Abstract
Enterococcus faecalis is a commensal bacterium and a major opportunistic human pathogen. In this study, we combined in silico predictions with a novel 5′RACE-derivative method coined ‘5′tagRACE’, to perform the first search for non-coding RNAs (ncRNAs) encoded on the E. faecalis chromosome. We used the 5′tagRACE to simultaneously probe and characterize primary transcripts, and demonstrate here the simplicity, the reliability and the sensitivity of the method. The 5′tagRACE is complementary to tiling arrays or RNA-sequencing methods, and is also directly applicable to deep RNA sequencing and should significantly improve functional studies of bacterial RNA landscapes. From 45 selected loci of the E. faecalis chromosome, we discovered and mapped 29 novel ncRNAs, 10 putative novel mRNAs and 16 antisense transcriptional organizations. We describe in more detail the oxygen-dependent expression of one ncRNA located in an E. faecalis pathogenicity island, the existence of an ncRNA that is antisense to the ncRNA modulator of the RNA polymerase, SsrS and provide evidences for the functional interplay between two distinct toxin–antitoxin modules.
INTRODUCTION
The ubiquitous Gram positive bacterium Enterococcus faecalis is a commensal of the intestinal tract and a major opportunistic pathogen responsible for nosocomial infections (1). Enterococcus faecalis has the ability to damage epithelial-cell DNA and may promote colonic inflammation and cancer by producing reactive oxygen species (2). The dual life style of E. faecalis hinges on a balance between host physiology, which imposes environmental constraints, and bacterial adaptation processes (1). About a dozen putative virulence genes have been reported in E. faecalis. However, the species features genomic diversity, as well as gene expression levels that vary between isolates, indicating that underlying regulatory processes are major determinants of the balance between commensalism and pathogenesis (3–5). Several chromosomal transcription regulators are involved in stress response, commensalism and/or virulence. Those identified to date affect antibiotic resistance, oxidative stress response, lysozyme resistance and tissue invasion, or may have more pleiotropic roles (6–14). In contrast, a sole example of RNA-dependent post-transcriptional control involved in commensalism and virulence has been described thus far, and concerns the utilization of ethanolamine, an abundant product of the gastrointestinal tract. Two RNA-dependent mechanisms participate in the regulation of this metabolic pathway: one involves an attenuation process in which the RNA-binding protein EutV acts in an ethanolamine-dependent manner; the other uses riboswitch elements (see below) that bind adenosylcobalamine, a cofactor required for ethanolamine degradation [(15) and references therein].
Bacteria reprogram their gene expression in response to environmental constraints partly via diverse categories of regulatory RNAs, mainly non-protein-coding RNAs (ncRNAs), antisense RNAs (asRNAs) and 5′-untranslated regions (5′-UTRs). Two classes of ncRNAs are defined according to the nature of their targets: those that bind proteins and change their activities, and those that pair to mRNA targets and affect their encoding capacity (16,17). The latter generally act via a short sequence partially complementary to their mRNA targets, which are transcribed at a different locus. ncRNAs can also act in cis when they are present in an antisense configuration with their mRNA targets (18). 5′-UTRs regulate the expression (transcription or translation) of their downstream mRNA sequence by mediating a switch in their folding. These conformational changes are induced by physical parameters (e.g. temperature or pH) or by binding to small ligands, such as metabolites in the case of riboswitches, e.g. (19–22). In addition, several mRNAs and riboswitches are known to exert regulatory functions by pairing to other RNAs, similarly to ncRNAs, e.g. (23–29). The key roles of regulatory RNAs in coordinating adaptation processes in response to environmental cues have been illustrated in many aspects of bacterial physiology, e.g. gene expression reprogramming during the exit from stationary phase, global stress response, virulence, carbon utilization, iron and envelope homeostasis, and antibiotic synthesis, e.g. (17,30–33).
The development of whole-transcriptome studies via tiling arrays or deep RNA-sequencing (RNA-seq) methods in the past two years has transformed our view on bacterial genomes, and RNA-based regulation has been revealed through the discovery of a plethora of unexpected transcripts and potential regulatory RNAs (34). To date, around fifteen ‘RNA landscapes’ have been inventoried in evolutionary distant species, revealing a general complexity of genomic organization due, in part, to the numerous promoters nested in open reading frames (ORFs), and to the existence of many ncRNAs and asRNAs (10–46% of annotated ORFs). Importantly, they establish that transcriptional organization on bacterial chromosomes cannot be deduced from the current annotation with ORFs alone [see (35) and references therein]. Despite the power of these methods, they remain demanding in terms of cost and expertise, and cannot be routinely performed in most laboratories. In addition, functional insights inferred from the majority of data published thus far are still limited as the full length RNA transcripts cannot be distinguished from the processed transcripts, which is a major issue in understanding gene expression control. Only recently, a differential RNA-seq method (dRNA-seq) has been applied to access primary transcripts by the enzymatic elimination of processed RNA forms (36–39). We present here a simple, reliable and sensitive 5′RACE-derivative method, coined ‘5′tagRACE’, in which 5′ RNA extremities are differentially tagged with two different sequences that distinguish full-length from processed RNA species. This method enables via a classical polymerase chain reaction (PCR) to specifically detect and map primary transcripts or processed ends in bacteria whose genome sequences are available. The 5′tagRACE may be used as a stand-alone method as presented here, or it may serve to generate material for high-throughput RNA sequencing to efficiently distinguish primary from processed transcripts. To benchmark the sensitivity and reliability of the 5′tagRACE, we applied the ‘classical’ approach to search for in silico predicted ncRNAs encoded by the E. faecalis chromosome. Out of 45 selected loci in the chromosome of the clinical isolate V583 (40), 28 were active for transcription. Among those, a total of 49 primary transcripts were characterized. Ten novel putative mRNAs and 29 novel ncRNAs were revealed, including 7 ncRNAs expressed from the described pathogenicity island of the E. faecalis strain (8). Remarkably, an antisense organization was demonstrated for a third of the 28 loci selected for study, among which we discovered an antisense ncRNA to the housekeeping ncRNA modulator of the vegetative RNA polymerase, SsrS (41). Results of this study present the first characterization of ncRNAs encoded by the chromosome of the human pathogen E. faecalis. They also provide the 5′tagRACE, a simple and reliable method which is generally applicable and should be of major interest for routine studies of bacterial gene expression.
MATERIALS AND METHODS
In silico predictions of ncRNAs and selection of chromosomal loci
In silico E. faecalis ncRNA predictions were done as we described previously (42), (Supplementary Figure S1). Our search was focused on chromosomal intergenic regions (IGRs) of the clinical E. faecalis isolate V583 (40,43). Positions and sequences of IGR (defined as DNA sequences between annotated ORFs) were downloaded from the TIGR website: (http://cmr.jcvi.org/cgi-bin/CMR/shared/MakeFrontPages.cgi?page=intergenicregion&crumbs=genomes). The analysis was restricted to IGRs of at least 150 bp; regions carrying rRNAs, tRNAs, known riboswitches and introns were excluded from further analysis. About 832 such loci were tallied and 236 were picked randomly for in silico predictions of ncRNAs, among which 45 IGRs were selected to be probed by 5′tagRACE (see Supplementary Data).
ORF predictions were performed for all the newly discovered transcripts, enabling us to discern non-coding RNAs (ncRNAs) from putative mRNAs (see Supplementary Data).
Construction of a 5′tag-cDNA library
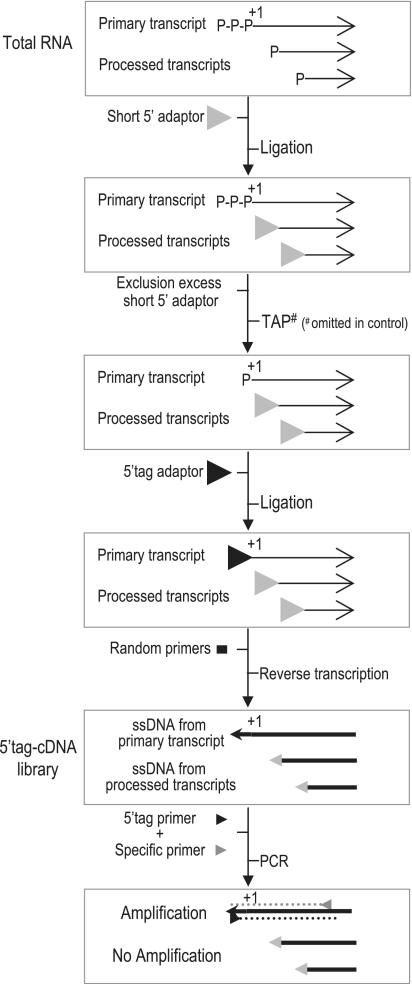
Total RNA was prepared from V583 grown in S and R conditions. Concentration and quality of RNA preparations were evaluated by measurements at 260 and 280 nm, and by denaturing agarose gel electrophoresis. A mixture of both RNA stocks (55 µg each) was used to detect ncRNAs expressed in at least one of the two conditions. DNA contamination of the RNA mixture was eliminated with 22 units of Turbo DNase, following the manufacturer’s protocol (AMBION-Applied Biosystems). About 10 µg of the RNA preparation was kept as PCR template to verify the absence of genomic DNA contamination. A 15 nt RNA adaptor (FW.5bt) was added to the remaining 100 µg of RNA at 50 pmol FW.5bt/µg bacterial RNA (Supplementary Table S1). The 5′-monophosphate groups carried by 5′-processed RNA ends or DNA oligonucleotides generated by the DNase treatment were ligated to FW.5bt using T4 RNA ligase (New England Biolabs) at 4 U/µg bacterial RNA; the ligation was performed for 18 h at 16°C in 10% dimethyl sulfoxide (DMSO). The excess of non-ligated adaptor FW.5bt was removed by exclusion chromatography using Micro Bio-Spin® 30 columns (BioRad). In a series of experiments, we previously determined that these columns do not selectively retain folded RNAs, nor affect their integrity, by determining recovery efficiency of synthesized RNA (data not shown). Half of the RNA mixture (∼50 µg) recovered from the column was then treated with the tobacco alkaline phosphatase (TAP; Epicenter Biotechnologies) at 2 U/µg RNA, transforming the 5′-triphosphate groups present at the 5′-ends of primary transcripts (‘transcription start site’ or ‘TSS’) into monophosphate groups (44). The TAP-treated RNA stock is referred to as ‘+T’. Identical treatments were applied to the other half of the RNA mixture, except that TAP was omitted, providing a negative control RNA mixture (−T) for the specific action of TAP on primary transcripts. RNA +T and −T stocks were ligated to a second RNA adaptor (FR.RNA5; Supplementary Table S1) at 75 pmol/µg of bacterial RNA. Note that the sequence and length of FR.RNA5 (38 nt long, Supplementary Table S1) may be adapted for specific uses such as high-throughput sequencing. The equivalent of 4 µg bacterial RNAs +T and −T RNAs were then reverse transcribed using the random primer NONAX and the SuperScriptTM III reverse transcriptase (ssIII), (Invitrogen), using 75 pmol NONAX/1 µg bacterial RNA and 50 U ssIII. Bacterial RNA and the NONAX random primer were mixed, incubated for 5 min at 65°C, and rapidly cooled on ice. The ssIII was then added and the annealing of NONAX to bacterial RNAs was allowed by incubating the mixture 5 min at room temperature. Reverse transcription was performed by four successive steps of 15 min at 42, 50, 55 and 60°C. The reaction was stopped by a 5 min shift to 85°C, and RNA was eliminated by adding RNase H (New England Biolabs) for 20 min at 37°C. Reverse transcribed samples +T and −T both contain single-stranded cDNA synthesized from RNA processed forms tagged at their 5′-ends by the oligonucleotide FW.5bt. The differential use of TAP generates specifically tagged TSSs by the oligonucleotide FR.RNA5 in the +T sample (this constitutes the ‘5′tag-cDNA library’), in contrast to −T which contains untagged TSSs (Figure 1).
Figure 1.
Principle of the 5′tagRACE method. 5′ monophosphate RNA extremities generated by processing or degradation are ligated to an excess of a 5′ RNA adaptor (‘Short 5′ adaptor’), resulting in inactive 5′-ends for subsequent ligation steps. The excess of 5′ RNA adaptor is eliminated via exclusion chromatography. 5′ triphosphate ends of primary transcripts (i.e. ‘+1’ or ‘TSS’) are then transformed into 5′ monophosphate by the TAP enzyme and ligated to a second 5′ RNA adaptor (‘5′tag adaptor’). Reverse transcription is performed using random primers to generate a cDNA library (5′tag-cDNA library) containing two types of 5′tagged single-stranded-DNA molecules: those ligated to the first 5′ adaptor (Short 5′ adaptor), i.e. processed ends, and those ligated to the 5′ tag adaptor, i.e. TSSs. The use of a specific primer to the 5′ tag adaptor and an oligonucleotide specific to the selected RNA (transformed into cDNA) will allow the specific amplification by PCR of the cDNA synthesized only from the primary RNA. An untreated TAP 5′tag-cDNA library is used as negative control (hash sign).
The 5′tag-RACE procedure
Within the 5′tag-cDNA library (+T), a given transcript was detected by PCR using a specific oligonucleotide and the primer FR.DNA5 which specifically hybridizes to the reverse transcribed sequence of FR.RNA5 ligated to TSSs (Supplementary Table S1). Amplifications were performed using 0.5 pmol/µl of specific primer, 1.25 pmol/µl FR.DNA5, 1.25 U Hot Gold Star DNA polymerase (Eurogentec, S.A.) and 0.5 mM dNTPs in final a 20 µl reaction. The amount of cDNA template used was arbitrarily defined as the concentration of reverse transcribed bacterial RNA utilized for the PCR reaction and named hereafter ‘Equivalent RNA’ (Eq.RNA). We determined a range of Eq.RNA values for which primary transcripts of the predicted ubiquitous ncRNAs (SsrS, SsrA, RnpB and Ffs) were specifically detected (data not shown). For our studies, Eq.RNA values ranged from 1.25 pg/µl to 1 ng/µl. PCR conditions applied were: 10 min at 94°C, and then 40 cycles (94°C, 40 s; 58°C, 40 s; 4 min at 72°C), and 10 min at 72°C. When using the +T sample as template, the specificity of amplified products was deduced by comparing bands to those obtained with the −T sample. The authenticity of amplicons was then confirmed by sequencing and the TSS was deduced as the first nucleotide of the targeted transcript ligated to the 3′-end of the FR.DNA5 oligonucleotide. Sequencing was performed directly on the amplicon when a single PCR product was obtained or after cloning of the PCR products in the pCRII-Topo vector when several amplicons were obtained.
3′-end mapping
The RNA samples used to generate the 5′tag-cDNA library were also used for 3′ mapping. Eighty micrograms of DNase-treated RNA was dephosphorylated with 40 U of calf intestinal alkaline phosphatase (New England Biolabs) for 1 h at 37°C. The 3′RNA adaptor FR.idT.R1 was then added to the treated RNA to a 75 pmol/µg RNA concentration (Supplementary Table S1). The RNA mixture was incubated for 5 min at 90°C and cooled on ice before adding T4 RNA ligase buffer and DMSO to a 10% final concentration. The RNA mixture was then split into two equal volumes each containing ∼40 µg RNA. Four units of T4 RNA ligase per µg of bacterial RNA was added to one of the samples and replaced by water in the second sample. Samples were then incubated for 18 h at 16°C, and then reverse transcribed using the random NONAX primer and the ssIII system in the same conditions as described for the 5′tag-cDNA library. The detection of specific 3′ extremities was performed by PCR using a specific primer for hybridization with a selected RNA and the oligonucleotide FR.3tail.D1 (Supplementary Table S1), as described for the 5′-end mapping. Specific amplicons obtained with the RNA samples treated here above with T4 RNA ligase were sequenced. The amounts of cDNA template employed for 3′-end mapping were in the same range as those used for the 5′tagRACE (from 1.25 pg/µl to 1 ng/µl Eq.RNA).
Nomenclature of RNAs characterized
Each primary transcript detected by 5′tagRACE and identified by sequencing was named ‘Ref’ for RNA in E. faecalis. Each ref gene was arbitrarily assigned to the number of the original IGR and the name of the specific primer used for the 5′tagRACE, e.g. ref1C corresponds to the gene found in the IGR FR1 and whose transcript was detected with the primer FR1C (Supplementary Table S1 and S2). When several TSSs were mapped with a single oligonucleotide, numbers were added with respect to genome coordinates; e.g. ref8C1 locates upstream ref8C4.
Standard procedures
Bacterial strains, growth conditions, plasmids, primers and RNA procedures are provided as supplementary data.
RESULTS AND DISCUSSION
Prediction of non-coding RNAs
The majority of known chromosomal bacterial ncRNAs are expressed from genes nested in sequences flanked by annotated ORFs (IGRs), e.g. (37,39,42,45–50). We therefore searched for ncRNAs in the IGRs of E. faecalis V583 chromosome (40). Out of 883 total IGRs, 236 were picked randomly and analyzed in silico for putative ORFs, RNA secondary structures and ‘orphan’ transcriptional terminators as described (42). Among them, 45 IGRs were selected as potentially containing ncRNAs genes and kept for further investigation based on the absence of putative translatable ORF and an empirical favorable defined RNA folding (42), thus excluding 191 IGRs (Supplementary Figure S1 and Table S4). Among the 45 selected IGRs, 4 were predicted in the Rfam database to correspond to the highly conserved bacterial ncRNAs, SsrS (6S), RnpB, Ffs (4.5S) and SsrA (tmRNA), [(42,46) and references therein], and another was flanked by syntenic genes pnpA (ef3064) and rpsO (ef3065), and shown in Escherichia coli and Listeria monocytogenes, two divergent bacterial species, to contain an asRNA to pnpA, SraG and RliD, respectively (42,45).
The 5′tagRACE procedure to probe for selected primary transcripts
Thus far, in silico predictions of ncRNAs have been experimentally tested by Northern blot [e.g. (42,45,46,48,51)], which confirms the existence, orientation and relative abundance of a given transcript. However, high numbers of predicted candidates render the method tedious, and detection may fail when RNA amounts are below a certain threshold (38). To systematically screen our predictions and as a more information-rich alternative to northern blotting, we developed a simple and sensitive 5′RACE-derivative method that we named 5′tagRACE, which allows the selective amplification of primary RNA 5′-ends (Figure 1). The 5′tagRACE was inspired from the detailed 5′RACE protocol (44), and its original description (52), and consists of: (i) ligation of a first 5′RNA adaptor (FW.5bt) to the 5′-monophosphate group present at the 5′ processed ends of RNA species, (ii) enzymatic modification of 5′-triphosphate groups of primary transcript ends (TSSs) into 5′-monophosphate groups, and (iii) ligation of resulting 5′-monophosphate ends to a second 5′-RNA adaptor (FR.RNA5), (Figure 1). The treated RNA is then reverse transcribed to generate a cDNA library (‘5′tag-cDNA library’) where 5′-RNA ends can be distinguished according to the sequence of the ligated 5′-RNA adaptor. By using primers specific to a selected RNA (in this study, RNAs predicted by bioinformatics) and to the 5′-adaptor ligated to ‘TSSs’, the PCR amplification step reveals the primary transcripts of interest (Figure 1). In contrast to the classical 5′RACE, only DNA strands which are complementary to single strand cDNAs synthesized from primary transcripts by reverse transcription are used as template by the Taq DNA polymerase in the 5′tagRACE procedure (Figure 1).
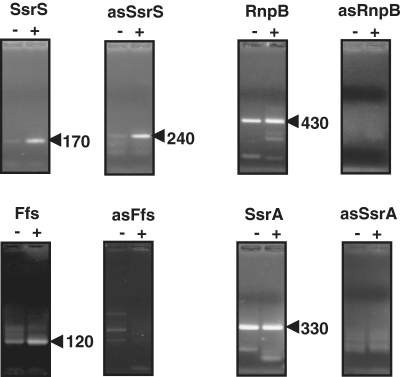
In a series of pilot experiments we applied the 5′tagRACE method to identify conserved housekeeping ncRNAs SsrS, SsrA, RnpB and Ffs, observed in diverse bacterial species to be highly abundant in their processed and active forms, but whose primary transcripts are present in low amounts [(53) and references therein]. For those ubiquitous ncRNAs, we determined that their detection was specific within Eq.RNA values ranging from 1 ng/µl to as low as 1.25 pg/µl (see ‘Materials and Methods’ section; Figure 2). Sequencing of amplicons confirmed the presence of selected primary transcripts in their predicted orientation and allowed us to map TSSs for ssrS, rnpB, ffs and ssrA in E. faecalis V583. Sequences and orientations obtained for these ncRNAs agreed with sequences predicted in the Rfam database. Note that 5′tagRACE uses at least 10-fold less total RNA than the classical 5′RACE method (44,52). To further confirm the validity of 5′tagRACE, we performed negative controls by searching for a priori non-existing RNAs; this was done by probing our 5′tag-cDNA library for RNAs transcribed in the opposite direction (antisense) to ssrS, rnpB, ffs and ssrA genes, using specific oligonucleotides nested in the sequences of the four known ncRNAs (Supplementary Table S1). Within the range of cDNA template defined above (from 1.25 pg/µl to 1 ng/µl of Eq.RNA) and as expected, no asRNAs were detected for RnpB, Ffs and SsrA (Figure 2), attesting to the reliability of the 5′tagRACE method. Unexpectedly, we detected an antisense transcript to SsrS, which was further confirmed by sequencing and northern blotting (see below).
Figure 2.
Probing for the housekeeping ncRNAs by 5′tag RACE. Names of RNAs probed are indicated at the top of each panel, antisense RNAs are denoted by ‘as’. Amplicons were separated on 2.5% agarose gels and stained with ethidium bromide. At the top of each lane, ‘−’ and ‘+’ refer to untreated and treated TAP RNA samples, respectively. The arrow heads show amplicons that were sequenced and their estimated lengths (nt) on gel. The amount of template used in these set of experiments was 0.05 ng Eq.RNA. With this amount of template, amplicons obtained for RnpB and SsrA with treated and untreated TAP samples were in equivalent amounts.
Taken together data obtained with the housekeeping ncRNAs SsrS, RnpB, Ffs and SsrA validate 5′tagRACE as a robust method to reveal the existence of unknown transcripts. It also lifts uncertainties associated with the classical 5′RACE method and northern blotting, in particular, when the RNA under study is predominant in its processed forms or when it competes with abundant non-specific RNAs, mainly ribosomal RNAs that constitute up to 80–90 % of total RNA (54).
Detection and characterization of predicted ncRNAs within selected IGRs
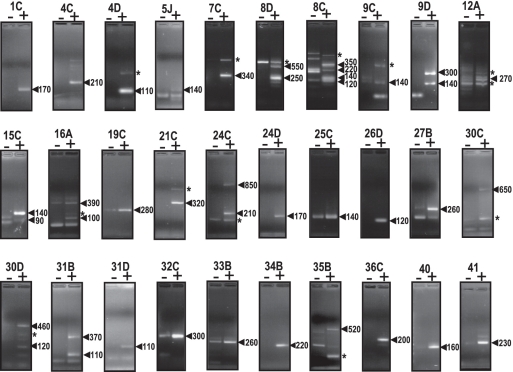
Primary transcripts encoded by in silico selected IGRs (Supplementary Table S2) were probed within the range of Eq.RNA values defined for SsrS, SsrA, RnpB and Ffs ncRNAs (from 1.25 pg/µl to 1 ng/µl). A single specific primer was used for each candidate with a predicted orientation, as deduced from detection of a transcription terminator (Supplementary Table S2). For candidates whose transcription orientation was uncertain, the two possibilities were probed by using two specific and complementary primers (Supplementary Table S1). In total, 66 transcripts were tested within the 45 IGRs (Supplementary Table S2; the 4 IGRs carrying ubiquitous ncRNAs are considered here). Seventeen of those IGRs either failed to give signals, or gave signals that were complex or non-specific, and were not analyzed further (Supplementary Table S2, Supplementary Figure S2). Among the remaining 28 IGRs, sequencing of resulting products enabled us to map a total of 45 TSSs (Figures 3 and 4 and Supplementary Figure S3). In this set of experiments, the asRNA detected for SsrS (Ref44 or ‘asSsrS’ in Figure 2), was also sequenced and confirmed (Figure 2 and Supplementary Figure S3). Thus, data obtained with the four housekeeping ncRNAs and the pool of predicted Ref candidates demonstrated the reliability and sensitivity of the 5′tagRACE method (Supplementary Figure S3). For comparison, we independently tested the feasibility of using the classical 5′RACE method to map TSSs for genes ssrS, ref44, ref25C and ref40 (44). Only Ref40 provided an unambiguous TSS signal identical to that obtained with the 5′tagRACE method, while data from the other samples were inconclusive (Supplementary Figure S4 and data not shown). To further confirm the reliability of 5′tagRACE, the end-mapping data was directly used as a guide for plasmid overexpression of selected Ref RNAs under control of their own promoters (e.g. ref25C, ref40, ref44 and ssrS). Results obtained confirmed that the characterized TSSs and promoter predictions were real (Supplementary Figure S5, and see below). The above analyses demonstrate the selectivity, sensitivity and reliability of the 5′tagRACE method.
Figure 3.
Primary transcripts detected by 5′tagRACE in IGR candidates. Names of oligonucleotides used to probe each RNA candidate are indicated at the top of each panel. Signs ‘+’ and ‘−’ refer to treated or untreated TAP RNA samples, respectively. Numbers on the right indicate lengths estimated for 5′tagRACE PCR products observed on 2.5% agarose gels. Amplicons below ∼80 bp were considered as aberrant products due to the average length of the specific primer (∼25–30 nt) and the 5′tag RNA adaptor (38 nt), (Supplementary Table S1). Asterisk: signals not sequenced or corresponding to an unrelated primary transcript as determined by sequencing. 5′tagRACE data obtained for rejected IGR candidates are shown in Supplementary Figure S2.
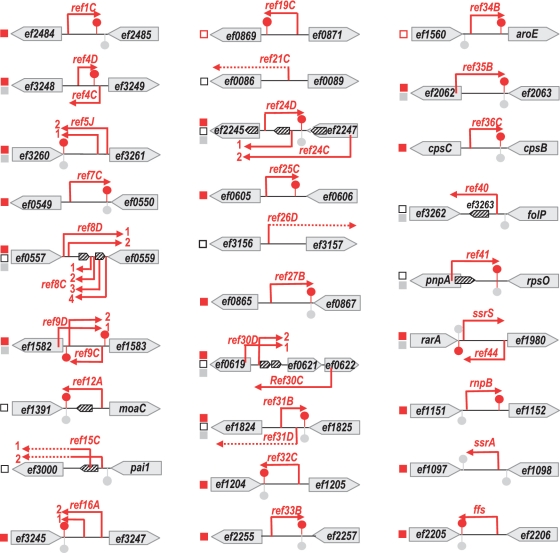
Figure 4.
ref loci and features of Ref RNAs. Squares are: ‘red filled’ for ncRNAs; ‘red empty’ for ncRNAs or riboswitches; ‘black empty’ for potential 5′-UTRs; and ‘gray’ for antisense organization. Red arrows symbolize characterized transcripts, dotted lines represent putative 5′-UTRs. Red and gray lollipops represent putative transcription terminators. Annotated ORFs in the V583 genome are shown by gray horizontal arrows, and predicted ORFs in Ref RNA sequences are symbolized by diagonally striped bar arrows. Representation is not at scale; sequences are provided as supplementary data (Supplementary Figure S3).
To further illustrate the general interest of the 5′tagRACE method, the 5′tag-cDNA library used in this study was submitted to deep sequencing. From our data set, we recovered identical TSSs as those mapped for several mRNAs by others, e.g. sodA, ace, glnQ, fsrB and gelE (55–58), attesting to the robustness of the 5′tagRACE. In depth analysis of whole genome data by combining 5′tagRACE and RNA-seq technology will be presented elsewhere.
3′-end mapping of unveiled primary transcripts
To get further insight on Ref RNAs, their 3′-ends were mapped using the 3′RACE method and specific primers complementary to those used for 5′tagRACE (Supplementary Table S1). In some cases, 3′-end mapping revealed long amplicons (>500 nt) and complex patterns (Refs 21C, 15C1-2, 26D and 31D), indicating that these transcripts most likely comprise downstream ORFs, and could correspond to 5′-UTRs; these were not analyzed further. In several cases (Refs 1C, 4D, 5J1-2, 19C, 25C, 27B, 32C and 34B, and RnpB), the mapped 3′-ends corresponded to the predicted rho-independent transcription terminators (Figure 4 and Supplementary Figure S3). For Ref44 and Ffs, the 3′-extremities we determined did not correspond to predicted transcription terminators, suggesting rapid processing between the transcription terminator and the specific primer used, and/or steric hindrance during the ligation step due to RNA secondary structures (Supplementary Figure S3). No transcriptional terminators were predicted for Refs 8C1-4, 8D1-2, 24C1-2, 30C and 30D1-2, and SsrS. The 3′-ends of these RNAs overlap with other transcripts expressed from the reverse complementary DNA strand, indicating an antisense organization (Figure 4 and Supplementary Figure S3). Interestingly, the absence of predictable transcription terminators for these Ref asRNAs evokes recent observations in Synechocystis sp., which suggested that asRNA counterpart genes may provoke transcriptional arrest or mutual attenuation without requiring intrinsic terminators (59).
Features of ref genes
Figures 4 and 5 summarize characteristics and positions of the 45 identified E. faecalis V583 Ref RNA sequences. Below we classify these RNAs according to their main features.
Figure 5.
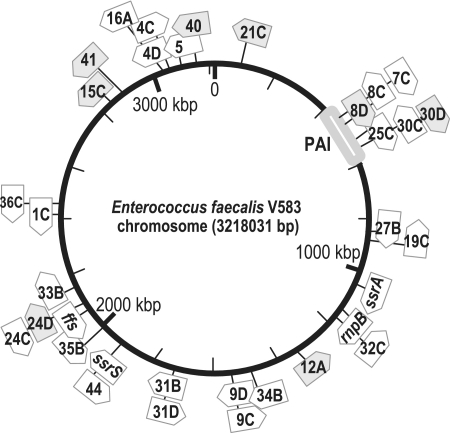
Distribution of Ref RNAs along the chromosome of V583 E. faecalis strain. ncRNAs characterized are shown in white; potential mRNA and 5′-UTRs are in gray. Arrows indicate transcription orientation. PAI: pathogenicity island.
Non-coding Ref RNAs
Among the characterized transcripts (including SsrS, RnpB, SsrA and Ffs), no translatable ORFs were found in sequences of 31 of them, allowing us to conclude that these transcripts are most certainly ncRNAs. Among those, 27 Ref RNAs were novel and their primary lengths ranged from 128 to 755 nt (Supplementary Table S2 and Figure S3, Figure 4). Two classes of Ref ncRNA genes were distinguished according to their transcriptional organization: (i) ncRNA genes embedded in annotated IGRs (e.g. ref7C, 8C1-4, 19C, 25C, 27B, rnpB), resembling the vast majority of ncRNA genes described to act on trans encoded mRNAs or proteins; (ii) ncRNA genes spanning protein-encoding genes transcribed in the opposite direction, which may act as antisense. Seven ref ncRNA genes (ref7C, 8C1-4, 25C, 30C) are located within the V583 pathogenicity island (PAI) and present in closely related E. faecalis isolates (Supplementary Table S3). To our knowledge, this is the first description of ncRNAs expressed from the E. faecalis chromosome. This collection, and the approach used for its identification, should open the way for characterizing RNA-based processes that govern the physiology of a major bacterial agent of nosocomial infections.
Non-coding RNAs or novel riboswitches?
Sequence analysis of Ref19C and Ref34B showed the absence of translatable ORFs, suggesting that these RNAs would also be non-coding transcripts. However, their transcription terminators are spaced by ∼100 nt from translation start codons of the downstream ORFs (Ef0869 and AroE, respectively) transcribed in the same direction, which resembles the organization described for riboswitches or regulatory 5′-UTRs (Figure 4), (21). Although Ref19C and Ref34B RNAs were not predicted within Rfam database as 5′-UTRs regulatory elements, we cannot fully rule out such a possibility (Figure 4 and Supplementary Figure S3), (60).
Ref RNAs encoding peptides
Ten characterized Ref RNAs were predicted to contain small translatable ORFs, ranging in size between 24 and 54 amino acids. None of them was predicted earlier (61). Properties and analysis of these putative ORFs are summarized in Supplementary Figure S3. Among these Ref mRNAs, remarkably Ref8D1 and Ref8D2, located on the PAI, are reminiscent of the group I toxin–antitoxin (TA) modules, since they encode a 35 amino acids peptide predicted to contain a trans-membrane domain (the toxin) and are transcribed from the complementary strand to ref8C1-4 genes expressing antisense ncRNAs (the antitoxins), (62). Besides putative encoded toxins, peptides are known to participate in regulatory processes related to cell–cell communication in Gram positive bacteria, such as virulence in Staphylococcus aureus, competence in Bacillus subtilis and Streptococcus pneumonia and ‘sex’ in E. faecalis (63–65). Functions of predicted small ORFs in Ref RNAs remain to be established, but as in other species, their occurrence likely reflects a variety of physiological aspects where they participate, e.g. (39,66).
Another peptide-encoding RNA was ref41, which bears a potential translatable 43 amino acids ORF transcribed on the opposite strand to the highly conserved pnpA gene, encoding the polynucleotide phosphorylase (40). A similar antisense gene was previously reported in two evolutionary distant species, E. coli and L. monocytogenes, respectively called sraG and rliD (42,45), indicating a functional relevance and the possible ubiquity of ref41 in the bacterial kingdom.
Antisense organization of ref loci
Ten of the 28 IGRs displayed transcriptional activity on both DNA strands, thus generating antisense organizations (Figure 4 and Supplementary Figure S3), an occurrence that correlates with the range of frequencies estimated in whole genome analysis using tiling arrays or RNA-seq methods [∼30 %; e.g. (39,49,59)]. Three classes of antisense organizations can be defined according to the nature of transcripts involved: (i) ncRNA antisenses to mRNAs, e.g. ref8C1-4/Ref8D1-2, Ref24D/Ref24C1-2, Ref30C/ef0621-22 and ref35B/ef2062; (ii) ncRNAs antisenses to ncRNAs, e.g. ref4D/ref4C, ref9D1-2/ref9C, and among those ref44 which encodes an asRNA to the RNA polymerase modulator ncRNA, SsrS (Figure 4 and Supplementary Figure S3); and (iii) mRNAs antisenses to mRNAs, e.g. ref40/ef3262 (see below), ref41/pnpA, ref8D1-2/ef0559 and ref30D1/ef0619. The diverse antisense transcriptional organizations found in this study for E. faecalis IGRs reflect those described in other bacterial species and in the eukaryotic kingdom (35,67). Antisense organization of genes can result in expression regulation at transcriptional and post-transcriptional levels. Generally, the expression of one gene hampers the expression of its counterpart by different, not necessarily exclusive, mechanisms, such as transcription initiation interference, premature transcription elongation arrest, translation repression and degradation of the formed RNA duplex (18,68). Our finding constitutes the first experimental characterization of antisense organizations in the opportunistic human pathogen E. faecalis chromosome. The apparent universality of gene expression control by asRNAs and the emerging examples of their contribution to bacterial virulence (69,70) provide to our results a significant importance towards the understanding of the versatile life style of E. faecalis.
Conservation of ref genes
ref genes can be distinguished according to their conservation in other bacterial genomes and their occurrence within the E. faecalis species. Besides the 4 housekeeping ncRNA genes (ssrS, rnpB, ssrA and ffs), only a few ref genes were found to be significantly conserved in closely related species (enterococcus sp.), (ref30C/D1-2, 34B, 35B, 41), and in other Firmicutes (ref7C in S. aureus and B. anthracis), (Supplementary Table S3). Among the 25 sequenced E. faecalis genomes (40,71,72), ref genes were either present in all sequenced isolates (ref1C, 9C/D, 19C, 27B, 32C, 34B, 35B, 41), or limited to a few E. faecalis isolates, including those localized in the PAI. Comparative genomic studies revealed that about two-thirds of the E. faecalis chromosome corresponds to the core-genome, and as in other bacteria, the pan-genome, defined by genomic islands and mobile elements, harbor virulence and antibiotic resistance traits, as well as accessory factors participating in adaptative functions and niche preferences (5,73). It is thus tempting to speculate that ref genes systematically present would ensure a central role in the maintenance of the E. faecalis species, while non-conserved ref genes would confer specific genetic traits, and each ‘ref repertoire’ would contribute to individual fitness capacities of isolates.
Expression of Ref RNAs in selected growth conditions
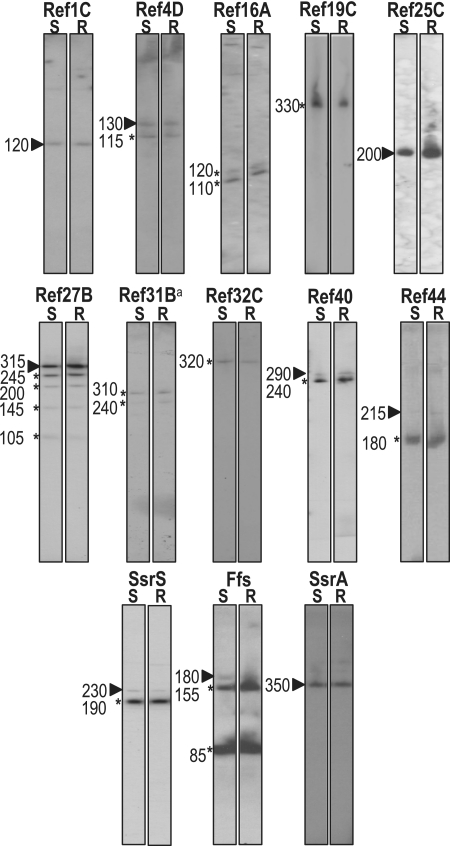
In the presence of oxygen, E. faecalis generates massive amounts of superoxide radicals, which can be damaging to the surrounding cellular host environment. However, in the presence of hemin and oxygen, metabolism is rerouted from fermentation to respiration. When this occurs, the production of superoxides is arrested, as reducing equivalents are routed to the respiration chain (74). These differential properties, which depend on hemin availability, are suspected to contribute to E. faecalis commensalism and pathogenicity (2,75,76). We pursued the characterization of Ref RNAs in northern blot experiments by visualizing their expression in bacterial growth conditions where oxygen and hemin were differentially available. Although northern blots are less sensitive than RT-PCR-based methods, e.g. (38), they provide information on processing and relative expression levels that is unavailable by the latter method. From the 28 IGRs expressing the Ref RNAs discovered via the 5′tagRACE method, 13 were readily visualized (Figure 6): Ref1C, 4D, 16A, 19C, 25C, 27B, 31B, 32C, 40, 44, SsrS, SsrA and Ffs. In most cases, lengths of RNA molecules estimated by northern blot were in good agreement with sizes deduced from the RACE data (Supplementary Table S2). The amounts of Ref RNAs were comparable in both growth conditions used here, except for Ref25C (see below). Complex expression patterns were observed for a few RNAs (Ref27B, 40, SsrS and Ffs), most likely due to specific processing of primary transcripts, e.g. (77–79). The unambiguous mapping of the TSSs via the 5′tagRACE method proved valuable for dissection of complex expression profiles observed on northern blots (e.g. Ref27B), and could be used in tiling array experiments when detecting operonic mRNAs, e.g. (50).
Figure 6.
Detection of Ref RNAs by northern blot. Names of detected transcripts are indicated at the top of each panel. Probes used were oligonucleotides designed for 5′tagRACE characterization (Supplementary Table S1). About 10 µg of total RNA was loaded in each lane: on left, static growth conditions (S), and on the right, respiratory (R) growth conditions. Arrow-heads show transcripts with an estimated length agreeing with sizes deduced from end-mapping of primary RNAs, asterisks indicate processed forms. Only 1 µg of total RNA was loaded for SsrS, SsrA and Ffs RNAs. (a): see comments and legend in Supplementary Table S2.
Ref44, an antisense of the RNA polymerase modulator SsrS
SsrS (or 6S) is a ncRNA that accumulates throughout growth and undergoes endoribonucleolytic processing. In E. coli, matured SsrS binds to the RNA polymerase holoenzyme Eσ70 and modulates its promoter usage, thereby defining a subset of genes transcriptionally dependent on the ncRNA. When bacteria enter stationary phase, ∼80% of Eσ70 is sequestered in a SsrS–Eσ70 complex. Upon outgrowth from stationary phase, SsrS–Eσ70 complex becomes transcriptionally active, and 14–20 nt RNA products (pRNA and pRNA*) are synthesized, leading to complex dissociation. Transcriptional activity of the SsrS–Eσ70 complex responds to nucleotide concentration and SsrS adjusts the bulk of Eσ70-dependent transcription to metabolic resources (39,41,80).
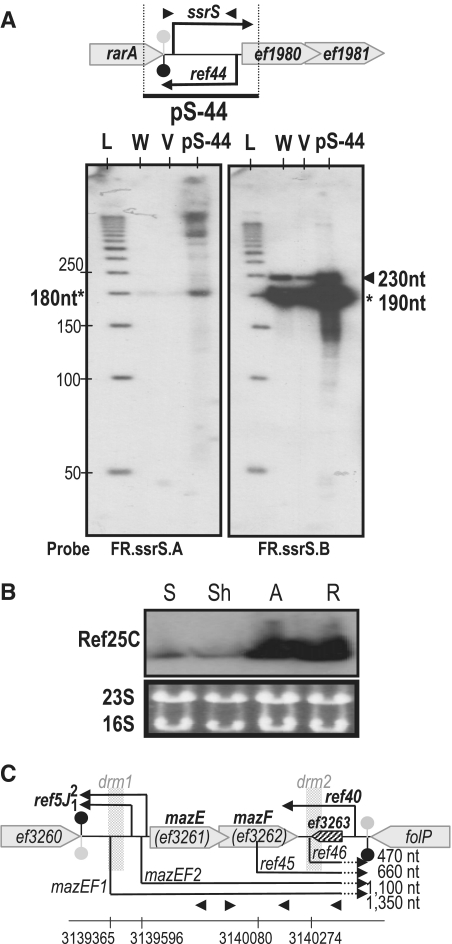
Unexpectedly, this study revealed the existence of Ref44, an antisense of SsrS. To confirm this finding, a genomic DNA fragment containing both genes was cloned in a multicopy plasmid vector, and introduced into V583. In this construct, Ref44 and SsrS should be expressed from their own promoters as deduced from 5′tagRACE mappings. Total RNA from this strain was separately and specifically probed for SsrS and Ref44. Both ncRNAs were overexpressed, in keeping with the TSS assignments and attesting that ref44 indeed encodes an antisense RNA to SsrS (Figure 7A). The function of ref44 is unknown, but as antisense of ssrS, these genes could interfere at transcriptional and post-transcriptional levels. The formation of an SsrS/Ref44 RNA duplex could participate in either degradation or maturation of SsrS, as suggested by our data showing that both ncRNAs are mainly present as processed products (Figure 7A and Supplementary Figure S3). In both scenarios, Ref44 may thus modulate the amount of SsrS available to affect Eσ70 activity. To date, we do not know whether such an asRNA is only present in E. faecalis species, or beyond, as no antisense to SsrS has been reported in whole-genome transcriptomic data. Our attempts to identify such RNA species by northern experiments in E. coli, B. subtilis and S. aureus did not reveal the existence of an SsrS antisense (our unpublished data).
Figure 7.
Characterization of three ref loci. (A) Expression of SsrS and its antisense Ref44. The genome organization of ssrS and ref44 genes deduced from 5′tag- and 3′-end mapping is shown in the upper part of the panel. Arrowheads indicate oligonucleotide used as probes for northern blots. ‘pS-44’ : plasmid construct based solely on mapping data and containing the genomic region represented by the black line. The existence of the Ref44 RNA is verified by overexpression-based 5′- and 3′-end mapping (lower part of panel 7A). About 5 µg total RNA was loaded in each lane when probing for Ref44 (right panel), and 1 µg was loaded for SsrS (left panel), respectively. Lanes noted: ‘W’, wild-type V583 strain; ‘V’, V583 strain harboring the plasmid vector; ‘pS-44’, V583 strain transformed by the plasmid pS-44; ‘L’, 50 base DNA ladder. Lengths indicated for Ref44 and SsrS correspond to the most abundant forms detected. Note that DNA (L) runs slightly faster than RNA. Exposure times were optimized for each panel and signal intensities do not indicate relative abundance between Ref44 and SsrS. Legend is otherwise as in Figure 6. (B) Oxygen-dependent expression of Ref25C ncRNA. Samples were taken at OD600 between 0.80 and 0.90. Growth conditions are indicated at the top of each lane: ‘S’ and ‘Sh’ correspond to static growth without and with hemin, respectively; ‘A’ and ‘R’ correspond to aeration growth without and with hemin, respectively. About 10 µg total RNA was loaded in each lane and the oligonucleotide FR.25C was used as probe for Ref25C. Ribosomal RNAs were used as loading controls (1 µg per lane). (C) Transcriptional organization of the overlapping group I and II TA modules. Location and orientation of specific oligonucleotides used for 5′tag- and 3′RACE analysis of mazEF1-2, Ref45 and Ref46 transcripts are indicated by arrowheads. Direct repeat motifs drm1 and drm2 are shown by gray boxes. A single and common transcription terminator for mazEF1-2, Ref45 and Ref46 RNAs was predicted (shown in black) and corresponds to the bidirectional terminator predicted for folP (in gray). Putative transcription termination ends for primary transcripts mazEF1-2, Ref45 and Ref46 RNAs are shown by dashed lines; numbers at the end of those transcripts are estimated length (see Supplementary Figure S3). Genome coordinates of TSSs mapped for mazEF1-2, Ref45 and Ref46 are given at the bottom of the panel.
Ref25C is induced in oxidative stress conditions
We observed that amounts of Ref25C were increased 5- to 10-fold when E. faecalis was grown in the presence of hemin and oxygen compared to static conditions without hemin (‘R’ and ‘S’ conditions, respectively; Figure 6), suggesting a functional relevance of this ncRNA in response to environmental oxygen and/or hemin. To determine whether oxygen, hemin or the combination was responsible for the observed variations, northern blots were performed on total RNAs extracted from bacteria grown in four different environments: static conditions without and with hemin, (‘S’ and ‘Sh, respectively), and aerated conditions without and with hemin (‘A’ and ‘R’, respectively), (Figure 7B). Ref25C showed a similar increased abundance in aerated conditions regardless of hemin, compared to static growth conditions, indicating that Ref25C is upregulated in conditions where oxygen is present in the medium (Figure 7B). ref25C localizes in the PAI originally described for the V583 isolate and other closely related strains of E. faecalis (Figure 5, Supplementary Tables S2 and S3), (8). This gene appears as a single transcriptional unit ending at a rho-independent transcription terminator and is flanked by genes ef0605 and ef0606, encoding a protein of unknown function and one of the two Dps-like proteins contained in the V583 chromosome, respectively, (Figure 4 and Supplementary Figure S3); the other dps gene is ef3233 (40). Dps plays a critical role in oxidative stress by protecting DNA from damage, and contributes to virulence in various bacterial pathogens (81). The location of ref25C within the PAI, its vicinity to a dps gene and its induction in high oxygen conditions, may suggest its involvement in oxidative stress response. We are currently investigating the functional role of ref25C in V583.
Antisense toxin–antitoxin modules I and II
ref5J1-2 and ref40 genes are transcribed in the same direction and flank the annotated bi-cistronic operon ef3261–ef3262 expressed from the reverse complementary strand. Remarkably, Ref5J1-2 and Ref40 RNA sequences contain a 167 nt direct repeat motif, called here ‘drm1′ and ‘drm2′, respectively (Figure 7C and Supplementary Figure S6). The intervening genes, ef3261 and ef3262, are annotated in the V583 genome as two transcriptional regulators of the broad AbrB- and PemK-protein superfamilies, respectively (40). However, closer inspection by PSI-BLAST revealed Ef3261 (76 amino acids) to be a member of the antitoxin DNA-binding protein MazE family, and Ef3262 (121 amino acids) to be a member of the MazE counterpart toxin protein family MazF. Thus, this operon constitutes a type II toxin–antitoxin module, as also suggested in a comparative-genome analysis of TA systems in prokaryotes (82), and will be here called ‘mazEF’. Intriguingly, the small ORF Ef3263 (27 amino acids) contained in Ref40 RNA, lies just downstream MazF and is transcribed from the complementary strand (Figure 7C). Ef3263 was recently described as a member of the TxpA toxin family, a type I TA module in which an asRNA counteracts synthesis of the peptide toxin (83). Thus in V583, the genetic locus flanked by TSSs of ref5J1-2 and ref40 genes, comprises type I and type II TA modules transcribed in opposite directions, which suggests that they may impact on the expression of each other. To get a first insight on the possible relationship between these TA systems, we performed in-depth mapping of the transcripts expressed from this locus by 5′tag- and 3′RACE. Data analysis revealed a complex transcriptional organization and rather unusual characteristics for the two distinct groups of TA modules. In addition to Ref5J1-2 and Ref40 RNAs, four other transcripts were found within this region and the complex pattern of overlapping RNA transcripts supports the above hypothesis that expressions of the two TA systems are inter-related (Figure 7C and Supplementary Figure S3).
Interestingly, in the E. faecalis OG1RF isolate, the mazEF operon is absent, and the duplicated drm is resolved into a single 167 bp sequence embedded in a classical ref40/ref46 group I TA module, making this locus variable among E. faecalis species (Supplementary Figure S6 and Supplementary Table S3). TA modules are widespread, including in commensal and pathogenic species where they impact on growth regulation and diverse physiological adjustments such as stress adaptation, antibiotic resistance, biofilm development and persistence in chronic infections (84–86). Biological functions of mazEF and ref40-46 TA modules are unknown in E. faecalis, but it may be assumed that these systems also contribute to host-adaptation processes faced by the bacterium when living in the commensal and/or pathogenic state (86). It is also remarkable that in contrast to the ref40-46 TA system which is conserved among E. faecalis strains, the mazEF module seems restricted to a limited number of isolates (Supplementary Table S3), and it is thus tempting to speculate that each TA system, separately, might offer a better survival in different niches, and when they cross-regulate, novel fitness capacities could be acquired. Our in-depth RNA mapping provides the basis for molecular and physiological studies of the functional interaction between the two types of TA systems in V583.
CONCLUSION
This study reports a simple, sensitive and general methodological strategy to identify and characterize transcripts, and provides the first data set of ncRNAs in the E. faecalis chromosome. Compared to the 5′RACE method (44,52) from which our method is derived, 5′tagRACE adds a single but crucial RNA ligation step that differentially tags primary and processed RNAs. 5′tagRACE can be utilized for same purposes as the 5′RACE, but in a more reliable manner as illustrated in this work. Most importantly 5′tagRACE can serve a double purpose that is not possible with 5′RACE: (i) simultaneous probing and terminus characterization of unknown transcripts, replacing the two classical successive steps, namely Northern blot and end mapping via 5′RACE; and (ii) a bacterial 5′tag-cDNA library can be used for high-throughput sequencing. By combining 5′tagRACE with RNA-seq, the differential RNA tagging should provide direct access to the ratio between primary and processed forms for a given transcript, thereby offering a dynamic view of bacterial RNA landscapes in terms of regulons through kinetic studies.
Transcriptional analysis of ref loci in E. faecalis unveiled major features of gene organization, as found with high-throughput methods in other bacterial species, and suggest the importance of RNA-mediated regulatory processes in this species. E. faecalis inhabits various ecological niches (soil, water, food, gut) and is able to switch from a normal and harmless commensal bacterium to a life-threatening pathogen (1), attesting to a broad range of regulatory pathways involved in adaptation processes where ncRNAs certainly contribute, as illustrated in other bacterial species, e.g. (17,31). Thus, Ref RNAs being the first ncRNAs described in the E. faecalis chromosome, their functional study should have implications in the deciphering of genetic adaptation programs used by the bacterium living in and interacting with the host.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut National de la Recherche Agronomique; EGIDE-PESSOA collaborative program (20014TA); transnational ERA-Net Pathogenomics program, the Agence Nationale de la Recherche (ANR-06-PATHO-008-02); Sweden Science Council (grant 2006-156). Funding for open access charge: UMR1319 Institut Micalis INRA, Domaine de Vilvert, F-78352 Jouy-en-Josas, France.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Many thanks to our colleagues, Philippe Palcy, Valérie Bourgogne and Patrick Régent, for handling our daily problems. We are grateful to the ‘Efa team’, especially Lionel Rigottier-Gois for numerous discussions and providing us with materials; Alexandra Gruss and Meriem El Karoui for scientific discussions and comments on the manuscript; Alexandra Gruss for her constant support and help with the English language. Thanks to our friend Massimo Vergassola who made possible the dialogue between Paris and Stockholm, and Alejandro Toledo-Arana to discuss on the original idea of the 5′tagRACE. We thank Ingemar Ernberg (Karolinska Institutet) for his generous hospitality by providing laboratory space and equipment for AFH.
REFERENCES
- 1.Gilmore MS, Ferretti JJ. Microbiology. The thin line between gut commensal and pathogen. Science. 2003;299:1999–2002. doi: 10.1126/science.1083534. [DOI] [PubMed] [Google Scholar]
- 2.Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp. Biol. Med. 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 3.Hew CM, Korakli M, Vogel RF. Expression of virulence-related genes by Enterococcus faecalis in response to different environments. Syst. Appl. Microbiol. 2006;30:257–267. doi: 10.1016/j.syapm.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Makhzami S, Quenee P, Akary E, Bach C, Aigle M, Delacroix-Buchet A, Ogier JC, Serror P. In situ gene expression in cheese matrices: Application to a set of enterococcal genes. J. Microbiol. Methods. 2008;75:485–490. doi: 10.1016/j.mimet.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 5.van Schaik W, Willems RJ. Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 2010;16:527–532. doi: 10.1111/j.1469-0691.2010.03201.x. [DOI] [PubMed] [Google Scholar]
- 6.Arthur M, Quintiliani R., Jr Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 2001;45:375–381. doi: 10.1128/AAC.45.2.375-381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002;415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 8.Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417:746–750. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- 9.Shankar N, Coburn P, Pillar C, Haas W, Gilmore M. Enterococcal cytolysin: activities and association with other virulence traits in a pathogenicity island. Int. J. Med. Microbiol. 2004;293:609–618. doi: 10.1078/1438-4221-00301. [DOI] [PubMed] [Google Scholar]
- 10.Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 2006;188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourgogne A, Singh KV, Fox KA, Pflughoeft KJ, Murray BE, Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol. 2007;189:6490–6493. doi: 10.1128/JB.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riboulet E, Verneuil N, La Carbona S, Sauvageot N, Auffray Y, Hartke A, Giard JC. Relationships between oxidative stress response and virulence in Enterococcus faecalis. J. Mol. Microbiol. Biotechnol. 2007;13:140–146. doi: 10.1159/000103605. [DOI] [PubMed] [Google Scholar]
- 13.Ballering KS, Kristich CJ, Grindle SM, Oromendia A, Beattie DT, Dunny GM. Functional genomics of Enterococcus faecalis: multiple novel genetic determinants for biofilm formation in the core genome. J. Bacteriol. 2009;191:2806–2814. doi: 10.1128/JB.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Jeune A, Torelli R, Sanguinetti M, Giard JC, Hartke A, Auffray Y, Benachour A. The extracytoplasmic function sigma factor SigV plays a key role in the original model of lysozyme resistance and virulence of Enterococcus faecalis. PLoS One. 2010;5:e9658. doi: 10.1371/journal.pone.0009658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–103. doi: 10.2217/17460913.4.1.85. [DOI] [PubMed] [Google Scholar]
- 17.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 18.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 20.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weel-Sneve R, Bjoras M, Kristiansen KI. Overexpression of the LexA-regulated tisAB RNA in E. coli inhibits SOS functions; implications for regulation of the SOS response. Nucleic Acids Res. 2008;36:6249–6259. doi: 10.1093/nar/gkn633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueroa-Bossi N, Valentini M, Malleret L, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P. Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 28.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 29.Gimpel M, Heidrich N, Mader U, Krugel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol. Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- 30.Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 31.Toledo-Arana A, Repoila F, Cossart P. Small non-coding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 32.D’Alia D, Nieselt K, Steigele S, Muller J, Verburg I, Takano E. Non-coding RNA of glutamine synthetase I modulates antibiotic production in Streptomyces coelicolor A3(2) J. Bacteriol. 2010;192:1160–1164. doi: 10.1128/JB.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol. Life Sci. 2010;67:217–237. doi: 10.1007/s00018-009-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 35.Toledo-Arana A, Solano C. Deciphering the physiological blueprint of a bacterial cell: revelations of unanticipated complexity in transcriptome and proteome. Bioessays. 2010;32:461–467. doi: 10.1002/bies.201000020. [DOI] [PubMed] [Google Scholar]
- 36.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezee-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 40.Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 41.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new non-coding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner EG, Vogel J. Approaches to identify novel non-messenger RNAs in Bacteria and to investigate their biological functions: functional analysis of identified non-mRNAs. In: Hartmann RK, Bindereif A, Schön A, and Westhof E, editors. Handbook of RNA Biochemistry. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2005. pp. 614–642. [Google Scholar]
- 45.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 46.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, et al. A search for small non-coding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 50.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 51.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all non-coding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bensing BA, Meyer BJ, Dunny GM. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Pandit S, Deutscher MP. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen MC, Nielsen AK, Molin S, Hammer K, Kilstrup M. Changes in rRNA levels during stress invalidates results from mRNA blotting: fluorescence in situ rRNA hybridization permits renormalization for estimation of cellular mRNA levels. J. Bacteriol. 2001;183:4747–4751. doi: 10.1128/JB.183.16.4747-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verneuil N, Maze A, Sanguinetti M, Laplace JM, Benachour A, Auffray Y, Giard JC, Hartke A. Implication of (Mn)superoxide dismutase of Enterococcus faecalis in oxidative stress responses and survival inside macrophages. Microbiology. 2006;152:2579–2589. doi: 10.1099/mic.0.28922-0. [DOI] [PubMed] [Google Scholar]
- 56.Le Breton Y, Muller C, Auffray Y, Rince A. New insights into the Enterococcus faecalis CroRS two-component system obtained using a differential-display random arbitrarily primed PCR approach. Appl. Environ. Microbiol. 2007;73:3738–3741. doi: 10.1128/AEM.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, Hartke A, Auffray Y, Giard JC. ace, Which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect Immun. 2009;77:2832–2839. doi: 10.1128/IAI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 2001;183:3372–3382. doi: 10.1128/JB.183.11.3372-3382.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ibrahim M, Nicolas P, Bessieres P, Bolotin A, Monnet V, Gardan R. A genome-wide survey of short coding sequences in streptococci. Microbiology. 2007;153:3631–3644. doi: 10.1099/mic.0.2007/006205-0. [DOI] [PubMed] [Google Scholar]
- 62.Gerdes K, Wagner EG. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 63.Chandler JR, Dunny GM. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides. 2004;25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 64.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 65.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008;42:541–64. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 66.Wassarman KM, Kiley PJ. Global approaches for finding small RNA and small open reading frame functions. J. Bacteriol. 2010;192:26–28. doi: 10.1128/JB.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giangrossi M, Prosseda G, Tran CN, Brandi A, Colonna B, Falconi M. A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 2010;38:3362–3375. doi: 10.1093/nar/gkq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stork M, Di Lorenzo M, Welch TJ, Crosa JH. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J. Bacteriol. 2007;189:3479–3488. doi: 10.1128/JB.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palmer KL, Carniol K, Manson JM, Heiman D, Shea T, Young S, Zeng Q, Gevers D, Feldgarden M, Birren B, et al. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 2010;192:2469–2470. doi: 10.1128/JB.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huycke MM, Moore D, Joyce W, Wise P, Shepard L, Kotake Y, Gilmore MS. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol. Microbiol. 2001;42:729–740. doi: 10.1046/j.1365-2958.2001.02638.x. [DOI] [PubMed] [Google Scholar]
- 75.Allen TD, Moore DR, Wang X, Casu V, May R, Lerner MR, Houchen C, Brackett DJ, Huycke MM. Dichotomous metabolism of Enterococcus faecalis induced by haematin starvation modulates colonic gene expression. J. Med. Microbiol. 2008;57:1193–1204. doi: 10.1099/jmm.0.47798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132:551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 77.Repoila F, Gottesman S. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 2001;183:4012–4023. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Urban JH, Papenfort K, Thomsen J, Schmitz RA, Vogel J. A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J. Mol. Biol. 2007;373:521–528. doi: 10.1016/j.jmb.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 79.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 2007;65:373–385. doi: 10.1111/j.1365-2958.2007.05796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beckmann BM, Grunweller A, Weber MH, Hartmann RK. Northern blot detection of endogenous small RNAs (∼14 nt) in bacterial total RNA extracts. Nucleic Acids Res. 2010;38:e147. doi: 10.1093/nar/gkq437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiancone E, Ceci P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 82.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin–antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin–antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Melderen L, Saavedra De Bast M. Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lewis K. Persister Cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.