapping of target mRNA cleavage sites by RNA ligase-mediated 5′ RACE. The arrows indicate the 5′ termini of miRNA-guided cleavage products, with the frequency of the clones sequenced shown above.
Most of the newly identified miRNAs have predicted targets (Supplementary Table S3), but a few miRNAs do not. The lack of predicted targets might be because these miRNAs have only recently evolved without actual targets (22) or because their target mRNAs have not been annotated in the rice genome. Unlike the conserved miRNAs whose targets are mainly transcription factors (13), the majority of new miRNAs are predicted to target mRNAs encoding important enzymes such as protein kinases, peroxidases and glyoxalases. This indicates that these miRNAs are widely involved in the regulation of diverse biological processes including signaling transduction and metabolic pathways. For example, miR812k-m and other miR812 family members are predicted to target a putative 1-aminocyclopropane-1-carboxylate (ACC) oxidase, which plays an important role in the regulation of ethylene production (43). Among the predicted targets of new miRNAs, some resemble targets of previously reported miRNAs. For example, one of the miR3979-3p targets is a putative NBS-LRR disease resistance protein, similar to one of the targets of miR167f (44), suggesting multiple families of miRNAs possibly regulate this target gene family. We also found that a unique subset of new miRNAs includes miR3980 and miR812n, of which the miRNA and its miRNA* have a common target. For these miRNAs, their precursors have relatively high levels of complementarity or similarity to target genes (data not shown), providing new evidence for the evolutionary origin of some miRNA loci through inverted duplication events from protein-coding gene sequences (22). We also noted that the target validation rate for novel miRNAs is rather low, as described in other studies for non-conserved miRNAs (22). This might be because the predicted target genes are nonexistent due to the imperfect annotation of the rice genome, the target genes are expressed at a very low level, the cleaved products of targets are very unstable, or there are other functional mechanisms such as translation inhibition reported recently (45).
A putative exonic miRNA found first in plants
Most known miRNA loci are located in intergenic regions of the genome, and are independently transcribed by RNA polymerase II (46). A few miRNAs are also located in the introns of protein-coding genes. For instance, miR1429.2 is reported to be a putative mitron in rice (26). However, it is very rare that the miRNA is located in the exon of a protein-coding transcript. One example is hsa-miR-935, whose hairpin is found in the last exon of the CACNG8 mRNA in human cervical cancer (47). In this study, a novel miRNA, miR3981, was found located in the last exon of the AK106348 mRNA encoding a putative glyoxalase (Figure 2A). This putative exonic miRNA is strongly supported by the evidence that most of the small RNAs in the MIR3981 locus mapped to the strand producing the mature miRNA and corresponded precisely to the miRNA and miRNA* (Figure 2B, Supplementary Figure S4). More interestingly, miR3981-5p and miR3981-3p are both predicted to target the AK106348 mRNA. By using 5′ RACE, the result showed that the cleavage site in the AK106348 transcript corresponded to miR3981-5p but not at position 10 (Figure 2C). Although some plant miRNAs were reported to direct self-cleavage of their pri-miRNAs (48), the cleavage of AK106348 might not be mediated predominantly by miR3981-5p because of the unexpected cleavage site and very low expression level of mi3981-5p (only 3 reads).
Figure 2.
(A) miR3981 is located in the last exon of AK106348. The hairpin represents the miR3981 stem-loop and the blue boxes indicate the exons. (B) Small RNAs are generated from the miR3981 precursor. miR3981-5p and miR3981-3p are shown in blue and red, respectively. Na, number of reads; Lb, length of sequences. (C) Mapping of the cleavage site in the AK106348 mRNA by RNA ligase-mediated 5′ RACE. The arrow indicates the cleavage site and the number shows the frequency of clones sequenced.
The presence of an exonic miRNA, miR3981, might suggests a new mechanism for posttranscriptional regulation of gene expression in plants. In animals, when the miRNA hairpin is located in the exonic region, Drosha processing can destabilize the transcript and reduces its protein synthesis (49). Likewise, the miR3981 biogenesis pathway might be involved in the regulation of glyoxalase expression (Supplementary Figure S5A). Excess AK106348 mRNA should cause an increase in DCL1 processing to produce more hairpins, and the cleaved hairpin (pre-miR3981) should then be further processed by DCL1 to produce the miR3981. Because AK106348 mRNA is also a potential target of miR3981, an increase in the miR3981 level might then lead to a decrease in the AK106348 mRNA level, avoiding the durable high expression of glyoxalase. Therefore, it is possible that competition or balance exists between glyoxalase protein output and miR3981 biogenesis, and the expression of glyoxalase is fine-tuned in rice. In this process, DCL1 might be the key regulatory factor, similar to Drosha in animals. First, miR3981 was expressed at a relatively low level, because it could only be demonstrated by sequencing data but not be detected by northern blots (data not shown). Second, the cleavage of AK106348 mRNA might predominantly result from the DCL1 processing. To confirm this hypothesis, we analyzed the transcriptional expression of AK106348 and DCL1 (Supplementary Figure S5B). AK106348 mRNA was upregulated in response to H2O2 stress, indicating that more glyoxalase was generated to participate in redox homeostasis, similar to our previous results on the protein level (4). In comparison, DCL1 was downregulated under H2O2 stress, and its expression was negatively correlated with AK106348 expression, consistent with its role as a negative regulator of AK106348. In addition to AK106348, other mRNAs encoding glyoxalase such as AK069814 also have similar hairpin like structures in their 3′ UTR regions. We confirmed that AK069814 mRNA was also cleaved in its hairpin (Supplementary Figure S5C). In plants, it can be assumed that glyoxalase-coding genes and other protein-coding genes with hairpin structures might be negatively regulated by DCL1 as well as by the miRNAs generated from their respective hairpins.
H2O2-responsive miRNAs in rice seedlings
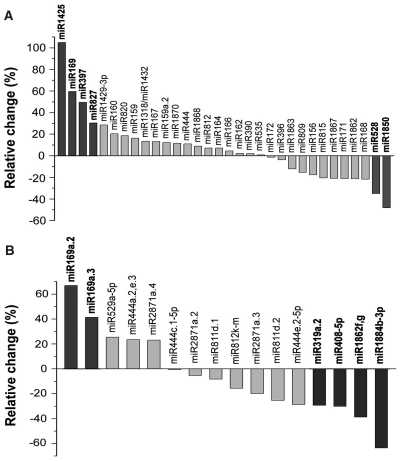
As reported earlier (22,26), a deep sequencing approach with good reproducibility can be used as a powerful tool for profiling miRNA expression. The change in the frequency of miRNAs between the H2O2-treated and control libraries might indicate that their expression is regulated in response to oxidative stress. To minimize noise and improve accuracy, we selected only the miRNAs with sequence reads over 100 in at least one library for comparison. As shown in Figure 3, comparison of the normalized sequence reads of the miRNAs between the two libraries indicated that six known rice miRNA families and six new miRNA sequences had relative changes greater than 30% and thus might be differentially expressed. Not surprisingly, miR398 sequence reads displayed no meaningful changes between two libraries (Supplementary Table S2) even though its expression had been reported to be affected by some oxidative stress treatments (21). The result indicated again that different oxidative stress conditions might evoke diverse plant responses and physiological adaption. In addition to miRNAs, the expression of ta-siRNAs in both libraries was also examined (Supplementary Table S5), but none of the functional ta-siRNAs had H2O2-regulated expression patterns based on the same comparison for miRNAs.
Figure 3.
Net increase or decrease of the normalized sequence reads of the known miRNA families (A) and the newly identified miRNAs (B) in the H2O2-treated library relative to the control library. Only the miRNAs with more than 100 sequence reads in at least one small RNA library were compared, and those with a relative change greater than 30% are highlighted in boldface.
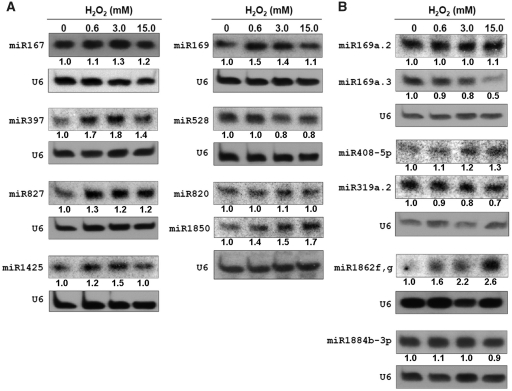
Northern blot analysis was performed to examine the differential expression of these miRNAs in rice seedlings exposed to different H2O2 concentrations (0, 0.6, 3.0 and 15.0 mM). The results revealed that five out of six known miRNA families had the expression pattern coincident with the sequencing data. As shown in Figure 4A, miR169, miR397, miR827, miR1425 were upregulated by at least two different concentrations of H2O2, while miR528 was downregulated under two different concentrations, both consistent with the relative change in sequence reads. The only exception was miR1850, whose expression in the northern blot analysis was upregulated by H2O2 exposure and was not in agreement with the estimation made by the sequencing data. In contrast, the expression of miR820 on the same blot showed an expression change similar to the sequencing data. Therefore, it is possible that the sequencing depth was not sufficient to reflect a genuine change in the two samples for miR1850 and other miRNAs with relatively low abundance (25). Moreover, we also confirmed the expression of miR167, whose expression pattern was slightly upregulated under H2O2 exposure both in the northern analysis and sequencing. Out of the six newly identified miRNAs, only miR319a.2 exhibited an expression pattern similar to the sequence frequency in the libraries (Figure 4B). Assessment of the sequencing data predicted that miR169a.2 and miR169a.3 would be upregulated, and miR408-5p, miR1862f,g and miR1884b-3p would be downregulated by H2O2. In reality, however, miR169a.2 and miR1884b-3p showed negligible expression differences under the H2O2 treatments, and miR169a.3, miR408-5p and miR1862f,g even displayed contrary changes in the northern blot analysis. The apparent discrepancy between the northern blot data and the sequencing data of the newly identified miRNAs might be due to the unreliability of the insufficient sequence reads, cloning preferences in high-throughput sequencing and/or a potential cross-hybridization problem in the northern blot analysis (21). Based on the above results, seven miRNAs (miR169, miR397, miR528, miR827, miR1425, miR319a.2 and miR408-5p) can be considered to be H2O2-responsive miRNAs because they are expressed differentially between H2O2-treated and control samples in northern blot analysis and their predicted targets all have specific function information (Table 2). Therefore, these H2O2-responsive miRNAs and their targets are worthwhile to be analyzed further.
Figure 4.
Northern blot analysis of the known miRNA families (A) and the newly identified miRNAs (B) in rice seedlings treated with different concentrations of H2O2 (0, 0.6, 3.0 and 15.0 mM) for 6 h, respectively. Except for miR167 and miR820, all of the selected miRNAs had a relative change greater than 30% in the normalized sequence reads between the two sequencing data sets. The numbers under the blots indicate the relative abundance of miRNAs, which were calculated by using U6 as the loading control.
Table 2.
H2O2-responsive miRNA familes and their targets
| miRNA family | SDa | NBb | Predicted targetc | Putative function of targets |
|---|---|---|---|---|
| miR169 | ↑ | ↑ | AK062135, AK069348, AK069854, AK071595, AK073742, AK101684 | HAP2 like transcription factor |
| miR397 | ↑ | ↑ | AK061816, AK065154, AK066513, AK068047, AK071929, AK105333, AK109431 | Laccase |
| miR528 | ↓ | ↓ | AK065478, AK070501, AK102279, AK105607 | F-box/LRR-repeat MAX2; IAA-alanine resistance protein 1 (IAR1) like |
| miR827 | ↑ | ↑ | AK066068 | SPX domain protein |
| miR1425 | ↑ | ↑ | OsIFCC031311, OsIFCC042756, OsIFCC044509 | Pentatricopeptide repeat (PPR) protein |
| miR319a.2 | ↓ | ↓ | OsIFCC006754 | Metacaspase |
| miR408-5p | ↓ | ↑ | AK069327, OsIFCC002332, OsIFCC002333 | Monosaccharide transport protein |
(↑) Upregulated and (↓) downregulated in response to H2O2 treatment.
aSD, in sequencing datasets.
bNB, in northern blots.
cValidated targets by RNA ligase-mediated 5′ RACE are shown in bold.
The targets of H2O2-responsive miRNAs
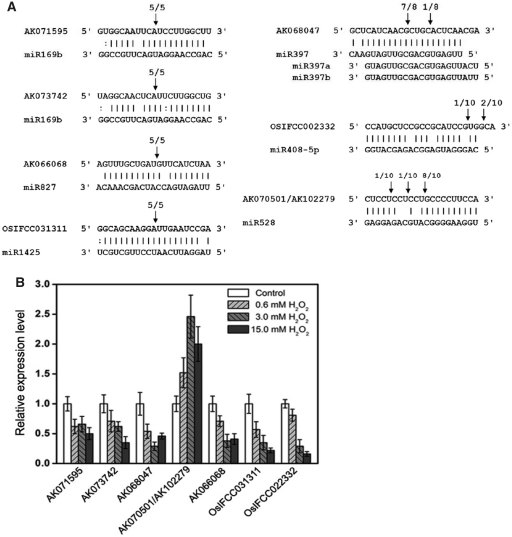
To understand the functions of H2O2-responsive miRNAs, the first step is to predict and experimentally validate their targets. The targets of seven H2O2-responsive miRNAs are listed in Table 2. Of these targets, miR169, miR397 and miR827 are highly conserved miRNA families and have been validated to target those mRNAs encoding HAP2 like transcription factors, laccases and SPX domain proteins, respectively, in Arabidopsis (13,22) and/or rice (39,41,44). In addition, miR1425 has been predicted and confirmed to cleave a class of mRNA targets that encode pentatricopeptide repeat protein (PPR) proteins (24,41). Importantly, the respective targets of miR169, miR397, miR827 and miR1425 were also confirmed for cleavage in vivo by 5′ RACE in this study (Figure 5A). According to the deep sequencing data, the 5′ end of the mature miR397 sequence is different from those of the annotated sequences in the miRbase (Supplementary Figure S3A), and this mature miR397 sequence is believed to play a primary role in rice because the predominant cleavage site of the miR397 target is located at its expected site (Figure 5A). For miR528, we found a new class of targets encoding putative IAA-alanine resistance protein 1 (IAR1), which contained a conserved ZIP domain and had a high identity (∼60%) to Arabidopsis IAR1 (50). miR319a.2 and miR408-5p have been newly identified in this study. miR408-5p is supported by 5′ RACE to target a monosaccharide transporter protein, although the supporting evidence is weak because the target was not cleaved at the canonical position. miR319a.2 is predicted to target a putative metacaspase, but the target confirmation was not successful, probably due to the very low expression of the target gene in rice seedlings (data not shown).
Figure 5.
(A) Mapping of the target mRNA cleavage sites by RNA ligase-mediated 5' RACE. The arrows indicate the cleavage sites and the numbers show the frequency of clones sequenced. (B) Quantitative real-time PCR analysis of the target mRNAs of H2O2-responsive miRNAs. The data represent the mean values ±SD of three replicates.
The expression of the validated targets of H2O2-responsive miRNAs was investigated by quantitative real-time PCR in response to H2O2 (Figure 5B). We found that there was a negative correlation, in most cases, in the expression profiles of the H2O2-responsive miRNAs and their validated targets, and this is consistent with miRNA function in guiding the cleavage of target mRNAs. For example, miR408-5p expression was upregulated with an increase in H2O2 concentration, and its target mRNA level was correspondingly downregulated. It should be noted that some miRNAs and their targets were both downregulated under 15.0 mM H2O2 (the highest concentration), which could be attributed to the cellular damage and nucleic acid degradation in rice seedlings that were exposed to high H2O2 concentration (1).
A putative functional network of H2O2-responsive miRNAs in rice seedlings
A precise balance of H2O2 homeostasis in the plants is necessary to avoid cellular injury and to maintain a suitable level of H2O2 for the perception and transmission of different developmental and environmental signals. However, no systematic investigation has been conducted to determine how plants maintain their survival and growth under H2O2 stress via miRNA-mediated regulation. In the present study, deep sequencing combined with northern blot analysis revealed seven H2O2-responsive miRNA families, and each of them has itself function-specific targets (Table 2, Figures 3 and 4). Furthermore, a connection of H2O2-responsive miRNAs and their targets was established convincingly through validation of target cleavage and by negative correlation of the miRNA and target expression profiles in rice seedlings (Figures 4 and 5). Our results indicate that H2O2 can downregulate or upregulate the expression of seven miRNA family genes, which ultimately direct their target genes to be expressed negatively in rice seedlings (Figure 6). Under H2O2 stress conditions, there is a triggered miRNA-mediated regulation network, which bridges H2O2-responsive miRNAs with their targets (Figure 6).
Figure 6.
A possible functional network of H2O2-responsive miRNAs in rice seedlings. The arrows indicate positive regulation and the nail shapes represents negative regulation. PPR: pentatricopeptide repeat protein; MTP: monosaccharide transport protein; SPX: SPX domain protein; IAR1: IAA-alanine resistance protein 1.
As illustrated in Figure 6, the first subgroup out of seven H2O2-responsive miRNAs consists of five H2O2-upregulated miRNAs including miR169, miR397, miR1425, miR408-5p and miR827. In rice, miR169 was reported to be induced by high salinity (17) and drought stress (39). The targets of miR169 include multiple HAP2-type transcription factor subunits, components of the CCAAT-box binding factor complex (CBF/NF-Y/HAP) (51). As a plant transcriptional regulator, HAP2 has been shown to play a vital role in cell differentiation (52), respiration (53) and drought resistance (54). Under H2O2 stress, the miR169-mediated downregulation of HAP2 mRNA would slow or weaken many processes of development and metabolism such as cell differentiation and respiration in rice seedlings. Interestingly, the respiration rate of rice seedlings declined markedly under H2O2 treatments (unpublished data), and this might have partially resulted from the suppression of HAP2 expression by oxidative stress. Previous studies have shown that the expression level of miR397 was very low in mature organs such as leaves and stems, while expression was high in undifferentiated embryogenic calli (55). The miR397 was also found to be responsive not only to H2O2 stress but also to diverse abiotic stresses including cold, dehydration, NaCl and ABA (16). The targets of miR397 have been validated to be laccases, which are encoded by multigene families in plants. Unlike those of fungal laccases (56), the roles of laccases in plants have never been clearly demonstrated and might be involved in lignin biosynthesis (57) and copper homeostasis (58). Therefore, the downregulation of laccases by miR397 might limit some nonessential biological processes to allow conservation of energy for defense purposes, and to save copper for the most essential functions under adverse stress conditions (59). Furthermore, miR1425 is a rice-specific miRNA that has evolved to suppress the PPR transcripts (25). The PPR genes form a large family that is particularly prevalent in higher plants and participates in many post-transcriptional processes such as splicing, editing, processing and translation (60). Most of the PPR proteins are targeted to organelles including the mitochondria and chloroplast, and they have been shown to play crucial roles in virtually all stages of organellar gene expression (61). Although miR1425 is only predicted to target a minor fraction of hundreds of rice PPR genes, it has potential to control organellar biogenesis to some extent. Induction of miR1425 expression caused the downregulation of PPR expression and this could slow organellar biogenesis and cell division, representing a specific adaptation of rice seedlings to oxidative stress conditions. miR827 and miR408-5p both target mRNAs encoding major facilitator superfamily (MFS) proteins, including an SPX domain protein (AK066805) and a monosaccharide transporter protein (OsIFCC022332). MFS proteins contain a large and diverse group of secondary transporters and facilitate the transport across cytoplasmic or internal membranes of a variety of substrates (62). SPX proteins in plants were found to be involved in the internal regulation of nutrition homeostasis. For example, Arabidopsis PHO1, an SPX domain protein, plays a role in loading inorganic phosphate into the xylem of roots (63). Thus, the upregulation of miR827 and miR408-5p is a response to oxidative stress and reduces the transport of nutrients such as monosaccharide and phosphate to limit the nutrient consumption of plants. Together, the miRNAs induced by H2O2 suppress the processes of growth and metabolism to help rice seedlings adapt to the oxidative stress.
In the second subgroup, there were two H2O2-downregulated miRNAs including miR319a.2 and miR528. The target of miR319a.2 was predicted to be a transcript encoding a putative metacaspase, which was reported to be involved in PCD in plants (64). Since oxidative stress could induce the PCD, the reduced expression of miR319a.2 under H2O2 stress might result in an increase in PCD, consistent with previous results that H2O2 is an inducer of cell apoptosis (65). The target of miR528 was first validated in this study to be mRNAs encoding putative IAR1 proteins that are highly similar to Arabidopsis IAR1. IAR1 proteins have sequence similarity and structurally resemble ZIP family metal transporters that are conserved between animals and plants (50), and play an important role in the control of cellular-free auxin (principally indole-3-acetic acid, IAA) levels, possibly by transporting zinc, copper or another inhibitory metal out of the ER and away from the IAA-amino acid conjugate hydrolases such as IAR3 and ILR1 (50). In plants, most IAA is found in conjugated forms, and is implicated in processes such as storage, transport and protection from oxidative degradation (66). Since IAA is sensitive to oxidation, the free IAA in plants might be readily damaged under oxidative stress. In our study, miR528 was downregulated by H2O2 stress, resulting in the increase of IAR1 mRNA abundance. The upregulation of IAR1 would in turn promote the hydrolysis of IAA-amino acid conjugates to sustain the free cellular IAA. Therefore, miR528 might be involved in the regulation of IAA homeostasis by controlling IAA release under H2O2 stress.
ACCESSION NUMBERS
The rice small RNA sequence data have been submitted to NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE23217. The novel miRNA sequences have been submitted to miRBase under accession numbers miR3979–miR3982.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
We thank the National Natural Science Foundation of China (grant 30870197); National Transgenic Animals and Plants Research Project (grant 2009ZX08009-069B); State Key Basic Research and Development Plan (grant 2006CB101706); Hi-Tech Research and Development Program of China (grant 2008AA02Z116) for research supports (to J.-Y.L.). Funding for open access charge: National Natural Science Foundation of China (30870197 to J.-Y.L.).
ACKNOWLEDGEMENTS
We are grateful to Dr Xuemei Chen of University of California (Riverside) for her critical reading and her invaluable suggestions.
REFERENCES
- 1.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 2.Mittler R, Vanderauwera S, Gollery M, Breusegem FV. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Vanderauwera S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al. A comprehensive analysis of hydrogen peroxide-regulated gene expression in tobacco. Proc. Natl Acad. Sci. USA. 2003;100:16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan XY, Liu JY. Comparative proteome analysis reveals an intimate protein network provoked by hydrogen peroxide stress in rice seedling leaves. Mol. Cell Proteomics. 2008;7:1469–1488. doi: 10.1074/mcp.M700488-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 9.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 12.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Vionnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 13.Jones-Rhoades MW, Bartel DP. Computational identification of plant miRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphatestarvation response in Arabidopsis. Curr. Biol. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;18:2051–2065. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y. Identification of drought induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007;354:585–590. doi: 10.1016/j.bbrc.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S, Sun YH, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang SQ, Peng J, Qiu CX, Yang ZM. Heavy metal-regulated new microRNAs from rice. J. Inorg. Biochem. 2009;103:282–287. doi: 10.1016/j.jinorgbio.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 21.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc. Natl Acad. Sci. USA. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue LJ, Zhang JJ, Xue HW. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2009;37:916–930. doi: 10.1093/nar/gkn998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao W, Wang J, He X, Huang X, Jiao Y, Dai M, Wei S, Fu J, Chen Y, Ren X, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang S, Buell CR. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner PJ, Daub J, Tate J, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- 35.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carington JC, Chen X, Green PJ, et al. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 37.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD. Real-time quantitative PCR. Methods. 2001;25:383–385. doi: 10.1006/meth.2001.1260. [DOI] [PubMed] [Google Scholar]
- 39.Zhao B, Ge L, Liang R, Li W, Ruan K, Lin H, Jin Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009;10:29. doi: 10.1186/1471-2199-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacombe S, Nagasaki H, Santi C, Duval D, Piegu B, Bangratz M, Breitler JC, Guiderdoni E, Brugidou C, Hirsch J, et al. Identification of precursor transcripts for 6 novel miRNAs expands the diversity on the genomic organisation and expression of miRNA genes in rice. BMC Plant Biol. 2008;8:123. doi: 10.1186/1471-2229-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 43.Vriezen WH, Hulzink R, Mariani C, Voesenek LACJ. 1-Aminocyclopropane-1-carboxylate oxidase activity limits ethylene biosynthesis in Rumex palustris during submergence. Plant Physiol. 1999;121:189–195. doi: 10.1104/pp.121.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, Chen SY, Zhou H, Qu LH, Chen YQ. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 2009;583:723–728. doi: 10.1016/j.febslet.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 45.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 48.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotech. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 49.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lasswell J, Rogg LE, Nelson DC, Rongey C, Bartel B. Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell. 2000;12:2395–2408. doi: 10.1105/tpc.12.12.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 52.Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinkham JL, Guarente L. Cloning and molecular analysis of the HAP2 Locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li WX, Oono Y, Zhu J, He XJ, Wu JM, Lida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo YC, Zhou H, Li Y, Chen JY, Yang JH, Chen YQ, Qu LH. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006;580:5111–5116. doi: 10.1016/j.febslet.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 56.Mayer AM, Christopher RC. Laccase: New functions for an old enzyme. Phytochemistry. 2002;60:551–556. doi: 10.1016/s0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- 57.Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, Goffner D. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 2002;129:145–155. doi: 10.1104/pp.010988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claus H. Laccases: structure, reactions, distribution. Micron. 2004;35:93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 59.Abdel-Ghany SE, Pilon M. MicroRNAs-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008;283:15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in posttranscriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 61.O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008;25:1120–1128. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 62.Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamburger D, Rezzonico E, Petétot JMC, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Rodriguez-Nieto S, Zhivotovsky B, Smertenko A. Cysteine protease mcll-Pa executes programmed cell death during plant embryogenesis. Proc. Natl Acad. Sci. USA. 2005;102:14463–14468. doi: 10.1073/pnas.0506948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner BA, Buettner GR, Oberley LW, Darby CJ, Burns CP. Myeloperoxidase is involved in H2O2-induced apoptosis of HL-60 human leukemia cells. J. Biol. Chem. 2000;275:22461–22469. doi: 10.1074/jbc.M001434200. [DOI] [PubMed] [Google Scholar]
- 66.Bartel B. Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.