Abstract
MicA is a trans-encoded small non-coding RNA, which downregulates porin-expression in stationary-phase. In this work, we focus on the role of endoribonucleases III and E on Salmonella typhimurium sRNA MicA regulation. RNase III is shown to regulate MicA in a target-coupled way, while RNase E is responsible for the control of free MicA levels in the cell. We purified both Salmonella enzymes and demonstrated that in vitro RNase III is only active over MicA when in complex with its targets (whether ompA or lamB mRNAs). In vivo, MicA is demonstrated to be cleaved by RNase III in a coupled way with ompA mRNA. On the other hand, RNase E is able to cleave unpaired MicA and does not show a marked dependence on its 5′ phosphorylation state. The main conclusion of this work is the existence of two independent pathways for MicA turnover. Each pathway involves a distinct endoribonuclease, having a different role in the context of the fine-tuned regulation of porin levels. Cleavage of MicA by RNase III in a target-dependent fashion, with the concomitant decay of the mRNA target, strongly resembles the eukaryotic RNAi system, where RNase III-like enzymes play a pivotal role.
INTRODUCTION
Small non-coding RNAs (sRNAs) play very important roles in post-transcriptional control of gene expression. MicF was the first trans-encoded antisense sRNA described and was discovered a little more than a quarter-century ago as a regulator of the Escherichia coli ompF mRNA (1). Following the advent of systematic genome wide sRNA searches, the total number of known sRNAs in E. coli and the model pathogen Salmonella enterica serovar Typhimurium has grown to well over a hundred (2).
An extensive network of trans-antisense sRNAs have been shown to downregulate the expression of several outer membrane proteins (OMPs). While in some cases the same sRNA regulates multiple omp mRNAs (3,4), in other cases the same omp mRNA is target of multiple sRNAs (4–6). OMPs are embedded within the outer membrane, which together with the peptidoglycan layer and the inner membrane form the bacterial cell envelope, the first barrier of defense against external aggressions. Coordination in the expression of omp genes seems critical for proper envelope assembly, and accounts for the existence of so many sRNAs to regulate OMP mRNAs.
To survive in a changing environment, bacteria must constantly adjust the nature and abundance of surface components. Any condition that unbalances OMP levels activates the response of the transcription factor σE (7,8) that triggers transcription of a set of genes, which collectively help the bacterium to recover from the stress condition. MicA and RybB are two of the σE activated genes in stationary phase, whose role is to immediately limit OMP synthesis (4,6,9). Both sRNAs act in the same fashion: they inhibit protein synthesis by base pairing to the translation initiation region of their mRNA targets in an Hfq-dependent manner, followed by the subsequent degradation of the mRNA. Although sRNAs generally modulate translational initiation by interfering with 30S ribosome loading, alterations of target mRNA levels are also often observed (10,11). A few studies performed in E. coli suggest that RNase cleavage of target mRNAs may be directly coupled to the degradation of the sRNA that is regulating the process, with both RNAs being degraded upon sRNA action (12–14).
RNases can have a major impact on sRNAs regulatory pathways by performing a key role in the biogenesis and processing of sRNAs, as well as in controlling their cellular levels through regulation of their turnover (12,15–19). In E. coli, and presumably in many other Gram-negative bacteria, including Salmonella, mRNA decay is normally initiated by an endonucleolytic cleavage mainly performed by RNase E (20) and, sometimes by RNase III (21), followed by exoribonucleolytic degradation (19,22). In E. coli, both endoribonucleases have also been implicated in the decay of sRNAs, upon translational silencing (23).
We have previously reported specific contributions of several Salmonella ribonucleases on the turnover of different sRNAs (17). In this work, we have cloned and purified for the first time Salmonella RNase III and RNase E and have demonstrated that both endoribonucleases are responsible for the control of MicA sRNA levels. The role of the double stranded-specific endoribonuclease III over MicA only occurs through a target-dependent pathway, whether in vitro or in vivo. By contrast, the single stranded-specific endoribonuclease E is able to efficiently degrade free MicA sRNA. A model is proposed to explain the cooperation of both enzymes in the cell in order to achieve the fine-tuned control of the post-transcriptional regulator MicA.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides used in this study are listed in the Supplementary Table S1 and were synthesized by STAB Vida, Portugal.
Bacterial strains
All bacterial strains and plasmids used in this study are listed in the Tables 1 and 2, respectively. All Salmonella strains used are isogenic derivates of the wild-type Salmonella enterica serovar Typhimurium strain SL1344. The OmpA− (CMA-552), LamB− (CMA-554) and MicA− (CMA-555) mutants were constructed using the primer pairs pSV-104/pSV-105, pSV-108/pSV-109 and pSV-146/pSV-147, respectively, and following the λ-red recombinase method (24), with few modifications, as previously described (17). All chromosomal mutations were subsequently transferred to a fresh SL1344 background by P22 HT105/1 int-201 transduction (25). The chloramphenicol-resistance cassette of plasmid pKD3 replaces nucleotides −190 to +1064 of the ompA gene, −20 to +1339 of lamB and +8 to +78 of micA. All gene deletions were verified by colony PCR using the primer pairs pSV-106/pSV-107 for ompA, pSV-110/pSV-111 for lamB and pSV-148/pSV-149 for micA. The S. typhimurium RNase III deficient strain (CMA-551) was obtained by P22 transduction from SA5303 strain (26) and is tetracycline resistant. The double mutants were constructed using the same transduction method.
Table 1.
List of strains used in this work
| Strain | Relevant markers/Genotype | Source/Reference |
|---|---|---|
| S. typhimurium, SL1344 | StrRhisG rpsL xyl | (56) |
| CMA-537 | SL1344 rne-537 (Δrne::CmR) | (17) |
| CMA-551 | SL1344 rnc-14::ΔTn10 (TcR) | This study |
| CMA-552 | SL1344 ompA (ΔompA::CmR) | This study |
| CMA-554 | SL1344 lamB (ΔlamB::CmR) | This study |
| CMA-555 | SL1344 micA (ΔmicA::CmR) | This study |
| CMA-556 | SL1344 rnc-14 micA (rnc-14::ΔTn10/ΔmicA::CmR) | This study |
| CMA-557 | SL1344 rnc-14 ompA (rnc-14::ΔTn10/ΔompA::CmR) | This study |
| CMA-558 | SL1344 rnc-14 rne-537 (rnc-14::ΔTn10/Δrne::CmR) | This study |
| E. coli BL21(DE3) | F−ompT hsd SB(rb−mb−) gal dcm (DE3) | (57) |
| E. coli BL21(DE3)recA rnc105 | F−ompT hsd SB(rb−mb−) gal dcm (DE3) recA rnc105 | (27) |
| E. coli DH5α | recA1 endA1 gyrA96 thi-hsdR17 | New England Biolabs |
| supE44 relA1 ΔlacZYA-arg FU169 f80dLacZDM15 |
Table 2.
List of plasmids used in this work
| Plasmid | Comments | Origin/Marker | Reference |
|---|---|---|---|
| pKD3 | Template for mutants construction; carries chloramphenicol-resistance cassete | oriRγ/AmpR | (23) |
| pKD46 | Temperature-sensitive λ-red recombinase expression plasmid | oriR101/AmpR | (23) |
| pCP20 | Temperature-sensitive FLP recombinase expression plasmid | AmpR, CmR | (23) |
| pET-15b | Inducible expression vector, N-terminal His Tag | AmpR | Novagen |
| pSVDA-01 | pET-15b encoding His-RNase III | AmpR | This study |
| pSVDA-02 | pET-15b encoding His-RNase E | AmpR | This study |
Bacterial growth
All strains were grown in Luria-Bertani (LB) broth at 37°C with agitation throughout this study. SOC medium (Super Optimal Broth with Catabolite Repression medium) was used to recover transformants after heat shock (in the case of E. coli) or electroporation (in the case of Salmonella), before plating. Growth medium was supplemented with the following antibiotics when appropriate: ampicillin (150 µg/ml), chloramphenicol (25 µg/ml), streptomycin (90 µg/ml) and tetracycline (25 µg/ml).
RNA extraction and northern blot analysis
Overnight cultures were diluted 1/100 in fresh LB medium and grown until 6 h after OD600 of 2 (OD2+6). Culture samples were collected, mixed with 1 volume of stop solution [10 mM Tris (pH 7.2), 25 mM NaNO3, 5 mM MgCl2, 500 µg/ml chloramphenicol] and harvested by centrifugation (10 min, 6000g, 4°C). For stability experiments, rifampicin (500 µg/ml) and nalidixic acid (20 µg/ml) were added to cells grown in LB at 37°C, with agitation, till OD2+6. Incubation was continued and culture aliquots were withdrawn at the time-points indicated in the respective figures. RNA was isolated using the phenol/chlorophorm extraction method, precipitated in ethanol, resuspended in water and quantified on a Nanodrop 1000 machine (NanoDrop Technologies).
For northern blot analysis, 15 µg of total RNA was separated under denaturating conditions either by 8.3 M urea/8% polyacrylamide gel in TBE buffer or by 1.3% agarose MOPS/formaldehyde gel. For polyacrylamide gels, transfer of RNA onto Hybond-N+ membranes (GE Healthcare) was performed by electroblotting (1 h 50 min, 24 V, 4°C) in TAE buffer. For agarose gels, RNA was transferred to Hybond-N+ membranes by capillarity using 20 × SSC as transfer buffer. In both cases, RNA was UV cross-linked to the membrane immediately after transfer. Membranes were then hybridized in RapidHyb Buffer (GE Healthcare) at 68°C for riboprobes and 43°C in the case of oligoprobes and DNA probes. After hybridization, membranes were washed as described (17). Signals were visualized by PhosphorImaging (Storm Gel and Blot Imaging System, Amersham Bioscience) and analyzed using the ImageQuant software (Molecular Dynamics).
Hybridization probes
Primers for templates amplification are listed in Supplementary Table S1. Labeling of the riboprobes and oligoprobes was performed as described (17). The riboprobes were obtained using the primer pair pSV-118/pSV-141 for MicA and pSV-142/pSV-143 for ompA. The DNA probe for 16S rRNA was generated using the primer pair pSV-144/pSV-145 and ‘Amersham MegaprimeTM DNA Labeling Systems’ (GE Healthcare), according to the supplier instructions.
Construction of recombinant proteins
To overexpress Salmonella RNase E and RNase III proteins, the rne and rnc coding regions were amplified with primer pairs pSV-124/pSV-125 and pSV-129/pSV-130, respectively. The N-terminal region (comprising residues 1–522), corresponding to the catalytic domain of RNase E, was purified. In E. coli, the N-terminal half of RNase E (residues 1–498) was reported to be sufficient for the ribonuclease activity (27). The purified PCR products were double digested with BamHI and NdeI and ligated to the pET-15b vector previously digested with the same enzymes, yielding plasmids pSVDA-01 (rnc) and pSVDA-02 (rne). These plasmids were first cloned into E. coli DH5α and were subsequently transformed into BL21(DE3) strain in the case of pSVDA-02, and BL21(DE3) rnc105 recA (28) in the case of pSVDA-01 construction. This derivative strain of BL21(DE3), carrying an RNase III mutation, was used because it blocks the autoregulation of Salmonella RNase III by the endogenous E. coli homologue, resulting in a higher yield of the enzyme upon overexpression. All constructs were confirmed by DNA sequencing at STAB Vida.
Overexpression and purification of Salmonella RNase E and RNase III proteins
The BL21 (DE3) strain and derivative, containing the recombinant plasmids of interest, were grown in 100 ml of LB medium supplemented with ampicillin (150 µg/ml) to an optical density at 600 nm of 0.5. At this point, protein expression was induced by addition of 1 mM of IPTG for 3 h at 37°C. Cells were then harvested by centrifugation and the pellets stored at −80°C. The culture pellets expressing RNase III or RNase E were resuspended in 3 ml of Buffer A (20 mM Tris–HCl pH 8, 500 mM NaCl, 20 mM imidazole pH 8). Suspensions were lysed using a French Press at 900 psi in the presence of 0.1 mM of PMSF. After lysis, the crude extracts were treated with 125 U of Benzonase (Sigma) to degrade the nucleic acids and clarified by a 30 min centrifugation at 10 000g, 4°C. The histidine tagged recombinant proteins were purified by affinity chromatography, using the ÄKTA FPLCTM System (GE Healthcare). The clarified extracts were loaded into a HisTrap HP Sepharose 1 ml column equilibrated in Buffer A. Protein elution was achieved in buffer A with a linear imidazole gradient (from 20 to 500 mM). The fractions containing mostly the protein of interest, free of contaminants, were pooled. Eluted proteins were buffer exchanged with Desalting Buffer [10 mM Tris–HCl (pH 8), 3 mM MgCl2, 100 mM NaCl, 1 mM DTT] and concentrated by centrifugation at 4°C with Amicon Ultra Centrifugal Filter Devices (Millipore), with a molecular mass cutoff of 10 kDa (RNase III) or 50 kDa (RNase E). Proteins were quantified using the Bradford Method (29) and stored at −20°C in Desalting Buffer containing 50% (v/v) glycerol. The purity of the enzymes was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and revealed >90% homogeneity.
In vitro transcription and activity assays
DNA templates for the in vitro transcription were generated by PCR using chromosomal DNA from S. typhimurium SL1344 strain. The phage T7 RNA polymerase promoter sequence was included in the forward primer sequences. micA was amplified with the primer pair pSV-116/pSV-117, ompA with pSV-122/pSV-123 and lamB with pSV-120/pSV-121. For the synthesis of the internally labeled 5′ triphosphate MicA, in vitro transcription was carried out using the purified PCR product as template in the presence of an excess of [32P]-α-UTP over unlabeled UTP with ‘Riboprobe in vitro Transcription System’ (Promega) and T7 RNA polymerase. MicA substrate bearing 5′ monophosphate was obtained by adding an 8-fold excess of GMP over the other ribonucleotides to the in vitro transcription reaction. Non-radioactive molecules were transcribed in the same conditions but using equimolar concentrations of all four ribonucleotides. MicA transcripts were purified by electrophoresis on an 8.3 M urea/10% polyacrylamide gel. The gel slice was crushed and the RNA eluted with elution buffer [3 M ammonium acetate pH 5.2, 1 mM EDTA, 2.5% (v/v) phenol pH 4.3], overnight at room temperature. The RNA was ethanol precipitated and resuspended in RNase free water. For the synthesis of the 5′-end-labeled MicA or ompA, in vitro transcription was carried out using the corresponding PCR product as template. MicA and ompA transcripts were run on a 10 or 6% polyacrylamide gel, respectively, identified by ethidium bromide (EtBr) staining and cut out from the gel. The RNA was eluted from the gel slice as described above. The RNA substrates were end-labeled with [32P]-γ-ATP at 37°C for 1 h, with 10 units of T4 polynucleotide kinase (Fermentas) using the supplier exchange buffer and again purified from gel as above. The yield of the labeled substrates (cpm/µl) was determined by scintillation counting.
The hybridization between labeled and unlabeled substrates was always performed in a 1:40 molar ratio in the Tris component of the activity buffer by incubation for 10 min at 80°C, followed by 45 min at 37°C.
The activity assays were done in a final volume of 50 µl containing the activity buffer {for RNase III [30 mM Tris–HCl pH 8, 160 mM NaCl and 0.1 mM DTT] and for RNase E [25 mM Tris–HCl pH 7.5, 5 mM MgCl2, 60 mM KCl, 100 mM NH4Cl, 0.1 mM DTT and 5% (v/v) glycerol]} and ∼10 000 cpm of substrate. In the case of the activity assays with RNase III, 10 mM of MgCl2 was added to the reaction mixture. As a control, prior to the beginning of each assay an aliquot was taken and was incubated until the end of the assay (without the enzyme). The reactions were started by the addition of the enzyme at a concentration of 500 nM, and further incubated at 37°C in the case of RNase III and 30°C for RNase E (30,31). Samples were withdrawn at the time-points indicated in the respective figures, and the reactions were stopped by the addition of formamide-containing dye supplemented with 10 mM EDTA. Reaction products were resolved in a 7 M urea/15% or 8 % polyacrylamide gel as indicated in the respective figure legends. Signals were visualized by PhosphorImaging and analyzed using ImageQuant software (Molecular Dynamics).
OMPs extraction and analysis
The membrane protein fraction from late stationary phase cultures (OD600 of 2 + 6 h) was extracted as described (32). OMPs were analyzed on 4% urea–SDS–12% polyacrylamide gel. Gels were stained overnight with Coomassie Brilliant Blue.
RESULTS
Detection of MicA sense transcripts in an RNase III− mutant
We have previously studied MicA sRNA turnover in S. typhimurium and have analyzed the particular contribution of several RNases to the decay of this sRNA. We have found that the dsRNA-specific endoribonuclease III has a remarkable impact on the stability of MicA sRNA. In the wild-type, MicA sRNA has a half-life of ∼6 min (17). In an RNase III− mutant, there was a dramatic stabilization of the sRNA (no significant decay in >2 h), with the concomitant accumulation of a degradation intermediate, very stable, which was absent in the wild-type (17). In an RNase E mutant MicA was also stabilized, but the small stable intermediate was not detected.
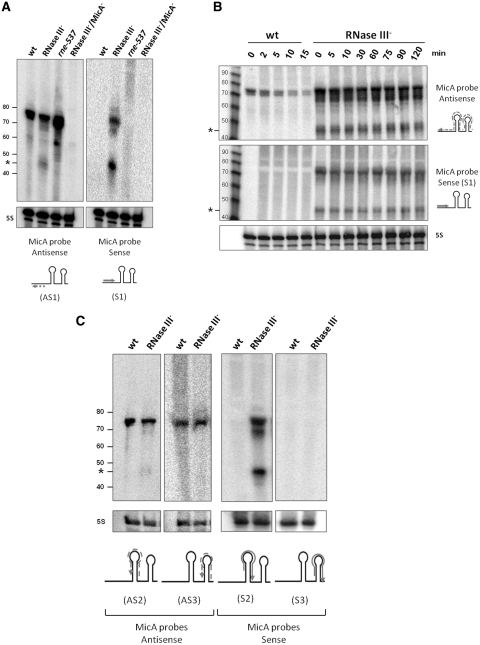
We were interested in clarifying the nature of this small intermediate. For this purpose we have compared the bands pattern of isogenic RNase III+ and RNase III− strains, by northern blot analysis, with different probes. We have used a probe antisense (AS1) or sense (S1) to the 5′-end of MicA, which corresponds to the region of interaction with its targets (33–35). The same short MicA sRNA stable intermediate of ∼45 nt (indicated by an asterisk in the figures) was detected with the antisense probe (AS1) only in the RNase III− strain (Figure 1A, left panel). When using the MicA sense probe (S1) we have also detected in this mutant a smaller transcript with approximately the same size (Figure 1A, right panel). Both species (sense and antisense) have a remarkably long half-life (see Figure 1B). It was also detected with the sense probe another band with the size corresponding to that of MicA full transcript (74 nt). None of the bands observed with the sense probe were visible in the wild-type or the RNase E mutant (rne-537). Moreover, the presence of these ‘sense transcripts’ is MicA dependent, since they were not detected in an RNase III−/MicA− strain.
Figure 1.
Analysis of MicA sense and antisense species in an RNase III− mutant. Total cellular RNA was extracted from the S. typhimurium strains indicated and analyzed by northern blot. 15 µg of RNA (each lane) were separated on an 8% PAA/8.3 M urea gel. The gel was then blotted to a Hybond-N+ membrane and hybridized with the corresponding probes. Details of RNA extraction and northern blot procedure are described in ‘Materials and Methods’ section. In each case, the membrane was stripped and then probed for 5S rRNA (pSV-139) as loading control. The radiolabeled marker 10bp DNA Step Ladder (Promega) is on the left side. The respective sizes are represented in nucleotides (full MicA is 74 nt long). The asterisk indicates the fragment that specifically accumulates in RNase III− strain. The probes used are indicated in the corresponding image. The arrow in each picture indicates the localization and direction of the probes in MicA sRNA: antisense (AS), represented by a dashed arrow; sense (S), represented by a solid arrow. The sequence of the probes is indicated in Supplementary Table S1. (A) Total RNA from S. typhimurium wild-type and mutant derivatives RNase III−, rne-537 and RNase III−/MicA− was hybridized with MicA antisense (AS1) and sense (S1) probes. The double mutant (RNase III−/MicA−) and the RNase E mutant (rne-537) were used as controls. (B) Comparison of the stability of both MicA sense and antisense species in the absence of RNase III. Total cellular RNA from wild-type and RNase III− mutant was extracted at the time-points (min) indicated on top, after transcription arrest. RNA samples were analyzed as described above using an antisense probe to the full MicA sequence (upper panel) or a sense probe (lower panel). (C) Total RNA from wild-type and RNase III− mutant strains was hybridized with two other differently located antisense and sense probes, as indicated in the pictures below each image.
The fact that the smaller transcript is equally present when using a sense or antisense probe and uniquely when RNase III is absent suggests that it is one strand of a stable dsRNA remnant of the MicA-target mRNA paired species. This smaller intermediate probably arises due to the previous activity of other degrading enzyme(s) but only accumulates in the absence of RNase III by virtue of its double stranded character. RNase E is probably a good candidate since the level of the smaller intermediate is decreased in an RNase III− mutant that is also impaired for degradosome formation - RNase III−/rne-537 (see Supplementary Figure S1). For instance, cleavage of MicA and ompA mRNA (a main target of MicA) by RNase E (33) together with exoribonucleolytic degradation of both RNAs may explain why the antisense and sense transcripts have approximately the same size.
In order to confirm that the smaller transcripts correspond to a stable dsRNA remnant of the MicA-target mRNA paired species, we have used two other MicA sense probes differently located along the MicA transcript (Figure 1C). For each sense probe used, the correspondent antisense probe, complementary to MicA, was also designed. The location of each of the probes in the MicA sequence is indicated in the figure below the respective images. A transcript having the same size was detected whether with the sense probes S1 and S2, or with the corresponding AS1 and AS2 antisense probes. However, we have not obtained any signal when using a sense probe located in the 3′-end of MicA (S3), while the MicA full transcript could still be detected with the corresponding antisense probe (AS3).
These results strongly indicate that the small degradation intermediate should correspond to a remnant of a duplex MicA-target mRNA. The lack of signal when using S3 (located in the 3′-end of MicA) further suggests that the duplex formation is confined to the 5′-end of MicA. This observation is in agreement with previous reports, which indicate that the interaction site is located in the 5′-end of the sRNA, at least for the two known targets of MicA (33–35).
MicA cleavage by RNase III is facilitated by base pairing with its mRNA target(s)
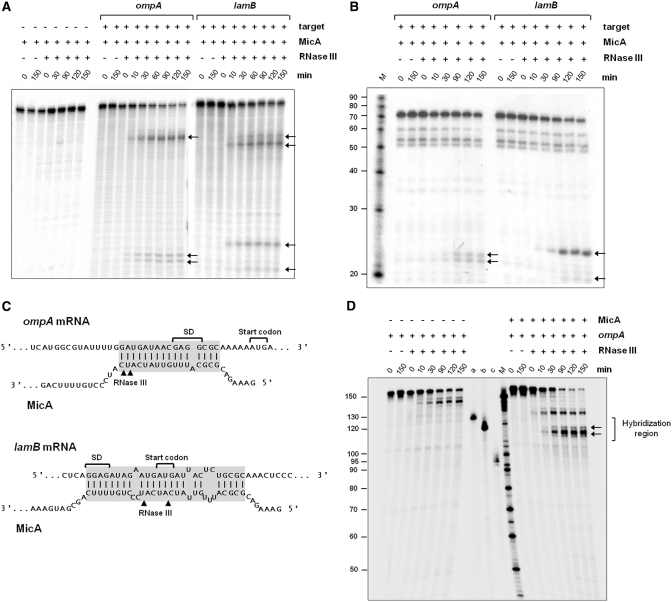
The results presented in Figure 1 suggest that the cleavage of MicA by RNase III occurs in a target- dependent fashion. Taking this into account, together with the fact that RNase III is a double-stranded-specific endoribonuclease, led us to compare in vitro the activity of the enzyme both over MicA transcript alone or in complex with its mRNA targets. Until now only two targets for this sRNA have been described in Salmonella, ompA and lamB mRNAs. MicA was reported to act over the translation initiation region of both molecules (33–35). In order to study the activity of Salmonella RNase III over MicA, we have cloned and purified the Salmonella enzyme as described in Material and Methods. The pure enzyme used in the in vitro experiments is shown in Supplementary Figure S2. Activity assays were performed by incubating the purified Salmonella RNase III with α32P-labeled MicA alone or in combination with the unlabeled 5′-untranslated region (UTR) of ompA or lamB mRNAs. Since it has been shown that MicA is also able to bind ompA mRNA without the help of Hfq (33), this protein was not included in the activity assays. MicA alone was found to be resistant to RNase III cleavage (Figure 2A). By contrast, in conditions favoring the hybridization of the sRNA transcript with each one of the target molecules, we could see the increasing accumulation of specific reaction products simultaneously with the disappearance of the substrate. This indicates that the formation of the sRNA-target mRNA complex promotes the RNase III cleavage of MicA.
Figure 2.
In vitro cleavage of sRNA MicA or ompA 5′-UTR by endoribonuclease III. The radioactivelly labeled substrate was incubated with 500 nM of Salmonella RNase III. Aliquots withdrawn at the time-points indicated above each lane were analyzed on a 7 M urea/15% or 8% PAA gel for MicA or ompA, respectively. The first two lanes of each reaction correspond to the controls without the protein at time zero (0) and at the end of the reaction time (150). The radiolabeled Decade Marker RNA (Ambion) is indicated by ‘M’. The arrows in the figure indicate specific degradation products. (A) Assays performed with internally labeled MicA in the absence (−) (left panel) or in the presence (+) of a molar excess of ompA (middle panel) or lamB (right panel) unlabeled transcripts (5′-UTR sequence). (B) Assays performed with 5′-end-labeled MicA in the presence (+) of a molar excess of ompA (left panel) or lamB (right panel) unlabeled transcripts (5′-UTR sequence). The bands that are already observed in the absence of the enzyme (control reactions) arise due to the radiolysis of the substrate. (C) Proposed interaction regions of ompA and lamB mRNAs with MicA [adapted from (33)]. The Shine–Dalgarno regions of ompA and lamB are indicated. The arrows indicate the RNase III cleavage sites on MicA as determined on A and B. (D) Assays performed with 5′-end-labeled ompA in the absence (−) (on the left) or in the presence (+) (right panel) of a molar excess of unlabeled MicA. On the left side of the marker (M) radiolabeled transcripts of known sizes were included (a) 130 nt; (b) 120 nt and (c) 95 nt. The arrows in the figure indicate the degradation products located inside the hybridization region.
The extension and location of MicA interaction with ompA or lamB mRNAs has been predicted to be slightly different (33,34). Since RNase III cleaves dsRNA, the different interaction between MicA and the two targets could be in the origin of the distinct cleavage pattern induced by ompA or lamB. In order to identify the cleavage points generated by RNase III on the MicA-ompA and MicA-lamB hybrids, in vitro assays were performed as described above, but using 5′-end-labeled MicA in combination either with the unlabeled 5′-UTR of ompA or lamB. The results are shown in Figure 2B. RNase III cleavage generates two main fragments of 22 and 23 nt on MicA-ompA hybrid, and 21 and 25 nt on MicA-lamB. Since in this experiment MicA was 5′-end-labeled, the size of these fragments indicates the distance from the cleavage point to the 5′-end of MicA. The higher molecular weight bands observed only when MicA was internally labeled (see Figure 2A) correspond to 3′-end fragments, since they are not detected in the cleavage of 5′-end-labeled MicA. A representation of the hybridization regions showing the RNase III cleavage positions in MicA sequence is presented in Figure 2C. All the cleavage positions are located inside the predicted region of interaction with each target, strongly supporting our hypothesis that RNase III is responsible for the coupled MicA-target degradation.
According to our proposal, cleavage of MicA is coupled with the mRNA target cleavage. In this sense the same kind of activity assays were carried out in order to check the direct activity of RNase III over the corresponding region of ompA mRNA. For this, the purified Salmonella RNase III was incubated with the 5′-end-labeled UTR of ompA (172 nt) alone or in combination with unlabeled MicA. As shown in Figure 2D although RNase III is able to cleave free ompA, a faster disappearance of the substrate when the hybrid ompA-MicA was used indicates that it is cleaved more efficiently. Moreover, the cleavage event gives rise to specific degradation products that were not observed after incubation with ompA alone (Figure 2D). Among these products, we could observe the accumulation of fragments in the range of 113–130 nt, which is the expected size of fragments generated by cleavage inside the hybridization region with MicA (see Figure 2C). The other products with a higher molecular weight probably arise due to alterations in the secondary structure of ompA after the duplex formation, which could generate a new dsRNA region suitable for RNase III. However, we cannot extrapolate to the in vivo situation, since these assays were performed with a truncated version of ompA. The ability of RNase III to preferentially cleave the hybrid ompA-MicA in the region corresponding to the hybridization between the two molecules is another evidence for the coupled degradation of the target and the sRNA.
Taken together, our results indicate that MicA decay in vivo is highly dependent on RNase III and its cleavage by this enzyme in vitro is triggered upon base pairing with its target mRNAs.
ompA expression is regulated by RNase III and is dependent on MicA
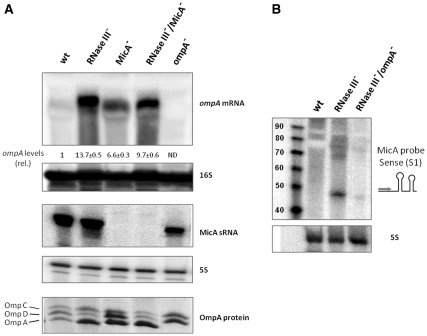
OmpA is a very abundant porin highly expressed in the exponential phase of growth. In stationary phase MicA is present at high levels and is the principal post-transcriptional downregulator of the ompA mRNA (33,35). Since our results indicate that MicA degradation by RNase III is target-dependent and we have observed the concomitant degradation of the ompA target mRNA in vitro, we analyzed the effect of an RNase III− mutation on the levels of ompA mRNA in stationary phase. The RNase III− mutant shows an increment of almost 14-fold in ompA mRNA level in comparison to the wild-type (Figure 3A). A strong increase in the OmpA protein level was also observed. This suggests that the reduced levels of ompA mRNA observed in stationary phase in the wild-type (RNase III+) are probably due to the cleavage and destabilization of the message by RNase III. This cleavage should be suppressed when RNase III is absent. This result strongly indicates that RNase III is implicated in the degradation of the ompA mRNA. On the other hand, the constant levels of OmpC and OmpD between the wild-type and RNase III−, further suggest that RNase III is not involved in the turnover of their messages and that the control of these proteins levels in the cell follows a different pathway. Interestingly, Paperfort et al. (4) have shown that MicA sRNA is also not involved in the control of ompC or ompD levels, while affecting ompA.
Figure 3.
Regulation of ompA and MicA expression in different mutant strains. Northern blot and SDS–PAGE analysis of RNA and protein samples extracted from wild-type and mutant strains as indicated on top of each lane. Details of experimental procedures are described in ‘Materials and Methods’ section. (A) (Upper panel) Analysis of steady-state ompA mRNA levels by northern blot. 15 µg of RNA (each lane) were resolved in a 1.3% formaldehyde-agarose gel. The gel was then blotted to a Hybond-N+ membrane and hybridized with the corresponding ompA riboprobe. Full-length transcripts were quantified using a Molecular Dynamics PhosphorImager. The amount of RNA found in wild-type was set as one. The ratio between each strain and the wild-type is depicted (relative levels). A representative membrane is shown and values indicated correspond to the average of several northern blot experiments with RNAs from at least two independent extractions. The membrane was stripped and then probed for 16S rRNA as loading control. (ND) Non-detectable. (Middle panel) MicA sRNA levels analysis by northern blot. 15 µg of RNA from the same mutants were separated on an 8% PAA/8.3 M urea. The gel was then blotted to a Hybond-N+ membrane and hybridized with the corresponding MicA riboprobe. The membrane was stripped and then probed for 5S rRNAs, as loading control. (Lower pannel) Outer membrane protein fraction analysis by 4% urea–SDS–12% polyacrylamide gel electrophoresis. The positions of the OmpC, OmpD and OmpA bands are indicated. An OmpA− mutant was used as control. (B) Comparison of the levels of MicA sense species on the wild-type, RNase III− and the double RNase III−/OmpA− strains. The experimental procedure was similar to the one described in (A). The arrow on MicA sRNA picture indicates the localization and direction of the probe (S1). Loading control of the RNA was done with 5S rRNA probe and is represented below. Sizes were estimated using the radiolabeled 10bp DNA Step Ladder (Promega), on the left side of the membrane.
Since the degradation of both the sRNA MicA and the ompA target mRNA is dependent on RNase III, we have checked whether the RNase III regulation of ompA expression was also MicA dependent. Therefore, we have analyzed the expression of ompA in a MicA− mutant strain. In stationary phase MicA basepairs with the 5′-UTR of ompA preventing ribosome binding and destabilizing the entire ompA mRNA (33,35). Accordingly, when the regulator is absent (MicA− mutant) the levels of ompA mRNA should be elevated. We observed an increase of about 7-fold in ompA mRNA levels when MicA is absent (Figure 3A). This result confirms that the control of ompA mRNA levels is dependent on MicA. However, the fact that in the absence of RNase III ompA mRNA levels are still higher than in the MicA− mutant indicates that RNase III may also have a role in ompA expression by an alternative pathway not involving MicA. In fact we show that RNase III is also able to cleave ompA in vitro in the absence of MicA. Additionally ompA mRNA may also be under the control of another sRNA in an RNase III dependent way.
If RNase III and MicA affect the ompA message through the same regulatory pathway, the combined absence of both would not result in a cumulative effect. In order to clarify this, we have constructed and tested the effect of the double mutant RNase III−/MicA− on the ompA mRNA levels. As shown in Figure 3A, in the double RNase III−/MicA− mutant the ompA levels are reduced in comparison with the RNase III− single mutant, demonstrating that MicA and RNase III act over ompA message through a common pathway.
Detection of the ‘sense transcripts’ in the RNase III− mutant depends on ompA
Taken together, the results presented here point out that ompA is subjected to MicA-coupled degradation by RNase III. Since we had indications that the ‘sense transcripts’ detected in the RNase III− mutant are remnants of the sRNA-target complex (see Figure 1), we have investigated if these ‘sense-transcripts’ corresponded to ompA mRNA fragments. Indeed, in the absence of both RNase III and ompA, the levels of these ‘sense transcripts’ are largely reduced when compared with those of the single RNase III− mutant (Figure 3B). This means that the detection of these ‘sense transcripts’ is related with the presence of ompA, strongly suggesting that this mRNA might be one of the targets degraded by RNase III in conjunction with MicA.
The fact that the ‘sense transcripts’ are still detectable in the absence of ompA indicates that this mRNA might not be the only candidate for the MicA-coupled degradation. However, in a LamB− mutant (the other known target of MicA) we did not observe, under our experimental conditions, a significant alteration in the level of the ‘sense transcripts’ (data not shown). Furthermore, in the absence of both ompA and lamB targets, the ‘sense transcripts’ could still be slightly observed (data not shown), indicating that other MicA targets subjected to the same type of regulation should exist in the cell.
RNase E cleaves ‘free MicA’ sRNA in vitro
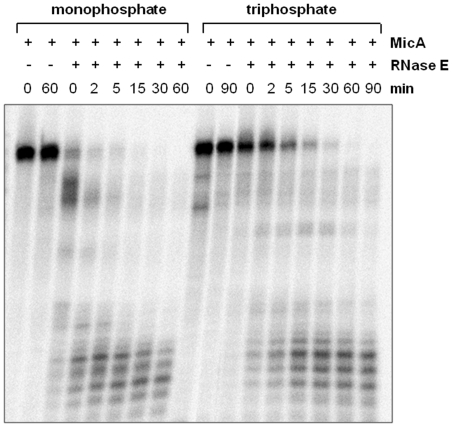
We have demonstrated that the sRNA MicA degradation is influenced by RNase III. However, this seems to happen only in the presence of the target mRNA. As we have shown in vitro, the enzyme was not able to cleave MicA alone. Thus, the question of how is free MicA degraded remains to be answered. In vivo experiments have shown a large impact of an RNase E mutant on the levels and stability of full MicA sRNA (17). However, these results concern studies undertaken with the rne-537 mutant derivative (17). Since this mutant only prevents degradosome formation, without totally abolishing the enzyme activity, we were also interested in clarifying the role of the catalytic activity of RNase E on the decay of MicA. Moreover, it has been shown that -A/U rich sequences together with adjacent stem-loop structures can comprise recognition sites for RNase E (36,37). The sequence of the sRNA MicA matches these characteristics. Therefore, we have analyzed the ability of this endoribonuclease to cleave MicA sRNA transcript, in vitro. For this purpose we have cloned and purified the amino-terminal region of Salmonella RNase E. The homologous region in E. coli RNase E is known to be responsible for the catalytic activity of the enzyme (27). The results of the purification of the N-terminal segment of Salmonella RNase E are shown in Supplementary Figure S2. In vitro assays with the purified protein were performed over uniformly labeled MicA transcript. It was seen before that RNase E preferentially cleaves RNAs with a 5′ monophosphate group over those endowed with a 5′ triphosphate (36,38–40). Thus, in the activity assays we have used as substrate both the monophosphate and the triphosphate MicA transcripts. Our results show that RNase E is able to cleave both substrates in vitro (Figure 4), though the efficiency of cleavage was superior over monophosphorylated MicA. This is in agreement with the recent report that E. coli RNase E is also active over some triphosphate substrates (41).
Figure 4.
In vitro study of MicA sRNA cleavage by RNase E. α-32P-labeled MicA transcript, 5′ monophosphate (left panel) or 5′ triphosphate (right panel), was incubated with 500 nM of purified Salmonella RNase E (residues 1–522) at 30°C. Aliquots withdrawn at the time-points indicated above each lane were analyzed on a 15% PAA/7M urea gel. The two first lanes of each reaction correspond to the controls without the protein both withdrawn at time zero and at the end of the reaction time.
We have previously shown that in cells in which the degradosome scaffold of RNase E was deleted the degradation of MicA is slower (>4-fold stabilization) (17). This suggests that, in vivo, RNase E may need the cooperation of other degradosome components in the decay of this transcript. Indeed it was previously shown in E. coli and Salmonella that the absence of PNPase, the exoribonucleolytic component of the degradosome, has a remarkable impact over the stability of MicA sRNA (16,17). However, the high ability of RNase E to cleave MicA in vitro indicates that the enzyme per se should importantly contribute for the in vivo degradation of free MicA.
DISCUSSION
Stress conditions that unbalance OMP levels activate the σE response, a complex set of changes normally devoted to protect the cell envelope from environmental challenges (42). The transcription factor σE triggers the synthesis of the sRNAs that control OMP levels (4,6). Upon downregulation of OMPs and the relief of membrane stress, the high sRNA levels have to be brought back to normal amounts. MicA sRNA is a σE-dependent porin downregulator whose transcription is activated in stationary-phase (4,6,9). Under this context, we were interested in studying the regulation of MicA cellular levels and determining the enzymes involved in this process.
MicA was previously found to be highly stabilized in cells lacking a functional RNase III (17). However, RNase III is not able to cleave MicA in vitro, suggesting that MicA alone is not a substrate for this enzyme. Indeed, we demonstrate that RNase III is only able to cleave MicA in vitro when it base pairs with its target(s). RNase III is a specific double stranded RNA endoribonuclease, which plays multiple roles in the processing of rRNA and mRNA (43) and its activity has also been demonstrated over several sRNA-target complexes formed by cis-antisense sRNAs (44–47). In these complexes, there is a perfect complementarity between the RNA partners, which constitutes a preferred substrate for RNase III, and avoids the need for Hfq. The limited complementary between trans-encoded sRNAs and their targets typically requires the help of the bacterial RNA chaperone Hfq. The trans-encoded sRNA MicA was shown to be dependent on Hfq both for stability and target degradation (17,33,35). It is generally assumed that Hfq binds both the regulator and the target RNA, favoring their interaction. Moreover, Hfq enhances the stability of many sRNAs in vivo, by protecting them from degradation (48). Curiously, IstR-1 from E. coli and RNAIII from Staphylococcus aureus are two trans-encoded sRNAs that act independently of Hfq and were also seen to be cleaved by RNase III in a target-coupled mechanism (13,49–51). Here we describe, in Salmonella, the first example of a system controlled by a Hfq-dependent trans-sRNA that involves the coupled degradation of the sRNA-target mRNA by RNase III.
In RNase III− cells, besides the high stabilization of MicA, a very stable smaller degradation intermediate is also observed. In agreement with the in vitro results, we have obtained several indications that this degradation intermediate corresponds to a remnant of a dsRNA complex formed by MicA and its target(s): (i) It is only detected in the RNase III− mutant (deficient for dsRNA degradation), where it is extremely stable by virtue of its double stranded character. When RNase III is present, dsRNA complexes are cleaved to products that are either further degraded or too small to be detected by northern blot; (ii) This small intermediate is visible with both antisense MicA probes and sense probes complementary to the targets and (iii) The level detected with sense probes is highly reduced in the absence of OmpA (a main MicA target). Thus, this remnant species should indicate the region of interaction between MicA and its targets. Since a strong signal is detected with the 5′ probes and no signal at all is obtained with the probes located in the 3′-end, this region corresponds to the 5′ half of MicA. In fact, it is known that MicA interacts through its 5′-end sequence with its two targets described till now (33–35). Accordingly, the RNase III cleavage sites determined in vitro on both MicA-ompA and MicA-lamB hybrids are located in the 5′ half of MicA, inside the respective predicted hybridization region, which strongly correlates with the in vivo observations. This result is further confirmed by the fact that RNase III also cleaves the ompA 5′-UTR inside the same region, demonstrating that both molecules are cleaved together.
In the absence of both ompA and RNase III the level of the ‘sense transcripts’ is strongly decreased (Figure 3B). The fact that these bands are still visible, despite at very low levels, may be related with the formation of complexes between MicA and other target(s), whose degradation should also be RNase III-dependent. We demonstrate that in vitro RNase III is also able to cleave the complex MicA-lamB. However, in vivo, whether in the absence of lamB or of both lamB and ompA we could still detect the sense transcripts referred above (data not shown). This means that probably besides lamB mRNA other Salmonella MicA targets (not yet identified) may exist in stationary-phase. In E. coli, expression of phoPQ has recently been shown to be repressed by MicA upon activation of σE (52). In fact, MicA is a trans-encoded sRNA highly conserved in Enterobacteriacea (53). Trans-encoded sRNAs generally establish short and imperfect interactions with its mRNA targets (∼10–25 nt) (11,54), allowing the regulation of multiple targets by the same sRNA. For example, RybB sRNA controls more than 17 mRNAs, 10 of which encode OMPs, including ompA.
Upon MicA accumulation in stationary phase, the sRNA binds to ompA mRNA blocking ribosome binding and translation initiation. This releases the mRNA from the ‘protection’ by the ribosomes and leads to degradation of the ribosome-free mRNA by the concerted action of endo- and exoribonucleases (19). In line with previous-work (55,56), RNase E is thought to be the endoribonuclease responsible for the decay of ompA mRNA after the blockage of ribosome loading caused by MicA binding (33,35). In this report, we show that additionally the binding of MicA to ompA renders both RNAs susceptible to RNase III cleavage. We have demonstrated that this endoribonuclease is essential for ompA repression and, in agreement with previous studies (33,35), we observed a relieve of ompA repression in the absence of MicA. Both effects were seen to occur in a concerted way. The RNase III pathway has the advantage of simultaneously controlling the levels of the sRNA, whose function after the repression of the target will no longer be necessary in the cell. In addition RNase III cleavage makes the repression irreversible. From a physiological point of view, the existence of two distinct pathways may enhance the cell response in stress conditions allowing a fine-tuned balance of OmpA levels needed to keep the envelop integrity. Moreover by having two alternative degradation pathways the cell warrants the metabolism of molecules no longer needed. This may be crucial in stationary phase, which is characterized by limited resources.
Interestingly, the ompA levels in the double mutant (RNase III−/MicA−) are not restored to those observed in the MicA− mutant. This can be due to a direct effect of RNase III over ompA. Indeed we show that in vitro RNase III is able to cleave ompA even in the absence of the sRNA. Alternatively, other(s) player(s) can be involved in the RNase III-mediated regulation of ompA. At least two other trans-encoded regulatory sRNAs (RseX and RybB) have been described as additional ompA regulators (4,5).
We have just described a pathway for degradation of MicA sRNA that involves target binding and is dependent on RNase III. Our results show that RNase III cleaves MicA when hybridized with the targets. The question then arises how the levels of free MicA are brought back to normal when the cell no longer needs it. Our previous results in vivo, showed a high stabilization of MicA in an RNase E deletion mutant lacking the C-terminal region (17). This may suggest that RNase E needs the cooperation of other degradosome components in the degradation of MicA. In fact, we have previously shown that PNPase, the exonucleolytic component of the degradosome, has a great impact over the stability of this sRNA in Salmonella. According to the results presented here, the catalytic domain of RNase E also shows, in vitro, a high efficiency in the cleavage of this sRNA. This single stranded endoribonuclease seems to play an important role in the regulation of the abundance of free MicA.
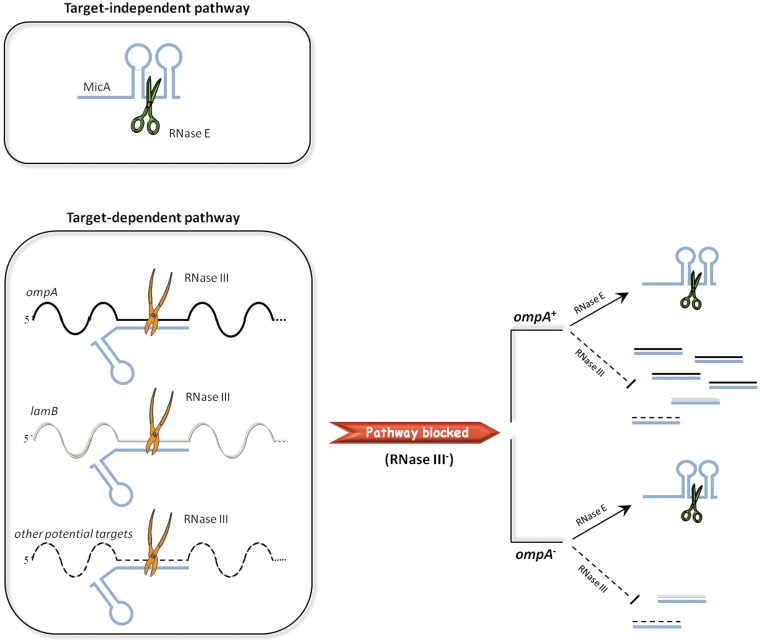
For each sRNA the characterization of its turnover has to be analyzed from two different perspectives: the independent, and the dependent of target interaction. The later can be similar or not, whether the sRNA decay is influenced or not by the respective target(s). Taken together, the results presented in this study indicate the existence of two different pathways for MicA sRNA turnover, each one involving a specific endoribonuclease. According to the model proposed in Figure 5, when MicA is free, RNase E seems to take the control by efficiently degrading the sRNA. However, if MicA is interacting with the targets the target-dependent pathway of degradation predominates. This mechanism involves a double stranded endoribonuclease that is able to degrade both the target and the sRNA, simultaneously.
Figure 5.
Schematic representation of the two degradation pathways followed by MicA. RNase E and RNase III are represented by scissors and pliers, respectively. The two different pathways for MicA degradation are shown on the left side. The possible associations of MicA with its targets are also depicted. In the wild-type, MicA and the targets should be fully degraded as a result of both degradation pathways in cooperation with the exoribonucleolytic activity. In the RNase III− mutant, the MicA-target dependent degradation by RNase III is blocked. As a result, some degradation intermediates are stabilized and can be detected, namely the target and MicA strands that have interacted but could not be cleaved by RNase III. ompA being the main target of MicA, its species are over-represented. When additionally the ompA target mRNA is absent, the respective degradation intermediate is no longer present in the cell and, as a consequence, there is a reduced level of transcripts detected with probes complementary to MicA-targets.
Cleavage by RNase III within the sRNA–mRNA duplex and the subsequent decay of the mRNA intermediate by the cell machinery could rather resemble the RNAi scenario in eukaryotic organisms. RNase III-like enzymes are known to have a pivotal role in eukaryotic small non-coding RNA function and biogenesis (19). Hence, it is not surprising that RNase III would also be a main player in the control of prokaryotic sRNA expression and function, broadening the enzyme’s global role in the regulation of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundação para a Ciência e Tecnologia (FCT, Portugal); FCT Post-Doctoral Fellowship (SFRH/BPD/30766/2006 to S.C.V. and SFRH/BPD/43390/2008 to S.D.); and Doctoral Fellowship (SFRH/BD/43211/2008 to I.J.S. and SFRH/BD/65607/2009 to M.S.). Funding for open access charge: Fundação para a Ciência e Tecnologia (FCT, Portugal).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors want to thank Prof. Allen W. Nicholson for providing BL21 (DE3) rnc105 recA strain.
REFERENCES
- 1.Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc. Natl Acad. Sci. USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 4.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 6.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 2004;52:613–619. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 10.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 2007;10:262–270. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viegas SC, Arraiano CM. Regulating the regulators: How ribonucleases dictate the rules in the control of small non-coding RNAs. RNA Biol. 2008;5:230–243. doi: 10.4161/rna.6915. [DOI] [PubMed] [Google Scholar]
- 19.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 20.Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog. Mol. Biol. Transl. Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 21.Conrad C, Rauhut R. Ribonuclease III: new sense from nuisance. Int. J. Biochem. Cell Biol. 2002;34:116–129. doi: 10.1016/s1357-2725(01)00112-1. [DOI] [PubMed] [Google Scholar]
- 22.Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3′-5′ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- 23.Kaberdin VR, Blasi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmieger H. The fate of the bacterial chromosome in P22-infected cells of Salmonella typhimurium. Mol. Gen. Genet. 1971;110:238–244. doi: 10.1007/BF00337836. [DOI] [PubMed] [Google Scholar]
- 26.Mattatall NR, Sanderson KE. RNase III deficient Salmonella typhimurium LT2 contains intervening sequences (IVSs) in its 23S rRNA. FEMS Microbiol. Lett. 1998;159:179–185. doi: 10.1111/j.1574-6968.1998.tb12858.x. [DOI] [PubMed] [Google Scholar]
- 27.McDowall KJ, Cohen SN. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 1996;255:349–355. doi: 10.1006/jmbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 28.Amarasinghe AK, Calin-Jageman I, Harmouch A, Sun W, Nicholson AW. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Chelladurai BS, Li H, Nicholson AW. A conserved sequence element in ribonuclease III processing signals is not required for accurate in vitro enzymatic cleavage. Nucleic Acids Res. 1991;19:1759–1766. doi: 10.1093/nar/19.8.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Diwa A, Belasco JG. Regions of RNase E important for 5′-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 2000;182:2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama S, Inokuchi K, Mizushima S. Promoter exchange between ompF and ompC, genes for osmoregulated major outer membrane proteins of Escherichia coli K-12. J. Bacteriol. 1984;158:1041–1047. doi: 10.1128/jb.158.3.1041-1047.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bossi L, Figueroa-Bossi N. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 2007;65:799–810. doi: 10.1111/j.1365-2958.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 36.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 37.Kaberdin VR, Walsh AP, Jakobsen T, McDowall KJ, von Gabain A. Enhanced cleavage of RNA mediated by an interaction between substrates and the arginine-rich domain of E. coli ribonuclease E. J. Mol. Biol. 2000;301:257–264. doi: 10.1006/jmbi.2000.3962. [DOI] [PubMed] [Google Scholar]
- 38.Lin-Chao S, Cohen SN. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell. 1991;65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- 39.Mackie GA. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J. Biol. Chem. 2000;275:25069–25072. doi: 10.1074/jbc.C000363200. [DOI] [PubMed] [Google Scholar]
- 40.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kime L, Jourdan SS, Stead JA, Hidalgo-Sastre A, McDowall KJ. Rapid cleavage of RNA by RNase E in the absence of 5′ monophosphate stimulation. Mol. Microbiol. 2010;76:590–604. doi: 10.1111/j.1365-2958.2009.06935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 2006;4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- 43.Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 44.Simons RW, Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu. Rev. Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- 45.Krinke L, Wulff DL. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987;1:1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- 46.Jerome LJ, van Biesen T, Frost LS. Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol. 1999;285:1457–1473. doi: 10.1006/jmbi.1998.2404. [DOI] [PubMed] [Google Scholar]
- 47.Gerdes K, Nielsen A, Thorsted P, Wagner EG. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 1992;226:637–649. doi: 10.1016/0022-2836(92)90621-p. [DOI] [PubMed] [Google Scholar]
- 48.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 49.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010;76:467–479. doi: 10.1111/j.1365-2958.2010.07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 55.Melefors O, von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988;52:893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- 56.Vytvytska O, Jakobsen JS, Balcunaite G, Andersen JS, Baccarini M, von Gabain A. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl Acad. Sci. USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 58.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.