Abstract
The explosion of scientific interest in protein kinase-mediated signaling networks has led to the infusion of new chemical methods and their applications related to the analysis of phosphorylation pathways. We highlight some of these chemical biology approaches across three areas. First, we discuss the development of chemical tools to modulate the activity of protein kinases to explore kinase mechanisms and their contributions to phosphorylation events and cellular processes. Second, we describe chemical techniques developed in the past few years to dissect the structural and functional effects of phosphate modifications at specific sites in proteins. Third, we cover newly developed molecular imaging approaches to elucidate the spatiotemporal aspects of phosphorylation cascades in live cells. Exciting advances in our understanding of protein phosphorylation have been obtained with these chemical biology approaches, but continuing opportunities for technological innovation remain.
Keywords: biosensors, chemical rescue, imaging, kinase, semisynthesis, signaling
INTRODUCTION
In the 1950s, it was discovered that the metabolic enzyme phosphorylase, responsible for the conversion of glycogen to glucose-1-phosphate, existed in two stable forms, an inactive (phosphorylase b) and an active (phosphorylase a) state (1). A search for the molecular mechanism responsible for the conversion of the inactive state to the active revealed that a protein kinase, phosphorylase kinase, could catalyze the attachment of a phosphate to phosphorylase and render it fully active (2). It was subsequently shown that phosphorylase kinase was itself activated by a protein kinase, protein kinase A (PKA, also called cyclic AMP-dependent protein kinase), and the concept of protein phosphorylation as a key regulatory mark was born (3). In the ensuing years, protein phosphorylation networks have been understood as undergirding most physiologic processes ranging from the cardiovascular system, gastrointestinal action, neurologic mechanisms and behavior, immune response, endocrine action, and musculoskeletal regulation (4). Moreover, the linkage of protein phosphorylation to pathogenic mechanisms, involving the aforementioned physiologic systems as well as cancer, have heightened interest in the phosphate posttranslational modification (PTM) within the academic and private sectors of the biomedical research community. The concept that protein kinases are “druggable” began to take hold by the mid-1990s and was placed on firm footing when it was shown that the Abl tyrosine kinase inhibitor imatinib (gleevec) (Figure 1) could effect dramatic remissions in more than 90% of preblast phase patients with chronic myelogenous leukemia (CML) (5). Moreover, patients treated with imatinib are generally free of serious side effects, refuting concerns of severe toxicity associated with targeting protein kinase enzymes considered to be essential to many cellular pathways. These findings have not gone unnoticed in the pharmaceutical industry, and over the past decade about a dozen protein phosphorylation-linked drugs have been launched, and many more are in the pipeline for a host of indications (6).
Figure 1.
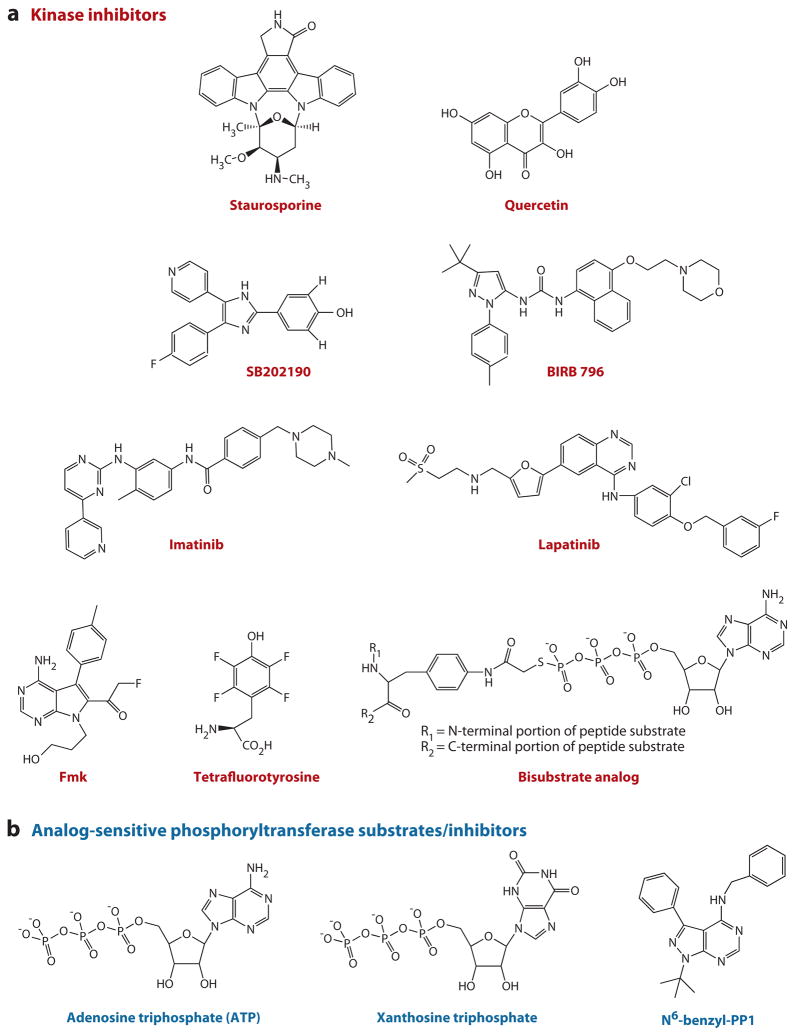
Molecules for protein kinase modulation. (a) Kinase inhibitors: staurosporine, quercetin, SB202190, BIRB 796, imatinib, lapatinib, fmk, tetrafluorotyrosine, and bisubstrate analog. (b) Analog-sensitive phosphoryltransferase substrates/inhibitors. The naturally occurring nucleotide triphosphate substrate, ATP (left), and its unnatural analog, xanthosine triphosphate (middle). The inhibitor N6-benzyl-PP1 (right), which is selective and specific for mutant kinases that can accommodate xanthosine triphosphate.
Despite these successes and the substantial enthusiasm for furthering protein kinase inhibitor development, many fundamental challenges remain in the protein phosphorylation field. Although imatinib has been a great success for controlling CML (5), most drugs that modulate protein phosphorylation have been less effective in disease treatment. It can be argued that we still have a primitive understanding of the function of protein kinases, phosphatases, and their substrates and effectors. The complexity of the phosphoproteome is daunting. There are about 500 mammalian protein kinases (7), 100 protein phosphatases (8), and hundreds of proteins containing domains (SH2, PTB, 14-3-3, BRCT, etc.), which interact with phosphorylated proteins (9, 10). We have also learned recently that precise timing (within minutes) and spatial aspects of protein phosphorylation are crucial to cell functioning. Genetic and conventional biochemical approaches, including the powerful RNAi methodologies (11, 12), have made enormous contributions to our understanding of protein kinase and phosphatase actions. However, these methods have had limitations in pinpointing kinase contributions to signaling, revealing rapid kinetic changes, clarifying functional effects of specific phosphorylation events, and relating cellular localization to kinase/phosphatase activity. In response to these obvious limitations, over the past 10–15 years a range of technologies have been introduced and applied to fill in the missing details in our understanding of phosphorylation networks. In this review, we outline a number of these approaches, which have at their heart the merging of chemistry and biology.
We discuss three general areas where chemical biological methods have been applied to sort out kinase action. First, we describe methods to modulate the action of kinases in vitro and cellular systems. Here, chemical design and screening have been used with wild-type and mutant kinases to gain specificity that has allowed insights into the rapid changes in signaling cascades. Second, we discuss methods to site specifically introduce phosphoamino acids or their mimics at known sites of phosphorylation in proteins. Protein semisynthesis and unnatural amino acid mutagenesis are the main vehicles for this new age protein engineering. Third, we highlight the recently introduced fluorescent reporters that allow for high-resolution imaging of phosphoryl transfer in cells and lysates. Such molecular imaging is providing unprecedented insights into the timing and cellular localization of signaling networks.
PROTEIN KINASE MODULATION
A hallmark of phosphosignaling is its rapid action. Changes in specific phosphorylation of protein targets can be detected within minutes after exposure to various stimuli, such as hormone exposure. This becomes apparent, for example, when epidermal growth factor (EGF) is added to the media of cells in culture and greater than 500 proteins undergo phosphorylation changes by 5 min, as detected using tandem mass spectrometry techniques (13). It is presumed, but in general not yet fully established, that the specific kinetic details of these cellular phosphoryl transfer reactions are critical to their macroscopic effects on gene regulation, cell shape, and cell growth, which occur over longer timescales. Molecular biology approaches, including knockout/knockdown, overexpression, and dominant negative tactics, offer major insights into functional effects of specific proteins and into chronic changes in cell biology but are limited in teasing out intimate details.
Nonselective and Selective Kinase Inhibitors
An attractive strategy for rapid analysis of kinase mechanism and function is to apply natural products or synthetic compounds that inhibit the enzymatic activity of kinases. Relatively early work using natural products, such as staurosporine and quercetin (Figure 1) (14), revealed that the ATP (adenosine triphosphate)-binding sites of protein kinases provide versatile binding sites for relatively hydrophobic molecules (15), and the negatively charged phosphosugar moiety interactions are not important components of affinity. Because the protein kinase nucleotide-binding sites are relatively conserved, broad kinase inhibition is possible with a variety of relatively nonselective inhibitors. Such inhibition experiments can provide general information indicating a role of protein phosphoryl transfer in a process but lack precision in pathway dissection.
Motivated in part by therapeutic need, more selective nucleotide site kinase inhibitors that exploit subtle differences in kinase active sites have been discovered. A relatively early example of a p38 MAP kinase-targeted inhibitor is the pyridinyl imidazole SB202190 (Figure 1) (16). Identified as an anti-inflammatory agent before its protein target was known, SB202190 was converted to a photoaffinity azidophenyl derivative used for cross-linking and target identification. SB202190 was determined to be a highly potent (low nanomolar) and rather selective inhibitor of MAP kinases (16). The basis of its selectivity for this kinase subfamily was shown to be related to certain conserved residues within the MAP kinase binding pocket, ultimately confirmed using X-ray crystallography in a complex structure of p38 plus compound (17). Although not completely specific, SB202190 and related compounds have been used widely in signaling studies to probe the role of MAP kinases in signaling action.
MAP Kinase Inhibition in a Novel Pocket
Another class of relatively selective MAP kinase inhibitors utilizing a diaryl urea scaffold, reported several years ago, targets a pocket adjacent to the ATP site that is created by a conformational change in the Asp-Phe-Gly (DFG) loop (18). Exemplified by BIRB 796 (Figure 1), this diaryl urea class shows highly potent (sub-nanomolar) action against p38 and selectivity versus many other protein kinases examined. An X-ray crystal structure revealed the basis for BIRB 796’s potency and selectivity, and kinetic analysis demonstrated that it is a slow, tight-binding inhibitor (18). Despite its apparent clinical promise, detailed investigation has revealed that BIRB 796 can block other protein kinases (19), limiting its ultimate utility in signaling analysis. These findings suggest that the DFG conformational switch is not unique to the MAP kinases.
Targeting the Inactive State of Protein Kinases
An alternate strategy for achieving protein kinase selectivity, which has met with some success, has involved targeting the inactive state of protein kinases. Most notably observed with the Abl tyrosine kinase inhibitor imatinib and the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor lapatinib (Figure 1), such inhibitors bind to the “off” state of kinases, which can achieve such a conformation (20, 21). The ability to access such inactive conformations is not limited to Abl and EGFR, and imatinib is known to inhibit a number of other kinases with significant potency. This lack of selectivity, while reducing its value in signaling research, allows imatinib to be used to treat patients with gastrointestinal stromal tumors by blocking Kit receptor tyrosine kinase.
Covalent Kinase Inhibitors
More recent tactics to obtain specific kinase inhibitors have involved developing covalent modification strategies also known as affinity labeling. It was first noted in the 1990s that a relatively specific EGFR tyrosine kinase family inhibitor alkylates a Cys near the ATP-binding site (22). Not essential for catalysis by EGFR, the Cys residue is not commonly found in a conserved location in other kinases, suggesting a basis for specificity. The PD168393 inhibitor contains a modestly electrophilic acrylamide functionality, which presumably modifies EGFR by Michael addition because of the proximity of the active-site thiol. Extending this strategy, it was recognized that other protein kinases have Cys proximal to the nucleotide-binding sites. A successfully designed approach to exploiting the Cys in p90 ribosomal protein S6 kinase (Rsk) has been achieved (23). A tepid α-fluoroketone electrophile was installed in the ATP-targeting pyrrolo-pyrimidine scaffold to produce a more selective Rsk kinase inhibitor, fmk (Figure 1) (24). Clever design strategies may allow this approach to be used for other Cys active-site protein kinases. However, the potential for nonspecific alkylation and the irreversibility of the inhibition may affect the scope of applications.
Peptide Inhibitors for Kinases
There has been a sustained interest in the identification and development of protein kinase inhibitors that occupy the protein substrate-binding site. In principle, this has the inherent advantage that kinases show much less conservation in their protein substrate-binding surfaces so that identification of kinase-specific inhibitor ligands should be possible. The plausibility of such an approach was made tangible with the identification of the naturally occurring peptide inhibitor of PKA, which shows nanomolar affinity and high specificity for this kinase. Indeed, an X-ray crystal structure suggests that PKA-bound PKI is likely to perfectly mimic peptide substrate binding but is not turned over because the Ser/Thr residue is replaced by an Ala (25). In contrast to the high potency of PKI, most other peptide inhibitors for various protein kinases composed of natural amino acids have been found to be of modest potency (IC50 ~ 20 μM-1 mM). In general, kinases achieve protein substrate specificity in multifaceted ways, including long-range tertiary structural interactions between enzyme and substrate, adaptor molecule participation, and cellular compartmentalization (26), accounting for the limited specificity imparted by local amino acids surrounding the phosphorylated residue. In contrast to the ATP-binding pocket, the kinase peptide-binding groove is often shallow, complicating identification of potent synthetic compounds (26). Peptides themselves often show poor bioavailability and pharmacokinetic properties, which also have limited progress in this area.
Despite these challenges, there has been noteworthy progress in recent years in the identification of more potent and specific peptide-based kinase inhibitors. One approach has relied on combinatorial chemistry and the incorporation of unnatural amino acids within the peptide motif. This method has led to an extremely potent protein kinase C subtype-specific inhibitor, as well as tight-binding peptide and selective inhibitors of Src and Akt (27–29). Although these compounds await structural analysis in complex with their cognate kinases, they appear to be linear competitive inhibitors versus peptide substrates, suggesting that these inhibitors bind the same pocket in the active site of the enzyme as the peptide substrates.
Mechanism-Based Kinase Inhibitors
A second strategy for the design of peptide inhibitors has involved consideration of enzyme mechanism. Early experiments using tetrafluorotyrosine (Figure 1) suggested that a peptide phenolate anion could be a transition state analog inhibitor for a tyrosine kinase (30). However, later studies revealed that proton transfer from the phenol occurs late in the phosphoryl transfer reaction and that a more dissociative catalytic mechanism is involved (31–33). In a dissociative mechanism, the bond between the γ-phosphorus and the ADP leaving group is largely cleaved in the transition state prior to significant bond formation with the entering tyrosine. This mechanism would be predicted to involve an extended initial reaction coordinate where the distance between the tyrosine and ATP is estimated to be separated by 4.9 Å or more (34). A novel class of ATP- peptide bisubstrate analogs (Figure 1) was designed (35) on the basis of this prediction, and these compounds have proved moderately to highly potent and selective inhibitors of a number of protein kinases, including insulin receptor kinase, Csk, PKA, EGFR, Abl, Cdk, and calmodulin-dependent II kinase, based on the peptide sequence linked to ATP (35–41). Although these protein kinase bisubstrate analogs have not yet been applied to studies in vivo, they have proved to be powerful structural tools. On the basis of kinase structures in complex with the ATP-peptide conjugates, these compounds have provided novel insights into the molecular recognition of peptide substrates by IRK, EGFR, Abl, and Cdk (35, 38–40). The complexes of the Abl and EGFR kinases with bisubstrate analogs also assisted in clarifying the allosteric interconversions of these enzymes between active and inactive states (38, 39). In particular, the Abl-bisubstrate structure showed a conformation that revealed how previously unexplained imatinib-resistant mutants lose sensitivity to this drug (39). The EGFR-bisubstrate structure provided a model for the basis of kinase activation through an asymmetric dimer (38). Additional applications of these bisubstrate analogs include examining the molecularity of autophosphorylation (42) and proteomic pull-down analyses (36).
Other bivalent kinase design approaches have led to a selective PKA blockade. These methods have involved helix design (43), use of oligo-Arg motifs (44), and phage display (45). The precise structural interactions for these compounds are still uncertain, but the kinetic analyses are consistent with the bisubstrate model. Although peptide modulators have generally not been used in vivo, examples of the use of cell-permeabilizing peptides linked to agonist motifs for protein kinase C and inhibitors of Jun kinase have demonstrated the potential for biomedical applications (46, 47).
Analog-Sensitive Kinases
Ultimately, the conserved nature of protein kinases and the large size of this superfamily present a difficult obstacle to achieving exquisite specificity by compound modulators. This difficulty is most apparent when distinguishing among the closely related homologs within individual families, such as the PKA paralogs, the PKC paralogs, and the Src family. This had inspired the use of bump-and-hole strategies for targeting specific enzymes. Early related studies on G proteins show that a replacement of an Asp residue in the GTPase-binding site with Asn changes their preferences for GTP to xanthosine triphosphate (Figure 1) (48). When substituted for the wild-type G proteins, these D/N mutant G proteins can be modulated in cells by the use of xanthosine nucleotide. Related engineered complementary protein-ligand pairs have been developed for proteases (49), cyclophilins (50), and, subsequently, kinases (51). A conserved gatekeeper ATP-site residue (Ile338 in Src), which is typically bulky in kinases (Thr, Ile, Phe, Met), is mutated to Gly, and these kinases are generally still quite active catalysts and show increased ability to utilize unnatural ATP analogs as substrates by in vitro assays and assays with cell lysates (52). These mutant kinases also display enhanced sensitivity to specifically designed inhibitors, most commonly N6-benzyl-PP1 (Figure 1) (53). These analog-sensitive kinases include Src, Cdk, Abl, PKA, Jun kinase, and many others (54–56).
This chemical complementation approach has been employed widely for studies in yeast, mammalian cells, and knockin mice and has provided a great range of insights into the function of these kinases in cell growth, differentiation, and signaling (54–56). The rapid temporal control (inhibition within 15 min in cells) has offered unexpected insights compared with standard genetic studies. One recent notable example was the finding that Cdk7 was required for the ordered assembly of Cdk1/cyclin B and Cdk 2 in mammalian cells (57). Chemical control of analog-sensitive kinases can give complementary results to gene knockout experiments, although the potential for unintended biological changes conferred by enzyme mutation has to be considered. Recent proteomic studies on Abl suggest that altered phosphotyrosine profiles can be seen with analog-sensitive kinases compared to wild type (58). However, gene knockout experiments are also subject to caveats so that a convergence of strategies is desirable when piecing together a cell signaling network.
Chemical Rescue of Mutant Tyrosine Kinases
An alternative complementation approach involves the chemical rescue of a mutant tyrosine kinase. This approach is grounded in the finding that, for many enzymes, mutation of an active-site residue, which can lead to catalytic reduction, can be complemented by a noncovalently bound small molecule, which mimics the lost side chain. Originally demonstrated for trypsin (59), transaminase (60), and carbonic anhydrase (61), it was shown that mutation of the catalytic Arg (388 in Src) to Ala in tyrosine kinases could be complemented with diamino compounds (62, 63). Interestingly, this conserved Arg is found either two or four residues upstream of the catalytic base Asp among tyrosine kinases.
When Csk or Src kinase double mutants were prepared, which moved the Arg to the alternate position, these enzymes showed catalytic activity in range of the wild-type kinases (62, 63). The most effective rescue agent for R/A Src and Csk is imidazole, and use of this small molecule allows for efficient rescue of transfected mutant enzymes in mammalian cell culture (63, 64). Imidazole rescue of mutant Csk has led to unexpected insights into Csk’s activity in controlling stress fiber formation and in guanylyl cyclase activity (64, 65). This chemical rescue approach has uncovered new targets of Src and revealed the precise temporal role of Src kinase action in signaling from growth factors to MAP kinase (63, 66). An advantage of this approach, which rapidly (<1 min) turns an intracellular enzyme on (rather than off), is that it provides a portrait of initial signaling events only indirectly observed when upstream events are triggered by hormones.
SITE-SPECIFIC INCORPORATION OF PHOSPHOAMINO ACIDS AND MIMICS IN PROTEINS
The effects of specific phosphorylation events on proteins can range from introducing an electrostatic point charge, which can influence protein conformation, to creating an intricate binding surface for adaptor protein interaction. For example, it is believed that the negative charge associated with glycogen phosphorylase Ser phosphorylation stimulates a conformational change and glucose-1-phosphate production by this enzyme (67). In contrast, phosphorylation of many proteins on Ser leads to 14-3-3 (10) recruitment, and phosphorylation of growth factor receptors on Tyr leads to the recruitment of Grb 2, an adaptor protein containing the SH2 domain (9). Typical analysis of the effects of protein phosphorylation at a particular site involves site-directed mutagenesis of the residue of interest. In particular, replacement of a Ser/Thr with Ala and mutation of a Tyr to Phe in transfected recombinant proteins prevent phosphorylation, and phenotype changes are interpreted as requiring a phosphate attachment at these sites. However, these experiments are associated with caveats. Although a conservative replacement, the deletion of an oxygen from the side chain changes the polarity and the hydrogen bonding capacity, which can alter the protein structure. Moreover, Ser/Thr can undergo alternative PTM (such as glycosylation) as can Tyr (such as sulfation).
For these reasons, it is especially attractive to substitute a phosphorylated residue with a mimic of phosphoamino acid, which would presumably render a more readily interpreted result. For phosphoSer/phosphoThr (pSer/pThr), replacement with Asp or Glu is sometimes used as a surrogate (Figure 2c) (68). There are considerable differences between the modified pSer/pThr side chains and the carboxylate functionality on the side chains of these acidic residues, including size (P bigger than C), number of oxygens (3 versus 2), and number of negative charges at neutral pH (2 versus 1). Yet, there are occasions when the Asp/Glu can partially or even faithfully mimic the pSer/pThr (69, 70). A very nice example is phosphorylation of Smad, which leads to trimerization and is similarly observed upon mutation to Glu (71). Nevertheless, there are many, probably the majority, of cases where the Asp/Glu mutation cannot recapitulate the pSer/pThr and is not useful for protein analysis. 14-3-3 adaptor recognition by pSer/pThr is not mimicked by protein with Glu substitution (72). PhosphoTyr (pTyr) shows even weaker resemblance to Asp/Glu, and it is remarkable (73) when such replacements show any ability to confer phosphorylation functionality.
Figure 2.
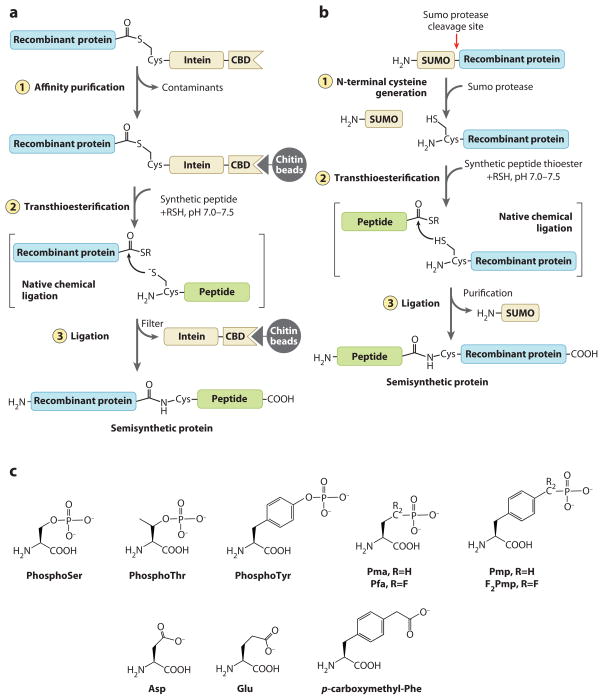
Site-specific incorporation of phosphoamino acids and mimics in proteins. (a,b) Strategies used for protein semisynthesis involving native chemical ligation. (a) Expressed protein ligation for attaching a synthetic peptide to the C terminus of a protein. (b) Proteolytic generation of N-terminal Cys for attaching a synthetic peptide thioester to the N terminus of a protein. (c) Phosphoamino acids and their mimics. (top) Phosphoamino acids and mimics that can be incorporated into proteins through semisynthesis. (bottom) Genetically encodable mimics for phosphoamino acids that can be incorporated into proteins through point mutations or by using nonsense codon suppression technology.
Classical approaches to studying phosphorylation events have involved the use of short synthetic phosphopeptides or the enzymatic phosphorylation of target proteins. The former method is inadequate for addressing phosphorylation events on intact proteins greater than 100 amino acids (aa) in size, and the latter method suffers from an inability to control site specificity and stoichiometry. However, recently developed methods in protein semisynthesis and unnatural amino acid mutagenesis offer new solutions to problems in protein phosphorylation.
Protein Semisynthesis
Over the past decade, protein semisynthesis, using the native chemical ligation procedure, has led to a versatile new technology to install phosphorylated residues and precise nonhydrolyzable mimics into large proteins (74). Such technology has produced exciting new insights into the function of site-specific protein phosphorylation. On the basis of the Wieland chemoselective reaction between a peptide/protein with N-terminal Cys and a peptide/protein C-terminal thioester, native chemical ligation involves initial transthioesterification, followed by an internal rearrangement to form a standard amide bond (75). Using this reaction, protein semisynthesis has been applied to a wide range of proteins where installation of unnatural amino acids, PTMs, biophysical probes, and isotopic labels is a desired objective. There are two general strategies commonly used for protein semisynthesis involving native chemical ligation (Figure 2a,b). In one, a recombinant protein containing an N-terminal Cys via proteolysis is ligated to a synthetic peptide with a C-terminal thioester (76). In the other, known as expressed protein ligation (EPL), a recombinant protein bearing a C-terminal thioester generated by intein action is ligated to an N-Cys-containing synthetic peptide (74). The latter approach has been more widely used because it avoids the chemically challenging task of synthesizing a thioester.
Expressed Protein Ligation and Csk
Protein semisynthesis is especially powerful when applied to proteins that are known to be regulated by phosphorylation near their N or C termini, where the synthetic moieties can be readily incorporated. Site-specific incorporation of one or more phosphorylated Ser/Thr/Tyr can be achieved within the synthetic peptide moiety, allowing for full stoichiometry. Moreover, when the role of the phosphorylation is analyzed in the presence of phosphatases, nonhydrolyzable phosphonate mimics can be used.
In the initial report of EPL, a pTyr was introduced into the C-terminal tail of tyrosine kinase Csk (74). Csk is the tyrosine kinase responsible for C-terminal phosphorylation of Src and Src family members. Tail tyrosine phosphorylation of Src by Csk leads to a closed conformation, involving an intramolecular interaction between the tail of Src and its SH2 domain (77). Csk is architecturally related to tyrosine kinase Src in that they have SH3 domains, SH2 domains, and kinase domains laid out in the same order from the N to C terminus. However, Csk differs from Src in that Csk lacks the C-terminal tail that can get Tyr phosphorylated and regulate conformation. To determine what effect a phosphotyrosine tail on Csk would have on its conformation and catalytic behavior, tail-phosphorylated semisynthetic Csk was prepared by fusing full-length Csk to an intein to generate a thioester and ligating a synthetic 12-mer tail peptide carrying the pTyr designed as a Csk SH2 ligand (74). In contrast to the inhibitory effect of tail phosphorylation of Src, semisynthetic Csk, containing a tail pTyr, shows enhanced activity toward its natural substrate Src compared with unphosphorylated Csk. This catalytic stimulation appeared to involve an intramolecular interaction between the Csk SH2 domain and this unnatural phosphotyrosine-containing tail. These unexpected differences in catalytic behavior are now better understood because NMR and X-ray crystal structures of Csk and Src show a different arrangement of the SH3, SH2, and catalytic domains between the proteins (78, 79).
Phosphonate Mimics and Protein Tyrosine Phosphatases
EPL has been especially valuable in analyzing C-tail tyrosine phosphorylation of protein tyrosine phosphatases (PTPases) (72, 80–83). This paradoxical modification is difficult to study because of the autoinstability of tyrosine phosphorylation of a PTPase. Two of the PTPases analyzed include SHP-1 and SHP-2. SHP-1 is involved as a negative regulator of signaling in hematopoietic cells (84). SHP-2 activity is associated with positive signaling in growth factor-induced pathways in a wide variety of cell types (85). Both SHP-1 and SHP-2 have two N-terminal SH2 domains, a C-terminal catalytic domain, and a C-terminal tail, which is subject to dual tyrosine phosphorylations. SHP-1 is phosphorylated at Tyr536 and Tyr564. SHP-2 is phosphorylated at Tyr542 and Tyr580. For both enzymes, the unliganded SH2 domains confer inhibitory effects on catalysis, and engagement of the N-SH2 domains by pTyr-containing peptides in trans was known to be activating. There was equivocal data about the effects of tyrosine phosphorylation on SHP-1 and SHP-2 function. The challenge of working this out was likely attributed to the short-lived phosphorylation of these enzymes because of self-cleavage.
EPL was used to replace the natural C termini of these PTPases with synthetic peptides carrying nonhydrolyzable phosphonate surrogates (81, 82). Both difluoro- and standard methylene phosphonates have been examined, and it is believed that the difluorophosphonates are better phosphate mimics because they show more closely matched pKas to the corresponding phosphates and have the potential to be hydrogen bond acceptors (Figure 2c). By analyzing semisynthetic SHP-1 with difluorophosphonates at the 536 and 564 positions, it was revealed that the 536 phosphonate had a large (eightfold) effect on catalytic stimulation resulting from the N-SH2 domain binding the phosphonate at this position, whereas the 564 phosphonate had a small (<twofold) effect on catalytic stimulation, resulting from its intramolecular interaction with the the C-SH2 domain (82). In SHP-2, incorporation of difluorophosphonates at the 542 and 580 positions each led to modest threefold enhanced catalysis. The difluorophosphonates at these positions were bound intramolecularly by the N-SH2 and C-SH2 domains, respectively, and the double phosphonate incorporation led to partially additive effects in stimulating catalysis of the SHP-2 enzyme (81). Using cellular microinjection, it was found that semisynthetic SHP-2, containing a 542 phosphonate, showed enhanced activation of the serum response element in rat embryonic fibroblasts, consistent with the catalytic activation observed in vitro (80).
Low-molecular-weight protein tyrosine phosphatase (LMWPTP) is also known to be subject to tyrosine phosphorylation (83, 84). LMWPTP is ubiquitously expressed, has been considered inhibitory to growth factor pathways, and is suggested to dephosphorylate platelet-derived growth factor receptor (PDGFR) and p190RhoGap. LMWPTP is tyrosine phosphorylated near its C terminus on Tyr131 and Tyr132 in response to growth factors, and these phosphorylations had been suggested, on the basis of limited biochemical data, to stimulate the enzymatic activity of LMWPTP (86, 87). LMWPTP is 157 aa, and introduction of phosphonate groups to mimic pTyr at 131 and 132 by EPL was achieved by replacement of the C-terminal 34 aa with a synthetic peptide (83). This semisynthesis required a denaturation and refolding step, which was feasible using the Gyr intein, and LMWPTP, like many small proteins, is tolerant to this procedure. In contrast to prior efforts on LMWPTP and studies discussed above on SHP-1 and SHP-2, both 131 and 132 phosphonates conferred marked (10–20-fold) reduction in LMWPTP catalytic activity toward pTyr-containing substrate peptides. This effect is rationalized from the crystal structure of LMWPTP as phosphates at Tyr131 and Tyr132 are envisaged to disrupt peptide substrate-enzyme interactions. The net functional effect of tyrosine phosphorylation of LMWPTP is now proposed to derail its ability to promote actin stress fiber formation and cell adhesion (83).
PhosphoSer Mimics and Semisynthesis
In addition to applications in assessing tyrosine phosphorylation, protein semisynthesis has been used to study the effects of Ser/Thr phosphorylation (42, 72, 88, 89). Serotonin N-acetyltransferase [arylkylamine N-acetyltransferase (AANAT)] is a key pineal regulatory enzyme governing melatonin biosynthesis in a circadian pattern (90). Activities and levels of AANAT rise at night and are rapidly reduced after light exposure. Correlating with the diurnal changes in AANAT is the activation of PKA and the phosphorylation of AANAT at two positions–Thr31 and Ser205 (91). It was hypothesized that modification of AANAT at these two positions was responsible for cellular stabilization of AANAT through 14-3-3 protein interaction, but direct evidence for this model was lacking. Mimicry of pThr31 with a phosphonate (Figure 2c) was achieved by replacing the N-terminal moiety with a synthetic peptide using native chemical ligation (72). This phosphono-AANAT showed high affinity for 14-3-3 in pull-down assays and greater stability in cells after microinjection compared to the unmodified protein. Interestingly, when AANAT was substituted with a Glu at the corresponding position, there was no difference in 14-3-3 binding or stability compared with wild type (72). This suggests that the carboxylate side chain of the Glu is unable to recapitulate the phosphate interactions related to 14-3-3 binding. Results similar to the Thr31-modified protein regarding AANAT interaction with 14-3-3 and cellular stabilization were obtained with a phosphonate introduced at Ser205 by EPL (88). More recent fluorescence anisotropy studies have shown that a doubly phosphorylated AANAT prepared by semisynthesis can bind to 14-3-3 in a bivalent manner compared with the monophosphorylated counterparts, which interact monovalently (89). Semisynthesis was also used to label AANAT with a fluorescein and rhodamine at its N and C termini, and this protein showed Forster (fluorescence) resonance energy transfer (FRET), which could be abolished by proteolytic cleavage (89). Using this microinjected FRET-active protein, it was demonstrated in a real-time assay by imaging live cells that activation of PKA or blockade of the proteasome was sufficient to stabilize AANAT, consistent with the phosphonate findings (89).
Semisynthesis has been elegantly applied to investigate the regulation of Smad Ser phosphorylation by TGFβ-receptor (TGFβR) kinase (92). TGFβR kinase is unusual as a receptor Ser/Thr kinase, and it is responsible for activating Smad proteins by C-terminal phosphorylation. TGFβR is activated by phosphorylation of Ser and Thr residues in a segment between its transmembrane domain and its kinase domain. Native chemical ligation was used to prepare the soluble TGFβR kinase fragment in site specifically tetraphosphorylated form within its N terminus (93). Studies on tetraphosphorylated TGFβR kinase revealed enhanced affinity of the phosphorylated kinase for its substrate Smad2, and in turn, the phosphorylated kinase shows reduced affinity for the inhibitory protein FKB12 (93). This dual mode of activation of the kinase for substrate, rather than inhibitor interaction, provided a new paradigm in signaling regulation. EPL has also been used to investigate the role of C-terminal phosphorylation of Smad2 at Ser465 and Ser467 (94, 95). By introducing phosphate modifications individually and combined at these sites, it was shown that Ser465 phosphorylation drives trimerization, although Ser467 phosphorylations plays an accessory role. Trimerization of SMAD2 is critical for nuclear localization and transcriptional activation. Moreover, Ser465 phosphorylation enhances phosphorylation by TGFβR kinase at the 467 position. By incorporating fluorescent probes and cross-linking reagents by EPL in addition to phosphates, more precise insights into the protein-protein interactions have been obtained (92, 96).
Nonsense Codon Suppression
Protein semisynthesis using chemical ligation techniques, although powerful, is difficult to use when PTMs are in the middle of large proteins. This would necessitate three-piece ligations, and problems of folding and solubility become more significant. The development of unnatural amino acid mutagenesis using ribosomal machinery (97) offers a potentially attractive alternative for site-specific introduction of posttranslationally modified residues and their mimics. This method involves the use of nonsense codon suppression with mischarged tRNAs, allowing transfer of a designer amino acid to a site of interest. Nonsense suppression mutagenesis was originally developed as an in vitro translation technique, which has been effective for production of a number of proteins and used to install phosphoamino acid residues and mimics site specifically (98, 99). The principal limitation of this in vitro approach has been that the protein amounts generated are typically in the 0.1 mg range, which restricts applications. Several years ago, it proved possible to select and engineer aminoacyl tRNA synthetases that could be used to deliver nonstandard amino acid residues to the growing polypeptide chain during in vivo translation in cells (100). Dozens of unnatural amino acids have now been introduced into a variety of proteins using custom-engineered aminoacyl-tRNA synthetases in Escherichia coli, yeast, and mammalian cells (101). In these cases, the unnatural amino acid of interest is placed in the cell culture medium, and after it gains entry into the cell, the aminoacyl-tRNA synthetases and ribosome take over.
Unfortunately, it has not yet been possible to use this approach to install phosphate or phosphonate amino acids in cells, presumably because they cannot penetrate cell membranes efficiently. However, a phosphotyrosine mimic, p-carboxymethyl-Phe (Figure 2c), has been introduced site specifically into recombinant Stat protein, facilitating its dimerization in a fashion that occurs with phosphorylation (102). It is expected that technical advances, such as the application of engineered amino acid transporters or esterase-sensitive protective groups, will ultimately allow phosphonate residues to be incorporated using in vivo nonsense suppression. The future promise of this approach is that it will enable effective in vivo study of signaling containing site specifically introduced PTMs. For this to be most effective, the toxicity of nonsense supression of endogenous genes by transgenic machinery will need to be tackled. However, given the pace of developments in this area, there is every reason to believe that this will some day be possible.
FLUORESCENT IMAGING OF KINASE ACTION
Correlating the activity of a particular protein kinase with a cellular event has been a challenge. Classical molecular biology approaches to measure kinase action have involved ex post facto analysis of enzymatic activities, using immuno-precipitation or immunocytochemistry methods. These delayed measurements suffer from problems of specificity and the inability to report on kinase actions in real time. The classical methods also lose key spatial information that can only be obtained in the live cell. To address these shortcomings, a variety of investigators have created fluorescent-based reporters to image kinase action in complex mixtures or living cells. Such fluorescent reporters have been prepared using chemical synthesis or sophisticated protein engineering. These reporters have been designed to be selective for a given kinase, provided specific substrate sequences can be identified, and have allowed for precise analysis of the timing of signaling events.
Peptide Biosensors
Peptide biosensors provide a useful tool for measuring phosphorylation events with very good temporal resolution in vitro and also have potential for live cell imaging using microinjection or cell-permeabilizing peptide sequences. The basic design of these biosensors involves synthetic fluorophore incorporation into peptides or protein interaction domains, where the fluorescent properties change upon phosphorylation (103). Such phosphorylation-induced fluorescent changes can involve shifts in the fluorophore’s emission wavelength, increases or decreases in its quantum yield, or both. Most of these biosensors can be classified into one of four fluorescence-related categories: environmentally sensitive, deep quench, self-reporting, or metal chelation enhanced (Figure 3).
Figure 3.
Peptide biosensors. (a) Environmentally sensitive biosensor. (b) Deep-quench biosensor. (c) Self-reporting biosensors. (d ) Metal chelation-enhanced fluoroscence biosensor.
Environmentally sensitive biosensors exploit phosphorylation-dependent changes in solvation (104). They typically employ a phosphospecific amino acid-binding domain such as SH2 or 14-3-3 to complex with the phosphorylated peptide, resulting in altered solvent polarity for the fluorophore. This type of peptide biosensor can be used for either Ser/Thr or Tyr phosphorylation. A Src tyrosine kinase peptide biosensor carrying a dapoxyl moiety was developed. In this strategy, the phosphorylated biosensor engaged an SH2 domain, enhancing fluorescence sevenfold (104).
The class of deep-quench biosensors employs a noncovalently attached quencher that interacts with the fluorophore in the peptide (105). Upon phosphorylation, these biosensors recruit a phosphospecific amino acid-binding domain that separates the fluorophore from the quencher, resulting in an increase in fluorescence. In one example, a PKA substrate peptide with a pyrene fluorophore was developed. The dye quencher suppressed fluorescence but was displaced upon phosphorylation and 14-3-3 recruitment. This displacement resulted in a dramatic 64-fold increase in fluorescence (105).
Self-reporting sensors are a domain-free sensor system that can be used to detect tyrosine phosphorylation (106). Aromatic amino acids such as tyrosine can be used to quench a fluorophore in the peptide biosensor through π-π stacking interactions. Upon phosphorylation of the tyrosine residue, it loses its ability to quench the fluorophore. Activities of Src and related tyrosine kinases have been analyzed in this way, providing fivefold fluorescence change upon phosphorylation (106).
Metal chelation-enhanced fluoroscence also avoids phosphoamino acid-binding domains. Using the nonnatural amino acid Sox, a chelation-enhanced fluorophore, a series of biosensors for protein kinase activities have been developed that respond to physiological Mg2+ levels (107). These biosensors for kinase activity consist of the critical kinase recognition elements, Ser/Thr residue, Sox, and a two-residue turn to preorganize Mg2+ binding between Sox and the newly introduced phosphate (107). Phosphorylation of the Ser/Thr residue in the peptide enhances the affinity of the peptide for Mg2+, and this chelation of Mg2+ generates the fluorescent signal. Probes for six dif- ferent kinases (PKC, Cdk2, PKA, Akt, MK2, Pim2) have been developed that show a three-to eightfold increase in fluorescence upon phosphorylation (108). A variant of the approach has been developed to detect tyrosine phosphorylation catalyzed by Abl (109).
These peptide-based biosensors have several applications. They are very practical for use in various in vitro kinase assays. They provide the temporal resolution desired for kinetic analysis. The continuous assay format allows for the detection of an initial lag phase in a biphasic progress curve for kinases that require activation through autophosphorylation. Peptide biosensors have also been used to simultaneously monitor the activities of several kinases. In general, live cell assays with these sensors are more difficult to perform, but examples have been reported with PKA, PKC, and Src reporters introduced by microinjection (110, 111). A variant of this approach, termed single-cell kinase assays, using a non-imaging based method with microinjected fluorescent substrates in which activities are measured after lysis, has been described (112, 113). Although useful, these microinjection methods for peptide biosensors have not yet been widely adopted because of their reliance on specialized equipment, the instability of the peptides in cells, and the cellular perturbation that can be induced by microinjection.
Conjugated Antibodies for Kinase Measurements
Immunocytochemistry has long been used to provide information about the localization of a protein of interest, and with the development of phosphospecific antibodies, this technique can be used to provide information about the cellular location of a kinase and phosphoprotein product. However, this technique suffers from several disadvantages, including the need to fix cells (eliminating live-cell and real-time imaging), poor specificity, and extremely limited spatial resolution. The use of phosphospecific antibodies in combination with other techniques can provide some additional advantages (114, 115). Multidimensional fluorescence-activated cell sorting analysis with a range of phosphospecific antibodies (conjugated to fluorophores) has been used to probe the activated states of states of kinases within single cells (116). This technique afforded information about the order of kinase activation in a signaling cascade. By combining phosphospecific antibodies with flow cytometry, the status of tyrosine phosphorylation and activation of tyrosine kinases was determined in a subpopulation of activated T cells (117). Despite these advances, temporal and spatial resolution are limited with these approaches.
The application of FRET between a fluorescently conjugated protein of interest and a phosphospecific antibody-conjugated fluorophore can significantly improve spatial resolution. In successful examples, FRET imaging is done by following the photo-chemical properties of the donor using FLIM (fluorescence lifetime imaging microscopy), rather than ratiometric imaging of FRET, which measures the fluorescence intensities of the donor and acceptor (118). FLIM detects the shortened fluorescence decay of the donor of the FRET pair in the presence of the acceptor. The use of FLIM to determine FRET provides several advantages. Lifetime (or the fluorescent decay time) measurements (unlike fluorescence intensity) are independent of the variations in fluorophore concentration and photobleaching. FLIM is more robust than fluorescent intensity-based FRET measurements because it can distinguish between actual FRET efficiency and probe concentration. FLIM also allows for the use of spectrally similar donor and acceptor molecules (118). For imaging of phosphorylation events by FRET with conjugated antibodies, the advantages of FLIM are crucial as one does not need to control for the amount of antibody added to the system, and any nonspecific binding of the antibody is virtually invisible. Only conjugated antibody bound within nanometers of the target fluorescently labeled protein is detected. The major disadvantages of this technique are the complex data interpretation needed for FLIM and the need for microinjection to get the conjugated antibodies into the cell.
In an early example of this approach, FLIM was employed to monitor the activation of PKCα in live cells (119). It was previously shown that the PKCα is phosphorylated in vivo on Thr250 upon activation. In this case, green fluorescent protein (GFP) (donor)-tagged PKCα and Cy3.5 (acceptor)-labeled antibodies were microinjected into COS-7 cells (119). Ng and coworkers generated an antibody specific for phosphorylated Thr250 on PKCα and conjugated it the Cy3.5 fluorophore. It was shown that upon treatment with a known PKC agonist, phorbol ester, there was a progressive increase in Thr250 phosphorylation (observed by the decrease in GFP lifetime). These results were confirmed in fixed culture cells, and a real-time assay for this pathway was established.
FLIM has also been exploited to image EGFR signaling (120). The phosphorylation and activation of expressed GFP-EGFR in response to EGF stimulation was imaged in MCF7 cells using microinjected Cy3-linked anti-pTyr antibodies. Based on the kinetics of the data, an activation mechanism involving transient rather than stable receptor dimers of EGFR was proposed. In a related study, FLIM was used to analyze the timing and localization of the PTPase PTP1B after EGFR or PDGFR stimulation using a dead-end mutant (121). Interestingly, it was found that PTP1B recruitment was coupled to receptor endocy-tosis. FLIM has also been applied successfully to study CD44 association with ezrin, directional cell motility regulated by PKC phosphorylation, and stimulation-dependent phosphorylation of lipoprotein receptor-related protein (LRP) in neurons (122, 123).
Translocation Assays
An interesting approach for the imaging of phosphorylation events within the cell employs a GFP-fused protein in a translocation assay. These assays are based on a phosphorylation-dependent movement conferred by a kinase activity of interest and are monitored by microscopic imaging of the fluorescence intensity. This technique has been applied to PKC and used to investigate diacylglycerol signaling in living cells (124) and correlations of PKC with the oscillations of Ca2+ in astrocytes (125). Although successful in these cases, the translocation assay approach is not generalizable to many systems because it depends on phosphorylation-dependent target movements, which have not been established for many kinases. It also suffers from limited time resolution and weak signal-to-noise ratio as the translocations are not always complete.
FRET-Based Genetically Encoded Biosensors
Most of the imaging techniques described above are faced with the challenge of getting the biosensor or some component of the detection system into living cells and/or have various limitations, which restrict their use. The genetically encoded FRET-based activity reporters largely overcome the cell entry problem by having the cell itself manufacture the complete biosensor (126). Because these reporters are genetically encoded, they can be either transiently or stably transfected into cells as DNA.
The discovery and extensive use of GFP, and its other color variants, allow for the genetically encoded reporter approach (127, 128). FRET is strongly dependent on the distance and orientation of the fluorophores. The dynamic changes in FRET can be read as changes in emission ratios between fluorophores or as a change in the donor fluorophore fluoroscence lifetime (129). Typically for these types of biosensors, the changes in FRET are measured as changes in emission ratios for the donor and acceptor fluorophores because the data interpretation for the lifetime of fluorescent proteins can be very complex (118). The basic design of these biosensors consists of the fluorescent protein FRET pair fused with a phosphoamino acid-binding domain and a substrate peptide between them (Figure 4). This simple design allows for a variation in several parameters, as described below, to optimize dynamic range, sensitivity, and selectivity.
Figure 4.
Forster (fluorescence) resonance energy transfer (FRET)-based genetically encoded biosensor. Basic design of biosensor consists of two fluorescent proteins (FPs) that are fused with a phosphoamino acid-binding domain (gray) and a substrate peptide (green) between them. Upon phosphorylation by a kinase or dephosphorylation of the substrate on the biosensor, there is a conformational change that results in a change in FRET.
The fluorescent protein FRET pair employed can be selected to generate a maximum dynamic range. The attachment site for the fluorescent proteins to the reporter construct can be adjusted (there are several circularly permuted variants of some of the fluorescent proteins that allow for this modification) (130). Swapping of the N- and C-terminal locations of the fluorescent proteins has also proven to increase dynamic range in some reporters (126). The choice of fluorescent protein pair should take into account that there must be sufficient spectral overlap between the emission of the donor fluorescent protein and the excitation of the acceptor fluorescent protein. Ideally there should be minimal overlap in the excitation spectra of the two fluorescent proteins to eliminate any bleed through or direct excitation of the acceptor fluorophore at the donor excitation wavelength.
The recognition of the phosphorylated substrate by the phosphoamino acid-binding domain is responsible for the conformational change in these biosensors that results in bringing together the two fluorescent proteins in close enough proximity to allow for FRET to occur (131). The domain is selected on the basis of the phosphoamino acid that is being detected. These biosensors can be developed for both tyrosine kinases as well as serine/threonine kinases. The different phosphoamino acid-binding domains have different preferences for recognition motifs (9, 10), and these need to be considered when selecting a domain, but there are multiple domains to select from if one does not work well. The substrate portion of the biosensor is what dictates the specificity of the reporter. As with the peptide biosensors, the substrate typically consists of a short consensus peptide that is designed to be specifically recognized and efficiently phosphorylated by the target kinase of interest and that is inert to nonspecific kinases. The substrate also needs to be designed such that, upon phosphorylation, it is efficiently recognized and bound by the phosphoamino acid-binding domain selected. However, it is also desirable for the binding interaction to be reversible, so that oscillations of phosphorylation attributed to kinase/phosphatase can be observed (131).
Numerous successful genetically encoded biosensors for reporting kinase activity have now been constructed with this basic design. Additional incorporated features of these reporters include sequences that allow for targeting to specific areas of the cell. Nuclear localization signals, domains for plasma membrane localization, and several other target domains have allowed for the generation of biosensors that provide further information about the spatial dynamics of phosphorylation in different cell subcompartments (132). Other kinase reporters omit the phosphoamino acid-binding domain. If the kinase itself has a conformational change upon phosphorylation or activation, the kinase can be tagged with the fluorescent protein pair, as was done for the ERK2 kinase reporter, which sandwiched ERK2 between CFP (cyan fluorescent protein) and YFP (yellow fluorescent protein) (126). Moreover, as also exemplified by ERK (extracellular signal-regulated kinase), if longer protein substrate rather than short peptide substrate sequences are incorporated in the reporter, the conformational change upon phosphorylation can be large enough to avoid the phosphoamino acid-binding domain (126). Additionally, the FRET signal does not necessarily need to derive from fluorophores in the same fusion protein. For measuring EGFR kinase activity, a FRET biosensor that detected the binding of a YFP-tagged phosphotyrosine binding domain and a CFP-tagged EGFR was employed (133).
Split-Protein Genetically Encoded Biosensors
Another class of the genetically encoded method involves split fluorescent protein complementation, which has been used for analyzing protein-protein interactions in signaling networks. Some fluorescent proteins can be split into two separate domains that are inactive until they are brought together in the proper orientation (134). The pieces of the split fluorescent protein can complement each other if one is fused to a kinase substrate and the other is fused to a phosphoamino-acid binding domain. Upon phosphorylation of the substrate motif, the split-protein fluorophore is reconstituted, and a fluorescent signal is obtained. Reporters utilizing this complementation technique have been performed with GFP, YFP, and Renilla luciferase (134).
A genetically encoded biosensor using bioluminescence as a readout has been developed for Akt and has been used for imaging of the Akt kinase activity within live cells and mice (135). This Akt kinase activity reporter contains an Akt consensus peptide and an FHA2 domain flanked by the N-terminal and C-terminal domains of firefly luciferase. Phosphorylation of the Akt consensus sequence stimulates interaction with the FHA2 domain, triggering a conformational change, which disrupts the split luciferase. Upon dephosphorylation of the Akt consensus sequence (or in the absence of Akt kinase activity), the reconstitution of the luciferase reporter molecule is allowed, and bioluminescence can be detected.
Applications of Genetically Encoded Reporters
There have now been a myriad of published applications of genetically encoded reporters, and we highlight just a few here (131, 136–142). The dynamic range (or percent change in signal) for the genetically encoded FRET-based biosensors is considerably smaller (only 10% to 70% signal change) (143) as compared to the severalfold increases in fluorescence for the peptide-based biosensors, but they show sufficient sensitivity to provide very useful information about the phosphorylation and regulation of signaling pathways. Using a PKA reporter, elegant studies revealed the unexpected delay between cyclic AMP increases and PKA activity (136). Through the development of a specific and reversible reporter for PKC activity in live cells, investigators were able to reveal the oscillations of phosphorylation by PKC in HeLa cells in response to histamine (131). A key aspect of this study involved targeting the PKC reporter to the plasma membrane. With a genetically encoded FRET biosensor for histone phosphorylation, investigators were able to demonstrate a correlation between the phosphorylation of Ser28 on H3 histone and the onset of nuclear-envelope breakdown, indicating that phosphorylation at this position may be involved in regulating changes during M phase (137).
The development and use of FRET-based biosensors for aurora B kinase activity permitted an analysis of the anaphase dynamics of phosphorylation by this key mitotic regulator (138). A phosphorylation gradient produced by aurora kinase B is generated in early anaphase and requires the localization of aurora B to the spindle midzone where it is activated. These observations indicate that events in anaphase are controlled primarily by spatial cues, such as this phosphorylation gradient rather than the timing of phosphorylation.
A Src tyrosine kinase reporter has been applied to analyze the role of Src activity in transmission of mechanical forces to the cytoskeleton. Using the Src reporter, rapid changes in activity in a spatiotemporal distribution near the plasma membrane were observed (139). This Src reporter was also employed to validate the rapid chemical rescue of a mutant tyrosine kinase, described above (63).
One of the more exciting applications of kinase reporters, which has pharmaceutical relevance, is in the area of high-throughput chemical screens. Typically, these screens are carried out under in vitro conditions, so that it is unclear if these compounds can target the intracellular kinase. In-cell kinase reporters can ensure compounds achieve efficient cell entry and provide rapid kinetic information on inhibition, which can establish that they are directly hitting the intended target. Although the FRET reporters are normally studied at the single-cell level, a PKA reporter, which had been optimized to have a high change in FRET, gave sufficient signal such that bulk fluorescence change could be measured in 96-well plate format with a standard fluorescent plate reader (144). Using this approach, a clinical compound library was screened, and several novel PKA inhibitors were identified.
SUMMARY AND EMERGING CHALLENGES
The rapid pace of chemical biology developments in the protein phosphorylation field, reviewed above, have led to great progress. New approaches that allow for specific modulation of wild-type or mutant kinases are offering exciting insights into the structures and functions of protein kinases. The ability to install phosphoamino acids and their mimics site specifically into proteins by semisynthesis or nonsense suppression strategies is providing unprecedented insights into how individual phosphorylation events can contribute to catalytic, structural, and cellular behaviors. Moreover, the new generation of fluorescent reporters of kinase activities are allowing for precise correlations between signaling and other cellular events.
However, challenges in the phosphorylation arena remain. It is estimated that 20% to 30% of proteins in the cell are phosphorylated (145), and sorting out which of the 500 kinases is responsible for a particular phosphorylation event remains a major challenge. Chemical and proteomic techniques are beginning to systematically uncover which kinase phosphorylates which proteins (146). Using engineered kinases with ATP analogs (51), it has been possible to link specific kinases directly responsible for a particular phosphorylation event. Protein chips, which consist of chemically immobilized proteins on a glass slide in a microarray format, also appear powerful (147). Chemical and photochemical cross-linking strategies are in development to pinpoint kinase-substrate relationships (148, 149). In addition, the use of bisubstrate analogs, consisting of protein-ATP conjugates, offers hope for identifying a kinase that can phosphorylate a particular protein site. Still lacking is an efficient in vivo approach along the lines of dead-end catalytic mutants, which have been used to link protein phosphatases to phosphoprotein substrates (150). Perhaps chemical complementation of an engineered mutant will provide a key for adapting this method to kinases.
Given the interest in developing small-molecule kinase inhibitors and the difficulty in achieving specificity, methods to characterize the cellular effects of potential kinase inhibitors are needed. Resistance mutants, pull-down assays, three-hybrid screens, mass spectrometry analysis, and activity-based proteomics may provide new answers (151–154). In addition, other PTMs, including acetylation, methylation, glycosylation, and ubiquitylation, are expected to have many overlapping but distinct influences on protein and cell functions, and these modifications will require the same detailed analyses as those performed for phosphorylation (155–160). In summary, the opportunities for chemical biologists to continue to make their mark in the thriving area of signaling will continue for many years ahead.
Acknowledgments
We thank our lab members and many collaborators and colleagues for advice and helpful discussions and the NIH for support.
Glossary
- PTM
posttranslational modification
- SH2 domain
a Src homology domain that binds phosphorylated Tyr residues
- pThr
phosphothreonine
- pTyr
phosphotyrosine
- Expressed protein ligation (EPL)
the ligation of a recombinant protein bearing C-terminal thiosester (generated by intein) to a N-Cys-containing synthetic peptide
- Native chemical ligation
a chemoselective reaction between a peptide/protein with N-terminal Cys and a peptide/protein C-terminal thioester, forming a standard amide bond
- SH3 domain
a Src homology domain that binds proline-rich peptides
- FRET
Forster (fluorescence) resonance energy transfer
- FLIM
fluorescence lifetime imaging microscopy
- YFP
yellow fluorescent protein
- Bioluminescence
the emission of visible light resulting from a chemical reaction that does not require external light
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Krebs EG, Fischer EH. Phosphorylase activity of skeletal muscle extracts. J Biol Chem. 1955;216:113–20. [PubMed] [Google Scholar]
- 2.Fischer EH, Krebs EG. Conversion of phosphorylase b to phosphorylase a in muscle extracts. J Biol Chem. 1955;216:121–32. [PubMed] [Google Scholar]
- 3.Hayes JS, Mayer SE. Regulation of guinea pig heart phosphorylase kinase by cAMP, protein kinase, and calcium. Am J Physiol. 1981;240:E340–49. doi: 10.1152/ajpendo.1981.240.3.E340. [DOI] [PubMed] [Google Scholar]
- 4.Hunter T. Signaling–2000 and beyond. Cell. 2000;100:113–27. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 5.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 6.Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–15. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 7.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 8.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol. 2002;3:177–86. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Curr Opin Cell Biol. 2001;13:131–38. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- 11.Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6:556–68. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 12.Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–49. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- 13.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Nakadate T, Jeng AY, Blumberg PM. Comparison of protein kinase C functional assays to clarify mechanisms of inhibitor action. Biochem Pharmacol. 1988;37:1541–45. doi: 10.1016/0006-2952(88)90016-0. [DOI] [PubMed] [Google Scholar]
- 15.Lawrie AM, Noble ME, Tunnah P, Brown NR, Johnson LN, Endicott JA. Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat Struct Biol. 1997;4:796–801. doi: 10.1038/nsb1097-796. [DOI] [PubMed] [Google Scholar]
- 16.Young PR, McLaughlin MM, Kumar S, Kassis S, Doyle ML, et al. Pyridinyl imidazole inhibitors of p38 mitogen-activated protein kinase bind in the ATP site. J Biol Chem. 1997;272:12116–21. doi: 10.1074/jbc.272.18.12116. [DOI] [PubMed] [Google Scholar]
- 17.Wilson KP, McCaffrey PG, Hsiao K, Pazhanisamy S, Galullo V, et al. The structural basis for the specificity of pyridinylimidazole inhibitors of p38 MAP kinase. Chem Biol. 1997;4:423–31. doi: 10.1016/s1074-5521(97)90194-0. [DOI] [PubMed] [Google Scholar]
- 18.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–72. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 19.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–36. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 20.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289:1938–42. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 21.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–59. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 22.Fry DW, Bridges AJ, Denny WA, Doherty A, Greis KD, et al. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. Proc Natl Acad Sci USA. 1998;95:12022–27. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3:156–60. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–21. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–20. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 26.Bose R, Holbert MA, Pickin KA, Cole PA. Protein tyrosine kinase-substrate interactions. Curr Opin Struct Biol. 2006;16:668–75. doi: 10.1016/j.sbi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Lee JH, Nandy SK, Lawrence DS. A highly potent and selective PKCα inhibitor generated via combinatorial modification of a peptide scaffold. J Am Chem Soc. 2004;126:3394–95. doi: 10.1021/ja037300b. [DOI] [PubMed] [Google Scholar]
- 28.Hah JM, Sharma V, Li H, Lawrence DS. Acquisition of a “Group A”-selective Src kinase inhibitor via a global targeting strategy. J Am Chem Soc. 2006;128:5996–97. doi: 10.1021/ja060136i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JH, Kumar S, Lawrence DS. Stepwise combinatorial evolution of Akt bisubstrate inhibitors. ChemBioChem. 2008;9:507–9. doi: 10.1002/cbic.200700583. [DOI] [PubMed] [Google Scholar]
- 30.Yuan CJ, Jakes S, Elliott S, Graves DJ. A rationale for the design of an inhibitor of tyrosyl kinase. J Biol Chem. 1990;265:16205–9. [PubMed] [Google Scholar]
- 31.Kim CU, Lew W, Williams MA, Liu H, Zhang L, et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc. 1997;119:681–90. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 32.Williams DM, Cole PA. Proton demand inversion in a mutant protein tyrosine kinase reaction. J Am Chem Soc. 2002;124:5956–57. doi: 10.1021/ja025993a. [DOI] [PubMed] [Google Scholar]
- 33.Kim KJ, Cole PA. Kinetic analysis of a protein tyrosine kinase reaction transition state in the forward and reverse directions. J Am Chem Soc. 1998;120:6851–58. [Google Scholar]
- 34.Mildvan AS. Mechanisms of signaling and related enzymes. Proteins. 1997;29:401–16. [PubMed] [Google Scholar]
- 35.Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Mechanism-based design of a protein kinase inhibitor. Nat Struct Biol. 2001;8:37–41. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]
- 36.Shen K, Cole PA. Conversion of a tyrosine kinase protein substrate to a high affinity ligand by ATP linkage. J Am Chem Soc. 2003;125:16172–73. doi: 10.1021/ja0380401. [DOI] [PubMed] [Google Scholar]
- 37.Hines AC, Cole PA. Design, synthesis, and characterization of an ATP-peptide conjugate inhibitor of protein kinase A. Bioorg Med Chem Lett. 2004;14:2951–54. doi: 10.1016/j.bmcl.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, et al. A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, et al. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem. 2006;281:23167–79. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
- 41.Ahn DR, Han KC, Kwon HS, Yang EG. ATP-conjugated peptide inhibitors for calmodulin-dependent protein kinase II. Bioorg Med Chem Lett. 2007;17:147–51. doi: 10.1016/j.bmcl.2006.09.070. [DOI] [PubMed] [Google Scholar]
- 42.Pickin KA, Chaudhury S, Dancy BC, Gray JJ, Cole PA. Analysis of protein kinase autophosphorylation using expressed protein ligation and computational modeling. J Am Chem Soc. 2008;130:5667–69. doi: 10.1021/ja711244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider TL, Mathew RS, Rice KP, Tamaki K, Wood JL, Schepartz A. Increasing the kinase specificity of K252a by protein surface recognition. Org Lett. 2005;7:1695–98. doi: 10.1021/ol050179o. [DOI] [PubMed] [Google Scholar]
- 44.Enkvist E, Lavogina D, Raidaru G, Vaasa A, Viil I, et al. Conjugation of adenosine and hexa-(D-arginine) leads to a nanomolar bisubstrate-analog inhibitor of basophilic protein kinases. J Med Chem. 2006;49:7150–59. doi: 10.1021/jm0605942. [DOI] [PubMed] [Google Scholar]
- 45.Meyer SC, Shomin CD, Gaj T, Ghosh I. Tethering small molecules to a phage display library: discovery of a selective bivalent inhibitor of protein kinase A. J Am Chem Soc. 2007;129:13812–13. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 46.Chen L, Wright LR, Chen CH, Oliver SF, Wender PA, Mochly-Rosen D. Molecular transporters for peptides: delivery of a cardioprotective εPKC agonist peptide into cells and intact ischemic heart using a transport system, R(7) Chem Biol. 2001;8:1123–29. doi: 10.1016/s1074-5521(01)00076-x. [DOI] [PubMed] [Google Scholar]
- 47.Borsello T, Clarke PG, Hirt L, Vercelli A, Repici M, et al. A peptide inhibitor of c-Jun N-terminal kinase protects against excitotoxicity and cerebral ischemia. Nat Med. 2003;9:1180–86. doi: 10.1038/nm911. [DOI] [PubMed] [Google Scholar]
- 48.Hwang YW, Miller DL. A mutation that alters the nucleotide specificity of elongation factor Tu, a GTP regulatory protein. J Biol Chem. 1987;262:13081–85. [PubMed] [Google Scholar]
- 49.Carter P, Wells JA. Engineering enzyme specificity by “substrate-assisted catalysis. Science. 1987;237:394–99. doi: 10.1126/science.3299704. [DOI] [PubMed] [Google Scholar]
- 50.Belshaw PJ, Schoepfer JG, Liu K, Morrison KL, Schreiber SL. Rational design of orthogonal receptor-ligand combinations. Angew Chem Int Ed Engl. 1995;34:2129–32. [Google Scholar]
- 51.Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci USA. 1997;94:3565–70. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- 53.Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–66. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 54.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 55.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–30. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 56.Zhang C, Kenski DM, Paulson JL, Bonshtien A, Sessa G, et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods. 2005;2:435–41. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 57.Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell. 2007;25:839–50. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci USA. 2006;103:19466–71. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Craik CS, Roczniak S, Largman C, Rutter WJ. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987;237:909–13. doi: 10.1126/science.3303334. [DOI] [PubMed] [Google Scholar]
- 60.Toney MD, Kirsch JF. Direct Bronsted analysis of the restoration of activity to a mutant enzyme by exogenous amines. Science. 1989;243:1485–88. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- 61.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28:7913–18. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 62.Williams DM, Wang D, Cole PA. Chemical rescue of a mutant protein-tyrosine kinase. J Biol Chem. 2000;275:38127–30. doi: 10.1074/jbc.C000606200. [DOI] [PubMed] [Google Scholar]
- 63.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–97. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 64.Lowry WE, Huang J, Ma YC, Ali S, Wang D, et al. Csk, a critical link of G protein signals to actin cytoskeletal reorganization. Dev Cell. 2002;2:733–44. doi: 10.1016/s1534-5807(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 65.Madhusoodanan KS, Guo D, McGarrigle DK, Maack T, Huang XY. Csk mediates G-protein-coupled lysophosphatidic acid receptor-induced inhibition of membrane-bound guanylyl cyclase activity. Biochemistry. 2006;45:3396–403. doi: 10.1021/bi052513u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Im E, Kazlauskas A. Src family kinases promote vessel stability by antagonizing the Rho/ROCK pathway. J Biol Chem. 2007;282:29122–29. doi: 10.1074/jbc.M702637200. [DOI] [PubMed] [Google Scholar]
- 67.Sprang SR, Acharya KR, Goldsmith EJ, Stuart DI, Varvill K, et al. Structural changes in glycogen phosphorylase induced by phosphorylation. Nature. 1988;336:215–21. doi: 10.1038/336215a0. [DOI] [PubMed] [Google Scholar]
- 68.Thorsness PE, Koshland DE., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987;262:10422–25. [PubMed] [Google Scholar]
- 69.Potter LR, Hunter T. Phosphorylation of the kinase homology domain is essential for activation of the A-type natriuretic peptide receptor. Mol Cell Biol. 1998;18:2164–72. doi: 10.1128/mcb.18.4.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao M, Lowy AM, Kapoor M, Deffie A, Liu G, Lozano G. Mutation of phosphoserine 389 affects p53 function in vivo. J Biol Chem. 1996;271:29380–85. doi: 10.1074/jbc.271.46.29380. [DOI] [PubMed] [Google Scholar]
- 71.Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol. 2001;8:248–53. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- 72.Zheng W, Zhang Z, Ganguly S, Weller JL, Klein DC, Cole PA. Cellular stabilization of the melatonin rhythm enzyme induced by nonhydrolyzable phosphonate incorporation. Nat Struct Biol. 2003;10:1054–57. doi: 10.1038/nsb1005. [DOI] [PubMed] [Google Scholar]
- 73.Xia F, Li J, Hickey GW, Tsurumi A, Larson K, et al. Raf activation is regulated by tyrosine 510 phosphorylation in Drosophila. PLoS Biol. 2008;6:e128. doi: 10.1371/journal.pbio.0060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–10. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–79. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 76.Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT. AP-1.DNA complex. Chem Biol. 1996;3:981–91. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 77.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–9. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 78.Shekhtman A, Ghose R, Wang D, Cole PA, Cowburn D. Novel mechanism of regulation of the non-receptor protein tyrosine kinase Csk: insights from NMR mapping studies and site-directed mutagenesis. J Mol Biol. 2001;314:129–38. doi: 10.1006/jmbi.2001.5126. [DOI] [PubMed] [Google Scholar]
- 79.Ogawa A, Takayama Y, Sakai H, Chong KT, Takeuchi S, et al. Structure of the carboxyl-terminal Src kinase, Csk. J Biol Chem. 2002;277:14351–54. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- 80.Lu W, Gong D, Bar-Sagi D, Cole PA. Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol Cell. 2001;8:759–69. doi: 10.1016/s1097-2765(01)00369-0. [DOI] [PubMed] [Google Scholar]
- 81.Lu W, Shen K, Cole PA. Chemical dissection of the effects of tyrosine phosphorylation of SHP-2. Biochemistry. 2003;42:5461–68. doi: 10.1021/bi0340144. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Z, Shen K, Lu W, Cole PA. The role of C-terminal tyrosine phosphorylation in the regulation of SHP-1 explored via expressed protein ligation. J Biol Chem. 2003;278:4668–74. doi: 10.1074/jbc.M210028200. [DOI] [PubMed] [Google Scholar]
- 83.Schwarzer D, Zhang Z, Zheng W, Cole PA. Negative regulation of a protein tyrosine phosphatase by tyrosine phosphorylation. J Am Chem Soc. 2006;128:4192–93. doi: 10.1021/ja0585174. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Somani AK, Siminovitch KA. Roles of the SHP-1 tyrosine phosphatase in the negative regulation of cell signalling. Semin Immunol. 2000;12:361–78. doi: 10.1006/smim.2000.0223. [DOI] [PubMed] [Google Scholar]
- 85.Feng GS. Shp-2 tyrosine phosphatase: signaling one cell or many. Exp Cell Res. 1999;253:47–54. doi: 10.1006/excr.1999.4668. [DOI] [PubMed] [Google Scholar]
- 86.Tailor P, Gilman J, Williams S, Couture C, Mustelin T. Regulation of the low molecular weight phosphotyrosine phosphatase by phosphorylation at tyrosines 131 and 132. J Biol Chem. 1997;272:5371–74. doi: 10.1074/jbc.272.9.5371. [DOI] [PubMed] [Google Scholar]
- 87.Bucciantini M, Chiarugi P, Cirri P, Taddei L, Stefani M, et al. The low Mr phosphotyrosine protein phosphatase behaves differently when phosphorylated at Tyr131 or Tyr132 by Src kinase. FEBS Lett. 1999;456:73–78. doi: 10.1016/s0014-5793(99)00828-5. [DOI] [PubMed] [Google Scholar]
- 88.Zheng W, Schwarzer D, Lebeau A, Weller JL, Klein DC, Cole PA. Cellular stability of serotonin N-acetyltransferase conferred by phosphonodifluoromethylene alanine (Pfa) substitution for Ser-205. J Biol Chem. 2005;280:10462–67. doi: 10.1074/jbc.M412283200. [DOI] [PubMed] [Google Scholar]
- 89.Szewczuk LM, Tarrant MK, Sample V, Drury WJ, Zhang J, Cole PA. Analysis of serotonin N-acetyltransferase regulation in vitro and in live cells using protein semisynthesis. Biochemistry. 2008;47:10407–19. doi: 10.1021/bi801189d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ganguly S, Coon SL, Klein DC. Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 2002;309:127–37. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- 91.Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci USA. 2005;102:1222–27. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hahn ME, Pellois JP, Vila-Perello M, Muir TW. Tunable photoactivation of a post-translationally modified signaling protein and its unmodified counterpart in live cells. ChemBioChem. 2007;8:2100–5. doi: 10.1002/cbic.200700404. [DOI] [PubMed] [Google Scholar]
- 93.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massague J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell. 2001;8:671–82. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 94.Ottesen JJ, Huse M, Sekedat MD, Muir TW. Semisynthesis of phosphovariants of Smad2 reveals a substrate preference of the activated T beta RI kinase. Biochemistry. 2004;43:5698–706. doi: 10.1021/bi0498407. [DOI] [PubMed] [Google Scholar]
- 95.Hahn ME, Muir TW. Photocontrol of Smad2, a multiphosphorylated cell-signaling protein, through caging of activating phosphoserines. Angew Chem Int Ed Engl. 2004;43:5800–3. doi: 10.1002/anie.200461141. [DOI] [PubMed] [Google Scholar]
- 96.Vila-Perello M, Pratt MR, Tulin F, Muir TW. Covalent capture of phospho-dependent protein oligomerization by site-specific incorporation of a diazirine photo-cross-linker. J Am Chem Soc. 2007;129:8068–69. doi: 10.1021/ja072013j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heckler TG, Roesser JR, Xu C, Chang PI, Hecht SM. Ribosomal binding and dipeptide formation by misacylated tRNA(Phe),S. Biochemistry. 1988;27:7254–62. doi: 10.1021/bi00419a012. [DOI] [PubMed] [Google Scholar]
- 98.Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–88. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 99.Xie J, Schultz PG. A chemical toolkit for proteins—an expanded genetic code. Nat Rev Mol Cell Biol. 2006;7:775–82. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 100.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 101.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–49. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 102.Xie J, Supekova L, Schultz PG. A genetically encoded metabolically stable analogue of phosphotyrosine in Escherichia coli. ACS Chem Biol. 2007;2:474–78. doi: 10.1021/cb700083w. [DOI] [PubMed] [Google Scholar]
- 103.Sharma V, Wang Q, Lawrence DS. Peptide-based fluorescent sensors of protein kinase activity: design and applications. Biochim Biophys Acta. 2008;1784:94–99. doi: 10.1016/j.bbapap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Q, Lawrence DS. Phosphorylation-driven protein-protein interactions: a protein kinase sensing system. J Am Chem Soc. 2005;127:7684–85. doi: 10.1021/ja050789j. [DOI] [PubMed] [Google Scholar]
- 105.Sharma V, Agnes RS, Lawrence DS. Deep quench: an expanded dynamic range for protein kinase sensors. J Am Chem Soc. 2007;129:2742–43. doi: 10.1021/ja068280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Q, Cahill SM, Blumenstein M, Lawrence DS. Self-reporting fluorescent substrates of protein tyrosine kinases. J Am Chem Soc. 2006;128:1808–9. doi: 10.1021/ja0577692. [DOI] [PubMed] [Google Scholar]
- 107.Shults MD, Pearce DA, Imperiali B. Modular and tunable chemosensor scaffold for divalent zinc. J Am Chem Soc. 2003;125:10591–97. doi: 10.1021/ja0355980. [DOI] [PubMed] [Google Scholar]
- 108.Shults MD, Carrico-Moniz D, Imperiali B. Optimal Sox-based fluorescent chemosensor design for serine/threonine protein kinases. Anal Biochem. 2006;352:198–207. doi: 10.1016/j.ab.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 109.Shults MD, Imperiali B. Versatile fluorescence probes of protein kinase activity. J Am Chem Soc. 2003;125:14248–49. doi: 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]
- 110.Yeh RH, Yan X, Cammer M, Bresnick AR, Lawrence DS. Real time visualization of protein kinase activity in living cells. J Biol Chem. 2002;277:11527–32. doi: 10.1074/jbc.M111300200. [DOI] [PubMed] [Google Scholar]
- 111.Wang Q, Dai Z, Cahill SM, Blumenstein M, Lawrence DS. Light-regulated sampling of protein tyrosine kinase activity. J Am Chem Soc. 2006;128:14016–17. doi: 10.1021/ja065852z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sims CE, Allbritton NL. Single-cell kinase assays: opening a window onto cell behavior. Curr Opin Biotechnol. 2003;14:23–28. doi: 10.1016/s0958-1669(02)00002-2. [DOI] [PubMed] [Google Scholar]
- 113.Li H, Sims CE, Kaluzova M, Stanbridge EJ, Allbritton NL. A quantitative single-cell assay for protein kinase B reveals important insights into the biochemical behavior of an intracellular substrate peptide. Biochemistry. 2004;43:1599–608. doi: 10.1021/bi035597k. [DOI] [PubMed] [Google Scholar]
- 114.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, et al. Single cell profiling of potentiated phosphoprotein networks in cancer cells. Cell. 2004;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 115.Krutzik PO, Irish JM, Nolan GP, Perez OD. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol. 2004;110:206–21. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 116.Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20:155–62. doi: 10.1038/nbt0202-155. [DOI] [PubMed] [Google Scholar]
- 117.Muller S, Demotz S, Bulliard C, Valitutti S. Kinetics and extent of protein tyrosine kinase activation in individual T cells upon antigenic stimulation. Immunology. 1999;97:287–93. doi: 10.1046/j.1365-2567.1999.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bastiaens PI, Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999;9:48–52. doi: 10.1016/s0962-8924(98)01410-x. [DOI] [PubMed] [Google Scholar]
- 119.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, et al. Imaging protein kinase Cα activation in cells. Science. 1999;283:2085–89. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 120.Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI. Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science. 2000;290:1567–70. doi: 10.1126/science.290.5496.1567. [DOI] [PubMed] [Google Scholar]
- 121.Haj FG, Verveer PJ, Squire A, Neel BG, Bastiaens PI. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science. 2002;295:1708–11. doi: 10.1126/science.1067566. [DOI] [PubMed] [Google Scholar]
- 122.Legg JW, Lewis CA, Parsons M, Ng T, Isacke CM. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat Cell Biol. 2002;4:399–407. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 123.Peltan ID, Thomas AV, Mikhailenko I, Strickland DK, Hyman BT, von Arnim CA. Fluorescence lifetime imaging microscopy (FLIM) detects stimulus-dependent phosphorylation of the low density lipoprotein receptor-related protein (LRP) in primary neurons. Biochem Biophys Res Commun. 2006;349:24–30. doi: 10.1016/j.bbrc.2006.07.212. [DOI] [PubMed] [Google Scholar]
- 124.Oancea E, Teruel MN, Quest AF, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J Cell Biol. 1998;140:485–98. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Codazzi F, Teruel MN, Meyer T. Control of astrocyte Ca(2+) oscillations and waves by oscillating translocation and activation of protein kinase C. Curr Biol. 2001;11:1089–97. doi: 10.1016/s0960-9822(01)00326-8. [DOI] [PubMed] [Google Scholar]
- 126.Zhang J, Allen MD. FRET-based biosensors for protein kinases: illuminating the kinome. Mol Biosyst. 2007;3:759–65. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 127.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–13. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 128.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–60. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 129.Suhling K, French PM, Phillips D. Time-resolved fluorescence microscopy. Photochem Photobiol Sci. 2005;4:13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]
- 130.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 131.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Allen MD, Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun. 2006;348:716–21. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 133.Offterdinger M, Georget V, Girod A, Bastiaens PI. Imaging phosphorylation dynamics of the epidermal growth factor receptor. J Biol Chem. 2004;279:36972–81. doi: 10.1074/jbc.M405830200. [DOI] [PubMed] [Google Scholar]
- 134.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–82. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 135.Zhang L, Lee KC, Bhojani MS, Khan AP, Shilman A, et al. Molecular imaging of Akt kinase activity. Nat Med. 2007;13:1114–19. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 136.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts β-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–73. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 137.Lin CW, Ting AY. A genetically encoded fluorescent reporter of histone phosphorylation in living cells. Angew Chem Int Ed Engl. 2004;43:2940–43. doi: 10.1002/anie.200353375. [DOI] [PubMed] [Google Scholar]
- 138.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–36. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–45. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 140.Green HM, Alberola-Ila J. Development of ERK activity sensor, an in vitro, FRET-based sensor of extracellular regulated kinase activity. BMC Chem Biol. 2005;5:1. doi: 10.1186/1472-6769-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sato M, Umezawa Y. Imaging protein phosphorylation by fluorescence in single living cells. Methods. 2004;32:451–55. doi: 10.1016/j.ymeth.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 142.Umezawa Y. Genetically encoded optical probes for imaging cellular signaling pathways. Biosens Bioelectron. 2005;20:2504–11. doi: 10.1016/j.bios.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 143.Rothman DM, Shults MD, Imperiali B. Chemical approaches for investigating phosphorylation in signal transduction networks. Trends Cell Biol. 2005;15:502–10. doi: 10.1016/j.tcb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 144.Allen MD, DiPilato LM, Rahdar M, Ren YR, Chong C, et al. Reading dynamic kinase activity in living cells for high-throughput screening. ACS Chem Biol. 2006;1:371–76. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- 145.Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 146.Bonilla L, Means G, Lee K, Patterson S. The evolution of tools for protein phosphorylation site analysis: from discovery to clinical application. BioTechniques. 2008;44:671–79. doi: 10.2144/000112800. [DOI] [PubMed] [Google Scholar]
- 147.Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, et al. Analysis of yeast protein kinases using protein chips. Nat Genet. 2000;26:283–89. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 148.Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal affinity purification of direct kinase substrates. J Am Chem Soc. 2005;127:5288–89. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, et al. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4:511–16. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1997;94:1680–85. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, et al. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–39. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Caligiuri M, Molz L, Liu Q, Kaplan F, Xu JP, et al. MASPIT: three-hybrid trap for quantitative proteome fingerprinting of small molecule-protein interactions in mammalian cells. Chem Biol. 2006;13:711–22. doi: 10.1016/j.chembiol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 153.Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci USA. 2006;103:9773–78. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–58. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 155.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–47. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 156.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–15. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 157.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–95. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 158.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–16. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lin CW, Jao CY, Ting AY. Genetically encoded fluorescent reporters of histone methylation in living cells. J Am Chem Soc. 2004;126:5982–83. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]