Abstract
The production of mature export-competent transcripts is under the surveillance of quality control steps where aberrant mRNP molecules resulting from inappropriate or inefficient processing and packaging reactions are subject to exosome-mediated degradation. Previously, we have shown that the heterologous expression of bacterial Rho factor in yeast interferes in normal mRNP biogenesis leading to the production of full-length yet aberrant transcripts that are degraded by the nuclear exosome with ensuing growth defect. Here, we took advantage of this new tool to investigate the molecular mechanisms by which an integrated system recognizes aberrancies at each step of mRNP biogenesis and targets the defective molecules for destruction. We show that the targeting and degradation of Rho-induced aberrant transcripts is associated with a large increase of Nrd1 recruitment to the transcription complex via its CID and RRM domains and a concomitant enrichment of exosome component Rrp6 association. The targeting and degradation of the aberrant transcripts is suppressed by the overproduction of Pcf11 or its isolated CID domain, through a competition with Nrd1 for recruitment by the transcription complex. Altogether, our results support a model in which a stimulation of Nrd1 co-transcriptional recruitment coordinates the recognition and removal of aberrant transcripts by promoting the attachment of the nuclear mRNA degradation machinery.
INTRODUCTION
During transcription elongation in the eukaryotic nucleus, the nascent mRNA molecule is sequentially coated with a variety of processing and binding proteins that mediate its transformation into an export-competent ribonucleoprotein particle (mRNP) ready for translation in the cytoplasm (1,2). The co-transcriptional maturation and assembly of export-competent mRNPs is facilitated by the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNAP II) that serves as a platform for sequential recruitment of the various factors (3). The CTD is formed by a tandem repetition of a heptapeptide motif (Tyr1–Ser2–Pro3–Thr4–Ser5–Pro6–Ser7) where a dynamic phosphorylation and dephosphorylation of Ser5 and Ser2 over the course of gene transcription adjusts the sequential recruitment of acting factors (4–7).
The production of export-competent transcripts is also under the surveillance of quality control steps that are interconnected with transcription elongation and mRNP biogenesis. Aberrant mRNP molecules resulting from inappropriate or inefficient processing and packaging reactions are targeted by the surveillance mechanisms leading to their retention at the transcription sites where they are degraded by the 3′–5′ exonuclease activity of the nuclear exosome (8–11). Insights into this process have come from studies of the budding yeast Saccharomyces cerevisiae mutant strains with defects in mRNA 3′-end formation or mRNP assembly and export machineries (12–15). It was shown that deletion or mutation of some components of the THO/Sub2 complex, which loads onto the nascent transcript and connects transcription to export, leads to a decrease in steady-state levels of several mRNAs. The normal levels of mRNAs can be recovered by inactivation of the nucleus-specific exosome component Rrp6 or components of the exosome-activating complex TRAMP (16,17). This Rrp6-dependent loss of mRNAs is also observed in strains carrying mutations in the mRNA 3′-end processing factors Rna14 and Rna15 (14,18).
The molecular mechanisms by which an integrated system recognizes aberrancies at each step of mRNP biogenesis and targets the defective molecules for destruction are still largely unknown. However, several lines of evidence point to a model in which the surveillance apparatus is recruited directly to the transcription complex, a position from which it can scrutinize all mRNP processing and packaging reactions looking for faulty events. For instance, components of Drosophila nuclear exosome have been shown to accompany transcribing RNAP II upon recruitment by transcription elongation factors (19,20). In yeast, genome-wide analyses indicated that the localization of some nuclear exosome subunits correlates with actively transcribed genes (21). Also, co-immunoprecipitation experiments in yeast suggested that components of both the exosome and TRAMP interact physically with the transcription complex. This interaction seems to be mediated by Nrd1, a protein involved in transcription termination of a subset of RNAs (22).
Two main pathways of terminating RNAP II-dependent transcripts have been described in yeast. Termination of poly(A)-containing mRNAs relies on the co-transcriptional recruitment and assembly of a large cleavage and polyadenylation complex that recognizes the termination signal and triggers the cleavage of the nascent transcript with subsequent addition of a poly(A) tail at the 3′-end (23–27). The highly conserved factor, Pcf11, is a prominent component of this complex that interacts with RNA as well as RNAP II via a CTD-interacting domain (CID) (28–32). The second termination pathway, used by RNAP II in transcription of the non-coding snRNAs and snoRNAs, is directed by another CID-containing protein, Nrd1, which binds RNA in a sequence-specific manner (33–35). Nrd1 is recruited to the transcription complex in association with another sequence-specific RNA binding protein, Nab3. Recently, the Nrd1–Nab3 complex was also implicated in termination and fast decay of cryptic unstable transcripts (CUTs); short non-coding RNAs that were first detected by genome-wide microarray analyses in strains defective for nuclear RNA degradation (36–38). Current models suggest that the co-transcriptional loading of Nrd1–Nab3 complex mediates the recruitment of the exosome and TRAMP which in the case of snRNAs and snoRNAs couples termination to maturation by trimming the 3′-ends of precursor RNAs until the encounter of a block formed by stable RNA structures or RNA-bound Nrd1–Nab3 (22,34). It was also suggested that the absence of such structural features in the case of the short-lived CUTs leads to their complete degradation following termination (36,37).
Recent studies ascribed the choice between the two termination modes to the stage of CTD phosphorylation which is dependent on the distance traveled by RNAP II from the promoter. Termination of short transcripts (non-coding RNAs and CUTs) appears to be associated with Ser5 phosphorylation and preferential recruitment of Nrd1 (39,40), whereas termination of longer transcripts relies on the subsequent phosphorylation at Ser2 sites that mediates recruitment of Pcf11 (23,41). However, chromatin immunoprecipitation (ChIP) experiments show clearly that Nrd1 cross-linking is not limited to the early transcribed region of a gene. Nrd1 recruitment within the body of the gene and at its 3′-end is only slightly decreased as compared to the 5′ part (23,40). Furthermore, a recent reevaluation indicates that the level of Ser5 phosphorylation, necessary for Nrd1 recruitment, persists throughout elongation (42,43). These results suggest that Nrd1 could play a pivotal role in the mRNP surveillance mechanism as a part of a protein network that travels with the transcription complex along the transcription unit.
Previously, we have implemented an assay in which mRNP biogenesis in yeast is disturbed by the heterologous expression of the bacterial transcription termination factor Rho. The expression of this RNA-dependent helicase/translocase induces the production of full-length yet aberrant transcripts that are targeted by the surveillance mechanism leading to their destruction by the nuclear exosome (44). Here, we took advantage of this new assay to explore the involvement of Nrd1 in the nuclear quality control system. We show that Nrd1 recruitment by the transcription complex via its CID and RRM domains is required for the targeting and degradation of the Rho-induced defective transcripts. Remarkably, overexpression of Pcf11 or its isolated CID domain suppresses the targeting and degradation of the Rho-induced aberrant transcripts through a competition with Nrd1 for recruitment by the transcription complex. Altogether, our results provide genetic and molecular evidence indicating that the recognition and targeting of aberrant transcripts is mediated by a large increase in Nrd1 recruitment to the transcription complex which helps to tether the nuclear mRNA degradation machinery.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains used in this study are listed in Supplementary Table S1. Similarly, all the plasmids are listed in Supplementary Table S2. The various overexpression plasmids with N- and C-terminal truncations in the PCF11 and NRD1 genes were constructed by amplification of genomic DNA with the gateway primers described in Supplementary Table S3. The M1, M2 and M3 mutations in pcf11-(1–145) were introduced by overlapping PCR mutagenesis with the primers described in Supplementary Table S3. The deletion of the Nrd1-RRM domain was done by PCR with two sets of partially overlapping primers designed to amplify the DNA sequence corresponding either to the first 339 amino acids or the last 165 amino acids of the Nrd1 protein. The two PCR products were re-amplified with the 5′- and 3′-end ‘NRD1’ gateway primers described in Supplementary Table S3 to create the ΔRRM plasmid.
RNA extraction and northern blotting
Yeast cells transformed with the appropriate plasmids were grown at 25°C to an OD600 of 0.6, then total RNAs were extracted by the hot phenol method (45). Northern blot and hybridization were described previously (44). Formaldehyde-containing 1.2% agarose gel was used to analyze PMA1 and PGK1 mRNAs and the 5.8S or 18S rRNAs which were detected sequentially using either a PMA1 random-primed probe or 5′-end labeled probes (see Supplementary Table S4). Hybridization signals were quantified with a Storm 860 phosphorimager (Molecular Dynamics) and the data were processed with Imagequant software version 2005.
Chromatin immunoprecipitation
Preparations of chromatin were performed essentially as described by (46). The NRD1-TAP (DLY1124) or RRP6-TAP (DLY269) strains were co-transformed with the empty vector (pCM185) or the Rho expression plasmid (pCM185-Rho) and either the pAG425GPD vector, the Nrd1 (1–448) or the Pcf11 (1–300) constructs. The ‘no TAG’ control strain is W303 transformed with pAG425GPD vector and either the pCM185 or the pCM185-Rho plasmids. Two hundred millilitre of cells grown for ∼20 h without doxycycline (OD600 ∼ 0.6) were fixed with 1% formaldehyde for 20 min. After glycine addition to stop the reaction, the cells were washed and lysed with glass beads to isolate chromatin. The cross-linked chromatin was sheared by sonication to reduce average fragment size to ∼500 bp. Immunoprecipitation of 800 µl of chromatin fractions was performed with 37 µl of IgG sepharose (GE healthcare) overnight at 4°C. After washing beads and chromatin elution, the eluted supernatants and the input controls were incubated with pronase (Sigma, 0.8 mg/ml final concentration) for 1 h at 37°C followed by 5 h at 65°C to reverse cross-linked DNA complexes. DNA was extracted using the Qiagen PCR-clean up columns. The immunoprecipitated DNAs (output) were quantitated by real time PCR (LightCycler 480 from Roche with the products and instructions recommended by the supplier) using three sets of primers located along the PMA1 or PGK1 coding sequence (described in Supplementary Table S5) and normalized to a 1/200 dilution of input DNA. Amplifications were done in duplicate for each sample, averages and standard deviations were calculated based on three independent experiments.
RESULTS
Bacterial Rho factor induces the production of aberrant transcripts in yeast
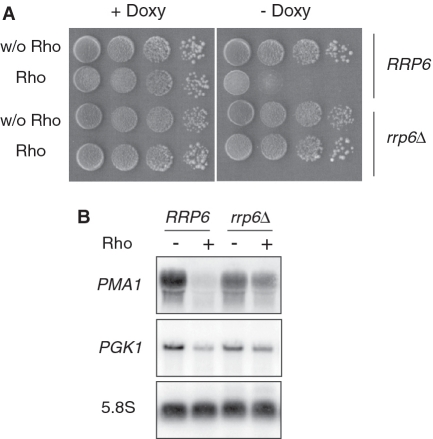
In a recent study (44), we reported that the conditional expression of the bacterial Rho factor in the yeast nucleus leads to a dose-dependent growth defect phenotype that stems from its interference in mRNP biogenesis. Rho helicase/translocase action induces the production of full-length but aberrant transcripts that are targeted and degraded by the nuclear exosome. This effect is illustrated in Figure 1 where the growth of yeast cells expressing Rho under the control of a Doxy-regulated TetO7 promoter within a centromeric plasmid is severely reduced under inducing conditions (-Doxy). Northern blot analysis of the steady-state abundance of a representative mRNA (PMA1) shows almost complete disappearance of the transcript under Rho expression conditions (Figure 1B). However, in a strain lacking the nuclear exosome component Rrp6, the Rho-induced growth defect is totally relieved (Figure 1A) and the steady-state abundance of PMA1 mRNA is now only slightly affected by Rho expression (Figure 1B), indicating that Rho action leads to the production of full length yet defective mRNPs that had been eliminated in the presence of active nuclear RNA degradation machinery. This conclusion was supported also by examination of the PMA1 mRNA 3′-ends following RNase H cleavage (44). The rescue of PMA1 mRNA was also obtained upon depletion of two components of the TRAMP complex, Air1 and Air2 [results not shown and (44)]. A similar trend was observed when the steady-state level of another mRNA (PGK1) was monitored (Figure 1B). Note, however, that the effect of Rho on PGK1 expression is less pronounced than for PMA1, as reported previously (44). Presently, the raison for this difference is still unclear, but it could be linked to the efficiency by which Rho loads onto the nascent transcripts.
Figure 1.
Depletion of the exosome component Rrp6 alleviates Rho-induced growth defect and restores the steady-state levels of mRNAs. (A) The rrp6Δ mutant and the corresponding wild-type isogenic strain (RRP6) were transformed with the empty vector (pCM185) or the Rho-expression construct harboring the Rho gene under the control of a Doxy-regulated TetO7 promoter (pCM185-Rho). Ten-fold serial dilutions of transformants grown selectively in liquid medium at 25°C with 1 µg/ml Doxy (non-induced conditions) were spotted on Rho-repressing (+Doxy) or Rho-inducing (−Doxy) plates. The plates were photographed after 3 days at 25°C. (B) Northern blot analyses of total RNAs extracted from cultures of cells transformed with the Rho-expression plasmid and grown with Doxy at a concentration of 1 µg/ml (−Rho) or 0.02 µg/ml (+Rho). Samples of total RNAs (12 µg) were fractionated on agarose gel, transferred to a membrane and the steady-state levels of specific RNAs were detected sequentially using a PMA1 random-primed probe or 5′-end labeled probes against PGK1 mRNA and 5.8S rRNA which serves as loading control (see Supplemental Table S4).
Our previous genetic and molecular analyses provided indirect evidence implying that the loading of Rho onto nascent transcripts and its subsequent translocation along the RNA chains counteract the deposition of the endogenous processing and packaging factors. This interference causes the production of mRNPs that are improperly processed or depleted of some crucial proteins and which will be recognized as export-incompetent and eliminated by the exosome-mediated nuclear quality control system. This conclusion led us to screen a yeast genomic library searching for dosage suppressors of the Rho-induced growth defect, with the idea that the overproduction of a protein partner important for mRNP integrity could shift its competition with Rho and thus rescue the transcript from degradation with the ensuing relief of the growth defect. The rational of this Rho-based screen was validated by the identification of numerous dosage-suppressor genes that encode proteins involved in mRNA 3′-end formation or mRNP assembly and export (44).
The suppression of Rho-induced defect by Pcf11 overproduction is mediated by its CID
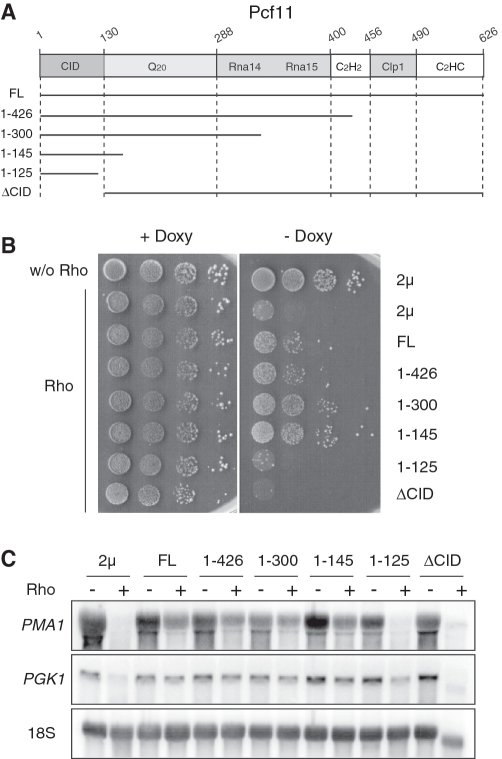
Among the dosage suppressors found in the Rho-based screen, Pcf11 is a prominent component of the cleavage factor CF1A that is recruited to the nascent transcript through the interaction of its well characterized CID with the CTD of RNAP II (28,29,31). To gain more insights into the mechanism by which the overproduction of Pcf11 suppresses the Rho-induced defect, we constructed a set of deletion mutants based on the known domain organization of the protein [Figure 2A and (29,31)]. The constructs expressed from a 2 µ vector were transformed into yeast cells harboring the Doxy-regulated Rho expression system and yeast growth was monitored by serial dilutions on agar plates. Remarkably, the suppressing effect of the Pcf11 variants increased significantly as the C-terminal part of the protein was gradually trimmed (Figure 2B). The best suppressing phenotype was obtained with a protein fragment containing the 145 N-terminal amino acids corresponding to the CID domain of Pcf11. A construct with a further truncation into the CID domain up to the last 125 N-terminal amino acids failed to suppress the Rho-induced growth defect. A similar failure to suppress the Rho-induced defect was also observed with the deletion variant in which only the CID domain was removed from Pcf11 (ΔCID). The lack of suppression is not due to instability of the overproduced protein fragments as was verified by Western blot analyses (see representative examples in Supplementary Figure S1). These growth results were substantiated by northern blot analyses showing a correlation between a partial rescue of the steady-state abundance of PMA1 mRNA and the growth defect suppression (Figure 2C). A similar trend was also observed when the steady-state level of another mRNA (PGK1) was monitored (Figure 2C). Taken together, these results indicate that Pcf11 is a dosage suppressor of the Rho-induced defect owing to the presence of its CID domain which is sufficient on its own to abrogate the defect. Moreover, the fact that the CID fragment is a better suppressor than full-length Pcf11 suggests that the dosage-suppression process is mediated by saturable physical interaction with the CTD. In effect, this interaction has been shown by two-hybrid experiments to be stronger when the CID is in isolation (31).
Figure 2.
Overexpression of the CID of Pcf11 is sufficient to suppress Rho-induced defect. (A) Schematic representation of the structural motifs of Pcf11. This diagram indicates the CID, the Q-rich segment (Q20), the Rna14/Rna15 and Clp1 interaction domains, and the two zinc-finger motifs. The numbers correspond to the amino acid sequence of Pcf11 and the black horizontal lines represent the protein fragments that were expressed from the 2 µ plasmid. (B) The two control strains were done by co-transformation of the 2 µ plasmid with either the empty vector (w/o Rho) or the plasmid expressing Rho into the BMA41 wild-type strain. BMA41 harboring the Rho expression vector was co-transformed with a 2 µ plasmid overexpressing the full-length Pcf11 (FL) or the various deletion variants represented in A. Strains grown selectively under repressing conditions were spotted for growth as described in Figure 1A. (C) Northern blot analyses were made as in Figure 1B and the membrane was hybridized sequentially with a PMA1 random-primed probe or 5′-end labeled oligonucleotides for PGK1 mRNA and 18S rRNA which serves as loading control.
Both CTD and RNA contacts within the CID of Pcf11 are required for dosage suppression of Rho-induced defect
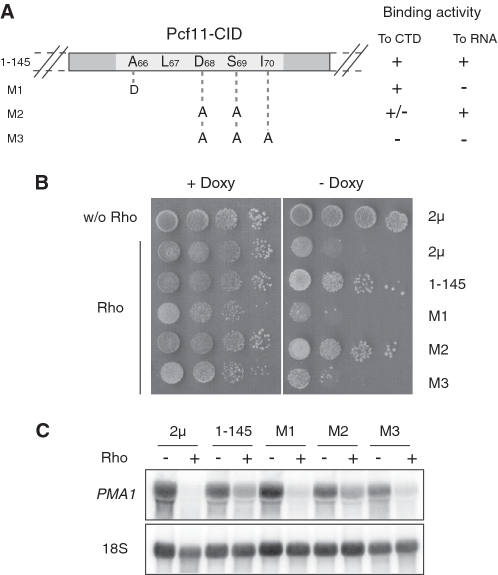
The ability of the isolated CID of Pcf11 to suppress the Rho-induced defect on its own led us to focus our investigations on this protein fragment (constructs 1–145). Amino acid mutations within a highly conserved region of the CID (between positions 66 and 70; Figure 3A) that impair correct transcript termination by Pcf11 have been characterized in vivo (31). These mutations were also studied in vitro in isolated CID to define their relative contributions to the interactions with the CTD and RNA (32). The CID-M3 mutant which has three consecutive amino acids substituted by alanines (D68A, S69A and I70A) lacks both CTD and RNA binding activities. The CID-M2 variant (D68A, S69A) has a slightly reduced CTD binding ability but retains full RNA binding activity. The third mutant, CID-M1 (A66D), has a full ability to interact with CTD but lacks RNA binding capacity. The constructs overproducing the CID variants were transformed into yeast cells harboring the Rho expression system and growth was monitored by serial dilutions on agar plates. As shown in Figure 3B, both M1 and M3 mutants have lost the ability to suppress the Rho-induced growth defect, whereas the M2 variant has a similar suppressing phenotype than the wild-type CID. The northern blot analyses are consistent with the growth results with a significant restoration of PMA1 mRNA abundance in the case of M2 overproduction as compared to M1 and M3 (Figure 3C). These results indicate that the relief of Rho-induced defect by overexpression of the CID domain of Pcf11 requires both CTD and RNA binding activities of the protein fragment. However, the lack of RNA binding activity seems to be critical for the suppressing process.
Figure 3.
The CTD and RNA binding activities of Pcf11-CID are required for dosage suppression of the Rho-induced defect. (A) Schematic representation of the amino acid substitutions introduced within the CID protein fragment (1–145) of Pcf11. The CTD and RNA binding abilities of the protein variants as described in ref. (32) are indicated on the right side. (B) BMA41 strains were co-transformed with pCM185-Rho and one of the 2µ plasmids overproducing the Pcf11-CID domain variants. Growth tests were carried out as described in Figure 1A. (C) Northern blots were performed as in Figure 2C.
The Rho-induced defect is aggravated in yeast cells with defective Pcf11
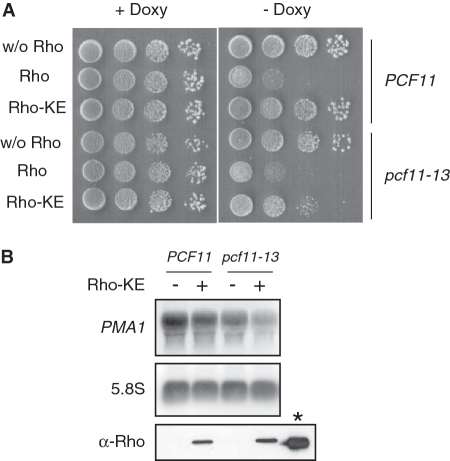
The results described above suggest that the suppression of Rho-induced defect by overproduction of Pcf11 or its CID domain could be mediated by a competition with a factor recruited to the transcription complex that orchestrates the targeting of aberrant transcripts with the ensuing growth defect. If such competition is already taking place under physiological expression of Pcf11, then a deficiency in its CID that impairs its CTD and RNA binding activities should exacerbate the Rho-induced defect. To evaluate this assumption, we tested the sensitivity to Rho expression of the yeast mutant strain pcf11–13 which has a defect in the CID of Pcf11 corresponding to the M3 substitutions described above (31). The sensitivity to the bacterial factor was monitored by expressing both the wild-type Rho and the Rho-KE variant which has a point mutation that reduces its helicase/translocase activity producing a mild growth defect phenotype in yeast (44). This intermediate effect of Rho-KE in yeast is particularly convenient to monitor synergistic sensitivity effects. As shown in Figure 4A, a similar growth inhibition was observed for PCF11 and pcf11–13 cells upon expression of wild-type Rho. In contrast, the expression of Rho-KE, which had virtually no effect on the growth of wild-type PCF11 cells under the present conditions, induced a significant growth defect for cells harboring the pcf11–13 allele. This different sensitivity of the two isogenic strains to Rho-KE expression was corroborated by northern blot analysis of PMA1 mRNA where the steady-state level was slightly affected by Rho-KE expression in pcf11–13 cells as compared to the wild-type strain (Figure 4B). Western blot analysis indicates that Rho-KE protein is expressed at a similar level in both strains. We also tested the mutant allele pcf11-2 which is defective in transcript cleavage and polyadenylation and in which the mutations are located exclusively outside the CID with no effect on CTD binding (28,31). In this case, the sensitivity to Rho and Rho-KE expression was similar to the wild-type strain (results not shown). These results support the idea that the recruitment of a factor that mediates the targeting and degradation of Rho-induced aberrant transcripts is favored in the presence of Pcf11 having a defective CID.
Figure 4.
A Pcf11 mutant strain with a defective CID due to impaired CTD and RNA binding activities is Rho hypersensitive. (A) The pcf11–13 mutant (DLY915) and the wild-type isogenic strain (DLY913) were transformed with an empty vector (pCM189) or a plasmid expressing Rho (pCM189-Rho) or the Rho-KE variant (pCM189-Rho-KE). Growth tests were carried out as described in Figure 1A. (B) Northern blot analyses of total RNAs extracted from the transformed strains expressing (+) or not (−) the Rho-KE protein variant were performed as described in Figure 1B. The lower panel (α-Rho) shows the western blot analysis of Rho-KE expression in wild-type and pcf11–13 mutant strains with the purified bacterial Rho protein (shown at the right side with an asterisk) as a Marker. The proteins were revealed using custom made anti-Rho polyclonal antibodies (Eurogentec, Belgium).
The Rho-induced defect is suppressed in yeast cells with defective Nrd1 or Nab3 as well as a shortened CTD
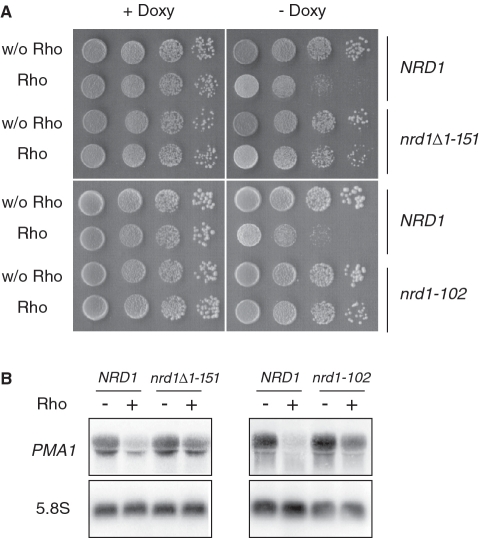
Nrd1 is known to interact both with the CTD through its CID domain which is homologous to that of Pcf11 (40) and with RNA by recognizing sequence elements within the nascent transcript in association with Nab3 (33,47). The Nrd1 complex was also shown to interact with the exosome and TRAMP (22). These properties led us to suspect Nrd1 as the possible factor that mediates the targeting and degradation of Rho-induced aberrant transcripts upon its recruitment to the transcription complex. This hypothesis was also supported by the fact that, in contrast to Pcf11, Nrd1 was not found as a dosage suppressor in our Rho-based screen and did not suppress the Rho-induced defect when tested directly following its cloning and overexpression (see the experiment described in Figure 7). As a first step to examine our prediction, we tested the sensitivity to Rho expression of a yeast strain harboring a CID deletion allele of NRD1 (nrd1Δ1–151). Previously, the Nrd1ΔCID protein was reported to have low affinity for binding RNAP II in cell extracts and its recruitment to two mRNA genes (including PMA1) was shown by ChIP experiments to be significantly reduced (40). The nrd1Δ1–151 strain was transformed with the Rho expression plasmid and the growth was monitored in parallel with an isogenic wild-type strain as a control. The transformant spotted on agar plates recovered a full capacity to grow under Rho-inducing conditions (Figure 5A). In agreement with the growth results, the steady-state level of PMA1 mRNA in the presence of Rho was rescued significantly in the nrd1Δ1–151 mutant strain (Figure 5B), indicating that Nrd1 recruitment to the transcription complex is required for the targeting and degradation of aberrant transcripts generated by Rho action. This assertion was further verified by analyzing the sensitivity to Rho expression of the strain nrd1–102 which has a single-point mutation in the RRM (RNA recognition motif) of Nrd1 leading to a deficiency in snoRNA termination as a result of a reduced RNA binding activity (47). Consistently, the Rho-induced growth defect was readily suppressed by the nrd1–102 mutation with the accompanying rescue of the PMA1 steady-state mRNA level (Figure 5). These results suggest that both CTD and RNA biding activities of Nrd1 are required to mediate the targeting and degradation of Rho-induced aberrant transcripts.
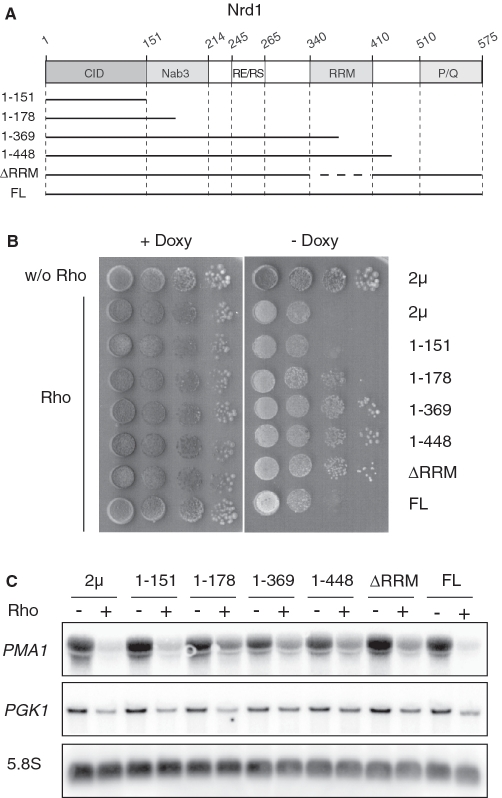
Figure 7.
Rho-induced defect is alleviated by overexpression of truncated forms of Nrd1. (A) Schematic representation of the domain organization of Nrd1. This diagram indicates the CID, the Nab3 interaction domain, the arginine–glutamine and arginine–serine rich region (RE/RS), the RNA recognition motif (RRM) and the proline- and glutamine-rich region (P/Q), respectively. The numbers correspond to the amino acid sequence of Nrd1 and the horizontal lines represent the protein regions which were expressed from the 2 µ vector. (B) The control strains were done as in Figure 2. The other strains were co-transformed with the Rho-expression vector and one of the 2 µ plasmids overexpressing the full-length Nrd1 (FL) or a truncated variant as indicated in (A). Growth tests were carried out as described in Figure 1A. (C) Northern blot analyses were made as in Figure 1B.
Figure 5.
Nrd1 mutant strains with deletion of the CID or mutation in the RRM are less sensitive to Rho. (A) The two Nrd1 mutant alleles, nrd1Δ1–151 (YNH026) and nrd1–102 (DLY1146), and the corresponding wild-type isogenic strains (NHY001 and DLY1144, respectively) were transformed with the empty vector or the plasmid expressing Rho. Cell cultures were spotted for growth tests as described in Figure 1A. (B) Northern blot analyses were performed as in Figure 1B.
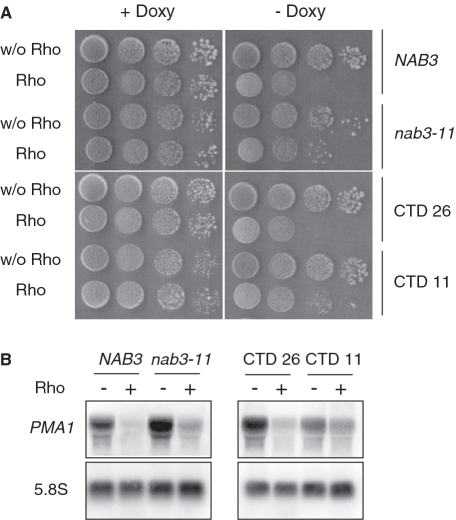
To test further the involvement of the Nrd1 complex in the targeting and degradation of Rho-induced aberrant transcripts, we examined the effect of alterations in two important partners (Nab3 and CTD) that contribute to Nrd1 recruitment. Nab3 forms a heterodimer with Nrd1 and binds to a specific RNA motif via its RRM domain (48). The effect of Rho expression was tested in the nab3–11 allele which has two point-mutations within the RRM region (47). As shown in Figure 6, a slight but significant relief of Rho-induced growth defect as well as a corresponding rescue of PMA1 mRNA was observed for the cells harboring the nab3–11 allele as compared to the isogenic wild-type strain. The growth defect suppression and rescue of PMA1 mRNA were even more pronounced in a yeast strain in which the CTD of RNAP II was truncated from 26 (wild-type) to 11 heptapeptide repeats (Figure 6), emphasizing the importance of the CTD as a platform for Nrd1 recruitment. Altogether, these results indicate that alterations in the different factors known to contribute to the co-transcriptional recruitment of Nrd1 counteract the targeting and degradation of Rho-induced aberrant transcripts, implying that the nuclear mRNA surveillance mechanism relies on efficient association of Nrd1 with the transcription machinery.
Figure 6.
Cells with a defective Nab3 protein or a truncated CTD become less sensitive to Rho expression. (A) The nab3–11 mutant (YPN102), the CTD 11 mutant (rpo21-Δ104) which lays 11 repetitions of the heptapeptide and the corresponding W303 wild-type isogenic strains (NAB3 and CTD 26, respectively) were transformed with the empty vector or the plasmid expressing Rho. Growth tests were carried out as described in Figure 1A. (B) Northern blot and hybridization were performed as in Figure 1B.
The Rho-induced defect is suppressed by overproduction of Nrd1 deletion mutants but not the full-length protein
If the mRNA surveillance mechanism relies on Nrd1 recruitment to the transcription machinery, then the overproduction of a mutant variant of the protein, that is functionally defective but still able to be recruited, should exert a dominant-negative effect by outcompeting the functional Nrd1 leading to a suppression of the Rho-induced defect. To test this hypothesis, we constructed a set of deletion mutants of Nrd1 based on the known domain organization [Figure 7A and (47)]. The constructs made within a high copy number 2µ plasmid were transformed into yeast cells harboring the Rho expression system. As shown in Figure 7B, the overproduction of a protein fragment containing the 151 N-terminal amino acids of Nrd1 corresponding to its CID domain (constructs 1–151) was unable to suppress the Rho-induced defect, indicating that additional contacts are probably needed for efficient competitive recruitment by the transcription complex. The possibility that the absence of suppression is due to instability of the overproduced protein was ruled out by western blot analysis (see representative examples in Supplementary Figure S1). The Rho-induced defect was partially relieved with a longer protein fragment that includes half of the Nab3-interacting region (constructs 1–178). The protein fragment encompassing the CID, the Nab3-interacting domain and a small portion of the RRM (constructs 1–369) was able to fully suppress the Rho-induced growth defect. A similar suppressing phenotype was observed with the deletion mutant containing the whole RRM region (constructs 1–448). These results suggest that Nab3 association with the mutant Nrd1 increases its competing efficiency, whereas the presence of the RRM region appears to be dispensable for an efficient competition. Consistent with this, the Nrd1 variant lacking only the RRM region (construct ΔRRM) was able to compete with functional Nrd1 leading to robust suppression of the Rho-induced defect. As expected, the overexpression of Nrd1 protein lacking the CID domain (Nrd1Δ1–151) failed to suppress the Rho-induced defect, reflecting the importance of CTD interactions for the competition process (results not shown). Similarly, the overproduction of the full-length Nrd1 protein did not show any suppressing phenotype. Indeed, in this case, the overexpressed plasmid-borne protein outcompetes the endogenous Nrd1 but it is functionally competent to mediate the targeting and degradation of Rho-induced aberrant transcripts leading to the growth defect. Northern blot measurements of the steady-state abundance of PMA1 mRNA and PGK1 mRNA (Figure 7C) corroborated the growth results. We conclude that Nrd1 recruitment to the transcription machinery coordinates the recognition and removal of the Rho-induced aberrant transcripts, probably by promoting the tethering of the exosome-mediated degradation apparatus (see below).
Rho expression stimulates the co-transcriptional recruitment of Nrd1 and Rrp6
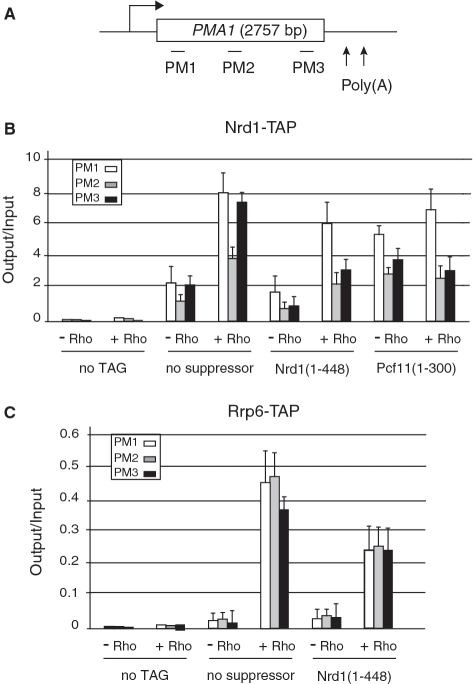
Previous chromatin immunoprecipitation (ChIP) experiments have shown Nrd1 cross-linking across the PMA1 gene with a slight enrichment toward the 5′ and 3′ regions of the locus (23,40,43). We wanted to see whether Rho action would lead to an increase of Nrd1 recruitment or a change in its cross-linking profile. We therefore analyzed the distribution of TAP-tagged Nrd1 along the PMA1 gene under Rho inducing or repressing conditions and in the absence or presence of competing suppressors. The Nrd1 occupancy was assessed by quantitative PCR (qPCR) using three primer-pairs that cover the 5′, the middle and the 3′ regions of the gene (Figure 8A). As shown in Figure 8B, the ChIP signals in the absence of Rho are consistent with previous reports (23,40,43) with occupancy slightly higher in the 5′ and 3′ regions as compared to the body of the gene. Remarkably, the expression of Rho leads to a 4-fold increase of Nrd1 recruitment across the entire locus with a similar enrichment toward both ends. Moreover, this Rho-induced stimulation of Nrd1 recruitment to the PMA1 gene is attenuated significantly by overexpression of competing suppressors such as the Nrd1 (1–448) or Pcf11 (1–300) protein fragments (Figure 8B), reflecting the competition for interaction with the transcription complex. Note, however, that the attenuation of Rho-induced Nrd1 recruitment by the competing suppressors is less pronounced for the 5′ region as compared to the 3′ region or the body of the gene, which could be linked to the stage of CTD phosphorylation and/or a more inherently robust recruitment of Nrd1 within the promoter-proximal region. Additionally, the basal level of Nrd1 occupancy in the absence of Rho is higher when the Pcf11 suppressor-derivative is expressed (for both PMA1 and PGK1 loci) which leads to a less efficient reduction of the effect of Rho on Nrd1 recruitment. The reason for this behavior is presently unclear. ChIP experiments conducted in parallel revealed that Rho expression induces a 15% decrease of RNAP II cross-linking along the PMA1 locus (results not shown). This is presumably due to a low level of premature Rho-dependent termination within promoter-proximal region, which is consistent with the fact that a fraction of full length transcripts is still missing upon inactivation of the degradation machinery [rrp6Δ in Figure 1 and (44)]. A similar trend was observed when Nrd1 recruitment across the PGK1 gene was monitored (Supplementary Figure S2). Again, these results provide a strong support for the involvement of Nrd1 in the targeting of Rho-induced aberrant transcripts through its association with the transcription machinery.
Figure 8.
Rho action stimulates Nrd1 and Rrp6 recruitment across the PMA1 gene. (A) Schematic representation of the PMA1 gene with the horizontal bars denoting the positions of the PCR products used in the ChIP analyses. (B) Quantifications of the fold enrichment for PMA1 DNA in Nrd1-TAP immunoprecipitates from cells expressing Rho (+Rho) or not (−Rho) and co-transformed with either the empty 2 µ vector for the assays without suppressor or the plasmids overexpressing the suppressors Nrd1 (1–448) or Pcf11 (1–300). The ‘no TAG’ control strain is W303 transformed with the empty 2 µ vector and either the (−Rho) or the (+Rho) plasmids. Immunoprecipitated samples (output) were normalized to input. The average of three independent experiments is shown with error bars representing standard deviations. (C) Quantifications of the fold enrichment for PMA1 DNA in Rrp6-TAP immunoprecipitates carried out as in (B).
We next asked whether the anchoring of Nrd1 to the transcription machinery promotes the association of the exosome-mediated degradation apparatus. We, therefore, performed ChIP experiments with a strain harboring TAP-tagged nuclear-specific exosome component Rrp6. As shown in Figure 8C, a large increase of ChIP signals across the PMA1 locus is observed under Rho-inducing conditions and this stimulation of Rrp6 recruitment is also readily reversed by overexpression of a competing-suppressor such as Nrd1 (1–448) protein fragment. Clearly, these results highlight a direct link between Nrd1 and Rrp6 recruitments to the transcription machinery upon Rho action. Interestingly, parallel experiments using TAP-tagged core exosomal components Rrp4 and Rrp41 did not show any significant ChIP signals both in the presence and the absence of Rho (results not shown), suggesting either a loose interaction between the anchored Rrp6 and the exosome or the possibility that Rrp6 may act independently from core exosome to target and degrade aberrant transcripts (see ‘Discussion’ section).
DISCUSSION
Eukaryotic cells have evolved an mRNP quality control system by which aberrant transcripts with processing and assembly defects are recognized as export-incompetent triggering their degradation by the nuclear exosome. Previous studies have suggested the interconnection of this surveillance process with transcription elongation and mRNP biogenesis. However, the molecular mechanisms by which a surveillance apparatus recognizes aberrancies at each step of mRNP formation and targets the defective molecules for destruction remained unknown. To address this question, we took advantage of a new assay in which the heterologous expression of a bacterial RNA-dependent helicase/translocase (transcription termination factor Rho) in yeast induces the production of full-length yet aberrant transcripts that are targeted and degraded by the nuclear exosome, causing a growth defect phenotype (44). In vitro, Rho functions as a powerful molecular motor that tracks along the RNA chain and can melt nucleic acid base pairs or disrupt RNA–protein complexes present on its path (49–51). We have postulated that Rho loading and subsequent translocation along nascent transcripts in yeast cells could antagonize the normal deposition of processing and packaging factors, yielding stripped mRNPs that are recognized as defective. This assumption was supported by the identification of several mRNP processing and packaging factors in a screen for dosage suppressors of the Rho-induced defect, suggesting that an overload of these RNA-interacting proteins can counteract the denuding effect of the bacterial helicase or neutralize the detection of the aberrant transcripts by the surveillance mechanisms. Additional support to our assumption is provided by a work to be published elsewhere in which decreased occupancy of some mRNP assembly proteins upon Rho action was revealed by ChIP experiments (R. H., C. M., N. C. and A. R. R., manuscript in preparation).
Among the dosage suppressors found in the Rho-based screen, Pcf11 is an essential factor for transcription termination of poly(A)-containing RNAs which is known to interact with the nascent transcript upon co-transcriptional recruitment by the CTD of RNAP II (28–32). These functional features incited us to use the Pcf11 suppressing ability as a starting point to investigate the mechanisms by which Rho-induced aberrant transcripts are recognized and targeted for destruction. A systematic deletion analysis of Pcf11 indicates that the rescue of Rho-induced aberrant transcripts and the accompanying relief of growth defect by overproduction of the protein stems from a physical interaction of its CID domain with the transcription complex. The interaction requires both CTD and RNA contacts and is more efficient with isolated CID fragment than the full-length protein. Similar rescue of Rho-induced aberrant transcripts and relief of growth defect were obtained when endogenous Nrd1, the other CID-containing protein involved in transcription termination of small non-coding RNAs, was altered in its CTD and RNA binding determinants by deletion of its CID or mutations within its RRM. Thus, overproduced Pcf11 or its CID-containing fragments exert their suppressing effect by outcompeting Nrd1 for recruitment by the transcription complex. Consistent with this conclusion, a similar competition-mediated suppression of Rho-induced defect is achieved by overproduction of functionally-deficient CID-containing Nrd1 fragments. Together, these results show that the recognition and targeting for destruction of Rho-induced aberrant transcripts is mediated by Nrd1 association with the transcription complex, a fact that was substantiated with the direct evaluation of Nrd1 occupancy by ChIP experiments. Indeed, Nrd1 recruitment across two representative genes, PMA1 and PGK1 (Figure 8 and Supplementary Figure S2), is shown to increase dramatically in the presence of Rho expression and this trend is alleviated by overproduction of competing suppressors.
What makes the transcription complex so attractive for Nrd1 association in the presence of Rho helicase/translocase activity? Co-transcriptional recruitment of Nrd1 was shown to be predominant at promoter-proximal regions matching the high level of CTD Ser5 phosphorylation. This was taken to explain the preferential involvement of Nrd1 complex in termination of short transcripts such as non-coding RNAs and CUTs (39,40). However, as detected with our ChIP experiments in the absence of Rho, recent data indicate that a basal level of Nrd1 association with transcribing RNAP II persists throughout elongation, apparently due to the persistence of a certain level of phosphorylated Ser5 residues (42,43). Moreover, this basal level of Nrd1 recruitment was shown to increase substantially and provoke termination at a site located >1.5 kb from a promoter when poly(A)-dependent termination is impaired (43). These results along with others suggest that the choice between the two termination modes is governed by a continuous competition between the two termination machineries for recruitment by the transcription complex, with the CID-containing proteins Nrd1 and Pcf11 being the prominent components of the two machineries (39,40).
We propose that a similar competition process, but in a biased way, is taking place for the recognition and targeting of Rho-induced aberrant transcripts. In effect, stripping of nascent transcripts from processing and packaging factors by Rho action should uncover specific RNA-binding sites promoting preferential recruitment of Nrd1 (Figure 9). For instance, this is well illustrated by our results with the CID-defective pcf11–13 strain in which the Rho-induced defect is aggravated, reflecting the unbalance in the competition between mutant Pcf11–13 and Nrd1. Conversely, the competition is counterbalanced at the expense of Nrd1 in the case of Pcf11 overproduction or in mutant strains with defective Nrd1 (nrd1Δ1–151 or nrd1–102 alleles). Work in progress in our laboratory indicates that a similar competition process is obtained by overexpression of other CTD-interacting proteins. In addition to Pcf11, Rna14 which is known to interact with the CTD was found as a dosage suppressor of the Rho-induced defect in our previous screen (44). In contrast, overexpression of the two other members of the CF1A complex, Rna15 and Clp1, for which CTD binding has not been found in two-hybrid experiments (31), did not confer any suppressing phenotype. A similar correlation between known CTD binding activity and dosage suppression of the Rho-induced defect is also found for some components of the CPF complex.
Figure 9.
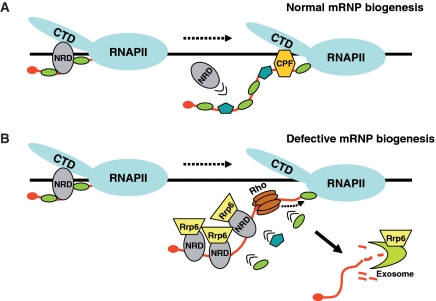
Model illustrating the Nrd1-mediated recognition and degradation of Rho-induced aberrant transcripts. (A) Normal conditions of mRNP biogenesis. Nrd1 complex is recruited to the transcription complex at early elongation then is outcompeted by the deposition of mRNA processing and binding proteins as RNAP II travels further downstream. (B) Rho-induced defective mRNP biogenesis. Rho loading and tracking along nascent transcript displaces processing and binding proteins uncovering specific RNA-binding sites that promote preferential recruitment of Nrd1 complex and association of nuclear RNA degradation machinery represented by Rrp6. Upon transcription completion, the aberrant transcript would be degraded by the exoribonuclease activity of Rrp6 either alone or in association with the core exosome.
In this biased competition model, Nrd1 might be weakly anchored to the transcription complex through interaction of its CID with the CTD, then the interaction is further stabilized by binding to specific RNA segments uncovered by Rho action. In this regard, our results suggest that Nab3 association with Nrd1 plays a key role in the stabilization process. First, a rescue of Rho-induced aberrant transcripts with accompanying relief of growth defect is observed in a strain with a defect in Nab3 RNA-binding domain (nab3–11 allele). Second, the dominant-negative effect induced by overexpression of Nrd1 deletion mutants is more robust when the protein fragments include the Nab3-interacting domain in addition to the CID. We note that the transcripts derived from the two representative genes studied in this work (PMA1 and PGK) contain much more Nab3 (UCUU) than Nrd1 (GUAA/G) binding sites (27 and 15 compared to 2 and 5, respectively) which should expend the opportunity for RNA stabilizing interactions and thus the recognition of a wide range of aberrant transcripts. These considerations are consistent with previous reports in which the formation of Nrd1–Nab3 heterodimer was shown to be the primary specificity determinant for transcription termination of small non-coding RNAs and CUTs. For instance, a CUT terminator containing only Nab3 binding sites functions efficiently in wild-type cells but does not function in the nab3–11 mutant strain, indicating that the Nab3 interaction with the transcript is sufficient to stabilize the recruitment of the heterodimer (36,48).
Previous co-immunoprecipitation studies have detected physical interactions between the exosome–TRAMP complex and Nrd1 (22). This was taken to suggest that co-transcriptional recruitment of Nrd1 could help to tether the exosome-dependent degradation machinery to the transcription complex. Our ChIP results provide a direct evidence for such suggestion by showing a correlation between the Rho-induced large increase of Nrd1 recruitment and the enrichment of nuclear-specific exosome component Rrp6 across the PMA1 gene. A general belief for nuclear mRNA surveillance is that Rrp6 exoribonuclease activity acts in concert with the core exosome through physical interactions. However, our ChIP experiments using tagged Rrp4 and Rrp41 did not reveal any association of these core exosomal subunits with the tethered surveillance machinery. This could be due to the fact that the core exosome does not have close contacts with RNA and DNA within the transcription complex and thus is less efficiently cross-linked to chromatin. A second possibility could be that the interactions of the core exosome with Nrd1 and Rrp6 are rather weak, which may explain the fact that in the Nrd1 co-immunoprecipitates, the core exosome components were significantly underrepresented as compared to Nab3, Rrp6 or the TRAMP component Air2 (22). A third possibility, which we favor, could be that co-transcriptional recruitment and RNA binding of Nrd1 followed by Rrp6 tethering constitutes the first step at which a defect is detected and the transcript is labeled as aberrant. Subsequently, the labeled transcript would be prevented from export then directed for destruction by facilitating a close coupling of Rrp6 with the core exosome scaffold (Figure 9). This second step is probably achieved upon transcription completion since exoribonuclease degradation by the exosome–Rrp6 complex requires exposed RNA 3′-end. However, an alternative scenario of this last model could be that Rrp6 exoribonuclease activity acts independently from core exosome to degrade aberrant transcripts. This scenario is supported by recent findings in which Rrp6 was found to carry out some of its exoribonuclease functions without physical association with the core exosome and to be stimulated by the TRAMP complex (52,53). Whatever the precise mechanism, this Nrd1–Rrp6-tagging model could explain how a general quality control system recognizes various aberrancies arising at different steps of mRNP biogenesis such as splicing, termination and 3′-end formation or assembly and export, then directs the defective molecules to the same outcome, destruction. In this regard, although our experimental approach uses an artificial system in which mRNPs are actively depleted from processing and packaging factors by Rho action, it gives a hint of what is likely to happen under naturally occurring circumstances that result in defective mRNP formation. However, it should be emphasized that the existence of alternative pathways for the mRNA surveillance mechanism cannot be excluded, especially in the case of transcripts in which Nrd1 and Nab3 binding sites are poorly represented or absent.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FRM (Loiret), la Ligue contre le Cancer Grand Ouest (Loiret and Sarthe) and the CNRS (program PEPS); Fellowship from the Ecole Doctorale Sciences et Technologies, Université d’Orléans (to R.H.). Funding for open access charge: FRM (Loiret), la Ligue contre le Cancer Grand Ouest (Loiret and Sarthe) and the CNRS (program PEPS).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank B. Seraphin and C. Saguez for helpful discussions and also J. Mouaikel and F. Jacquinot for technical assistance. We are indebted to Pierre Thuriaux and F. Lacroute for generous gifts of yeast strains.
REFERENCES
- 1.Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 3.Perales R, Bentley D. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 8.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 9.Fasken MB, Corbett AH. Process or perish: quality control in mRNA biogenesis. Nat. Struct. Mol. Biol. 2005;12:482–488. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- 10.Saguez C, Olesen JR, Jensen TH. Formation of export-competent mRNP: escaping nuclear destruction. Curr. Opin. Cell Biol. 2005;17:287–293. doi: 10.1016/j.ceb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Villa T, Rougemaille M, Libri D. Nuclear quality control of RNA polymerase II ribonucleoproteins in yeast: tilting the balance to shape the transcriptome. Biochim. Biophys. Acta. 2008;1779:524–531. doi: 10.1016/j.bbagrm.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 13.Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell Biol. 2005;25:9996–10004. doi: 10.1128/MCB.25.22.9996-10004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 19.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 20.Hessle V, Bjork P, Sokolowski M, de Valdivia EG, Silverstein R, Artemenko K, Tyagi A, Maddalo G, Ilag L, Helbig R, et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol. Biol. Cell. 2009;20:3459–3470. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 24.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 28.Amrani N, Minet M, Wyers F, Dufour ME, Aggerbeck LP, Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 31.Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl Acad. Sci. USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 36.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 39.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 40.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 42.Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-Safe Transcriptional Termination for Protein-Coding Genes in S. cerevisiae. Mol. Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mosrin-Huaman C, Honorine R, Rahmouni AR. Expression of bacterial Rho factor in yeast identifies new factors involved in the functional interplay between transcription and mRNP biogenesis. Mol. Cell Biol. 2009;29:4033–4044. doi: 10.1128/MCB.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 47.Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz A, Margeat E, Rahmouni AR, Boudvillain M. Transcription termination factor rho can displace streptavidin from biotinylated RNA. J. Biol. Chem. 2007;282:31469–31476. doi: 10.1074/jbc.M706935200. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz A, Rabhi M, Jacquinot F, Margeat E, Rahmouni AR, Boudvillain M. A stepwise 2′-hydroxyl activation mechanism for the bacterial transcription termination factor Rho helicase. Nat. Struct. Mol. Biol. 2009;16:1309–1316. doi: 10.1038/nsmb.1711. [DOI] [PubMed] [Google Scholar]
- 51.Walmacq C, Rahmouni AR, Boudvillain M. Testing the steric exclusion model for hexameric helicases: substrate features that alter RNA-DNA unwinding by the transcription termination factor Rho. Biochemistry. 2006;45:5885–5895. doi: 10.1021/bi0600648. [DOI] [PubMed] [Google Scholar]
- 52.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J. Biol. Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.