Abstract
Footprinting data for 33 open promoter complexes with Escherichia coli RNA polymerase, as well as 17 ternary complexes with different regulators, have been compiled using a computer program FUTPR. The typical and individual properties of their structural organization are analyzed. Promoters are subgrouped according to the extent of the polymerase contact area. A set of alternative sequence elements that could be responsible for RNA polymerase attachment in different promoter groups is suggested on the basis of their sequence homology near the hyperreactive sites. The model of alternative pathways used for promoter activation is discussed.
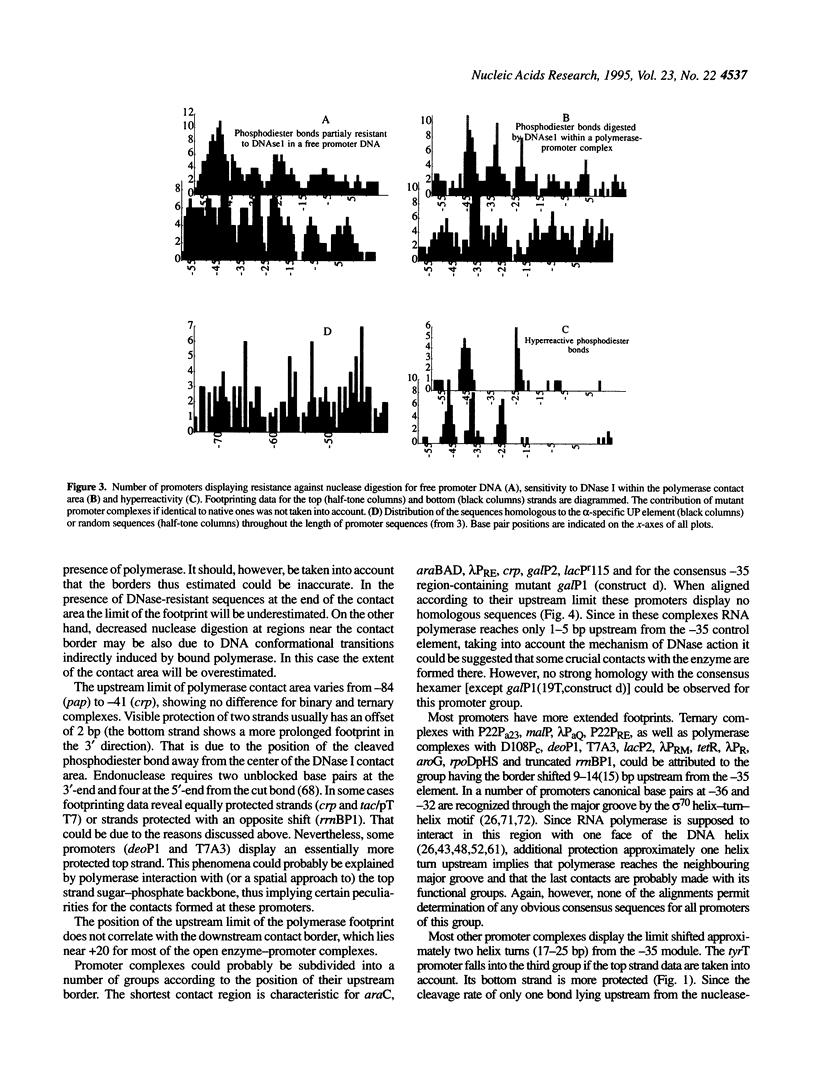
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983 Jan;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Attey A., Belyaeva T., Savery N., Hoggett J., Fujita N., Ishihama A., Busby S. Interactions between the cyclic AMP receptor protein and the alpha subunit of RNA polymerase at the Escherichia coli galactose operon P1 promoter. Nucleic Acids Res. 1994 Oct 25;22(21):4375–4380. doi: 10.1093/nar/22.21.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseggio N., Davies W. D., Davidson B. E. Identification of the promoter, operator, and 5' and 3' ends of the mRNA of the Escherichia coli K-12 gene aroG. J Bacteriol. 1990 May;172(5):2547–2557. doi: 10.1128/jb.172.5.2547-2557.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A., Gaston K., Williams R., Chapman K., Kolb A., Buc H., Minchin S., Williams J., Busby S. Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res. 1990 Dec 25;18(24):7243–7250. doi: 10.1093/nar/18.24.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva T., Griffiths L., Minchin S., Cole J., Busby S. The Escherichia coli cysG promoter belongs to the 'extended -10' class of bacterial promoters. Biochem J. 1993 Dec 15;296(Pt 3):851–857. doi: 10.1042/bj2960851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand-Burggraf E., Dunand J., Fuchs R. P., Lefèvre J. F. Kinetic studies of the modulation of ada promoter activity by upstream elements. EMBO J. 1990 Jul;9(7):2265–2271. doi: 10.1002/j.1460-2075.1990.tb07397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutel B. A., Record M. T., Jr E. coli promoter spacer regions contain nonrandom sequences which correlate to spacer length. Nucleic Acids Res. 1990 Jun 25;18(12):3597–3603. doi: 10.1093/nar/18.12.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Brunner M., Bujard H. Promoter recognition and promoter strength in the Escherichia coli system. EMBO J. 1987 Oct;6(10):3139–3144. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S., West D., Lawes M., Webster C., Ishihama A., Kolb A. Transcription activation by the Escherichia coli cyclic AMP receptor protein. Receptors bound in tandem at promoters can interact synergistically. J Mol Biol. 1994 Aug 19;241(3):341–352. doi: 10.1006/jmbi.1994.1511. [DOI] [PubMed] [Google Scholar]
- Chan B., Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989 Dec 14;84(2):227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- Chan B., Spassky A., Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus -35 region sequences. Biochem J. 1990 Aug 15;270(1):141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. T., Lebowitz J. The coupled use of 'footprinting' and exonuclease III methodology for RNA polymerase binding and initiation. Application for the analysis of three tandem promoters at the control region of colicin El. Nucleic Acids Res. 1983 Feb 25;11(4):1099–1116. doi: 10.1093/nar/11.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowing D. W., Mecsas J., Record M. T., Jr, Gross C. A. Intermediates in the formation of the open complex by RNA polymerase holoenzyme containing the sigma factor sigma 32 at the groE promoter. J Mol Biol. 1989 Dec 5;210(3):521–530. doi: 10.1016/0022-2836(89)90128-9. [DOI] [PubMed] [Google Scholar]
- Danot O., Raibaud O. Multiple protein-DNA and protein-protein interactions are involved in transcriptional activation by MalT. Mol Microbiol. 1994 Oct;14(2):335–346. doi: 10.1111/j.1365-2958.1994.tb01294.x. [DOI] [PubMed] [Google Scholar]
- Deuschle U., Kammerer W., Gentz R., Bujard H. Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J. 1986 Nov;5(11):2987–2994. doi: 10.1002/j.1460-2075.1986.tb04596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Duval-Valentin G., Ehrlich R. Interaction between E. coli RNA polymerase and the tetR promoter from pSC101: homologies and differences with other E. coli promoter systems from close contact point studies. Nucleic Acids Res. 1986 Mar 11;14(5):1967–1983. doi: 10.1093/nar/14.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval-Valentin G., Reiss C. How Escherichia coli RNA polymerase can negatively regulate transcription from a constitutive promoter. Mol Microbiol. 1990 Sep;4(9):1465–1475. doi: 10.1111/j.1365-2958.1990.tb02057.x. [DOI] [PubMed] [Google Scholar]
- Gardella T., Moyle H., Susskind M. M. A mutant Escherichia coli sigma 70 subunit of RNA polymerase with altered promoter specificity. J Mol Biol. 1989 Apr 20;206(4):579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- Gourse R. L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988 Oct 25;16(20):9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., de Boer H. A., Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986 Jan 17;44(1):197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger P. A., deHaseth P. L. RNA polymerase bound to the PR promoter of bacteriophage lambda inhibits open complex formation at the divergently transcribed PRM promoter. Implications for an indirect mechanism of transcriptional activation by lambda repressor. J Mol Biol. 1991 Dec 5;222(3):479–494. doi: 10.1016/0022-2836(91)90491-n. [DOI] [PubMed] [Google Scholar]
- Ho Y. S., Pfarr D., Strickler J., Rosenberg M. Characterization of the transcription activator protein C1 of bacteriophage P22. J Biol Chem. 1992 Jul 15;267(20):14388–14397. [PubMed] [Google Scholar]
- Ho Y. S., Rosenberg M. Characterization of a third, cII-dependent, coordinately activated promoter on phage lambda involved in lysogenic development. J Biol Chem. 1985 Sep 25;260(21):11838–11844. [PubMed] [Google Scholar]
- Ho Y. S., Wulff D. L., Rosenberg M. Bacteriophage lambda protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature. 1983 Aug 25;304(5928):703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- Hofer B., Müller D., Köster H. The pathway of E. coli RNA polymerase-promoter complex formation as visualized by footprinting. Nucleic Acids Res. 1985 Aug 26;13(16):5995–6013. doi: 10.1093/nar/13.16.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz A. H., Morandi C., Wilcox G. Deoxyribonucleic acid sequence of araBAD promoter mutants of Escherichia coli. J Bacteriol. 1980 May;142(2):659–667. doi: 10.1128/jb.142.2.659-667.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen C., Buchardt O., Henriksen U., Nielsen P. E. Photocleavage of DNA and photofootprinting of E. coli RNA polymerase bound to promoter DNA by azido-9-acridinylamines. Nucleic Acids Res. 1988 Jul 11;16(13):5755–5770. doi: 10.1093/nar/16.13.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa H., Mabuchi K., Futai M. Nucleotide sequence of the promoter region of the gene cluster for proton-translocating ATPase from Escherichia coli and identification of the active promotor. Biochem Biophys Res Commun. 1982 Jul 30;107(2):568–575. doi: 10.1016/0006-291x(82)91529-7. [DOI] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- Kobayashi M., Nagata K., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: effect of base substitutions in the promoter -35 region on promoter strength. Nucleic Acids Res. 1990 Dec 25;18(24):7367–7372. doi: 10.1093/nar/18.24.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Igarashi K., Ishihama A., Lavigne M., Buckle M., Buc H. E. coli RNA polymerase, deleted in the C-terminal part of its alpha-subunit, interacts differently with the cAMP-CRP complex at the lacP1 and at the galP1 promoter. Nucleic Acids Res. 1993 Jan 25;21(2):319–326. doi: 10.1093/nar/21.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic R. T. The 0 degree C closed complexes between Escherichia coli RNA polymerase and two promoters, T7-A3 and lacUV5. J Biol Chem. 1987 Oct 5;262(28):13654–13661. [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry. 1989 Sep 19;28(19):7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- Kukolj G., DuBow M. S. Integration host factor activates the Ner-repressed early promoter of transposable Mu-like phage D108. J Biol Chem. 1992 Sep 5;267(25):17827–17835. [PubMed] [Google Scholar]
- Lahm A., Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991 Dec 5;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- Lavigne M., Kolb A., Buc H. Transcription activation by cAMP receptor protein (CRP) at the Escherichia coli gal P1 promoter. Crucial role for the spacing between the CRP binding site and the -10 region. Biochemistry. 1992 Oct 13;31(40):9647–9656. doi: 10.1021/bi00155a018. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Matzura H. Binding of RNA polymerase and the catabolite gene activator protein within the cat promoter in Escherichia coli. J Mol Biol. 1981 Aug 5;150(2):185–196. doi: 10.1016/0022-2836(81)90448-4. [DOI] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisser S., Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993 Apr 11;21(7):1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister C. F., Achberger E. C. Effect of polyadenine-containing curved DNA on promoter utilization in Bacillus subtilis. J Biol Chem. 1988 Aug 25;263(24):11743–11749. [PubMed] [Google Scholar]
- Mecsas J., Cowing D. W., Gross C. A. Development of RNA polymerase-promoter contacts during open complex formation. J Mol Biol. 1991 Aug 5;220(3):585–597. doi: 10.1016/0022-2836(91)90102-c. [DOI] [PubMed] [Google Scholar]
- Melançon P., Burgess R. R., Record M. T., Jr Nitrocellulose filter binding studies of the interactions of Escherichia coli RNA polymerase holoenzyme with deoxyribonucleic acid restriction fragments: evidence for multiple classes of nonpromoter interactions, some of which display promoter-like properties. Biochemistry. 1982 Aug 31;21(18):4318–4331. doi: 10.1021/bi00261a022. [DOI] [PubMed] [Google Scholar]
- Newlands J. T., Ross W., Gosink K. K., Gourse R. L. Factor-independent activation of Escherichia coli rRNA transcription. II. characterization of complexes of rrnB P1 promoters containing or lacking the upstream activator region with Escherichia coli RNA polymerase. J Mol Biol. 1991 Aug 5;220(3):569–583. doi: 10.1016/0022-2836(91)90101-b. [DOI] [PubMed] [Google Scholar]
- O'Halloran T. V., Frantz B., Shin M. K., Ralston D. M., Wright J. G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989 Jan 13;56(1):119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- O'Neill M. C., Chiafari F. Escherichia coli promoters. II. A spacing class-dependent promoter search protocol. J Biol Chem. 1989 Apr 5;264(10):5531–5534. [PubMed] [Google Scholar]
- O'Neill M. C. Escherichia coli promoters. I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J Biol Chem. 1989 Apr 5;264(10):5522–5530. [PubMed] [Google Scholar]
- Ozoline O. N., Uteshev T. A., Masulis I. S., Kamzolova S. G. Interaction of bacterial RNA-polymerase with two different promoters of phage T7 DNA. Conformational analysis. Biochim Biophys Acta. 1993 Mar 20;1172(3):251–261. doi: 10.1016/0167-4781(93)90211-u. [DOI] [PubMed] [Google Scholar]
- Peterson M. L., Reznikoff W. S. Lactose promoter mutation Pr115 activates an overlapping promoter within the lactose control region. J Mol Biol. 1985 Oct 5;185(3):525–533. doi: 10.1016/0022-2836(85)90069-5. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S., Spassky A., Busby S. Studies with the Escherichia coli galactose operon regulatory region carrying a point mutation that simultaneously inactivates the two overlapping promoters. Interactions with RNA polymerase and the cyclic AMP receptor protein. FEBS Lett. 1987 Jul 13;219(1):189–196. doi: 10.1016/0014-5793(87)81214-0. [DOI] [PubMed] [Google Scholar]
- Ren Y. L., Garges S., Adhya S., Krakow J. S. Cooperative DNA binding of heterologous proteins: evidence for contact between the cyclic AMP receptor protein and RNA polymerase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4138–4142. doi: 10.1073/pnas.85.12.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Russell D. R., Bennett G. N. Characterization of the beta-lactamase promoter of pBR322. Nucleic Acids Res. 1981 Jun 11;9(11):2517–2533. doi: 10.1093/nar/9.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele D. A., Hu J. C., Walter W. A., Gross C. A. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J Mol Biol. 1989 Apr 20;206(4):591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- Spassky A., Kirkegaard K., Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985 May 21;24(11):2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987 Jan 20;193(2):267–278. doi: 10.1016/0022-2836(87)90218-x. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Straney S. B., Crothers D. M. Synergy between Escherichia coli CAP protein and RNA polymerase in the lac promoter open complex. J Mol Biol. 1989 Mar 5;206(1):41–57. doi: 10.1016/0022-2836(89)90522-6. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Suck D., Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986 Jun 5;321(6070):620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- Szoke P. A., Allen T. L., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase: effects of base substitutions in the -10 and -35 regions. Biochemistry. 1987 Sep 22;26(19):6188–6194. doi: 10.1021/bi00393a035. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., de Crombrugghe B. Interactions of RNA polymerase and the cyclic AMP receptor protein on DNA of the E. coli galactose operon. Nucleic Acids Res. 1983 Aug 11;11(15):5165–5180. doi: 10.1093/nar/11.15.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Fujita N., Ishihama A. Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol Microbiol. 1993 Mar;7(6):859–864. doi: 10.1111/j.1365-2958.1993.tb01176.x. [DOI] [PubMed] [Google Scholar]
- Travers A. A., Lamond A. I., Mace H. A., Berman M. L. RNA polymerase interactions with the upstream region of the E. coli tyrT promoter. Cell. 1983 Nov;35(1):265–273. doi: 10.1016/0092-8674(83)90229-5. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Wishart W. L., Machida C., Ohtsubo H., Ohtsubo E. Escherichia coli RNA polymerase binding sites and transcription initiation sites in the transposon Tn3. Gene. 1983 Sep;24(1):99–113. doi: 10.1016/0378-1119(83)90135-x. [DOI] [PubMed] [Google Scholar]
- Woody S. T., Fong R. S., Gussin G. N. Effects of a single base-pair deletion in the bacteriophage lambda PRM promoter. Repression of PRM by repressor bound at OR2 and by RNA polymerase bound at PR. J Mol Biol. 1993 Jan 5;229(1):37–51. doi: 10.1006/jmbi.1993.1006. [DOI] [PubMed] [Google Scholar]


