Abstract
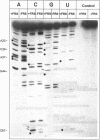
The phosphates of the tRNA(Phe) transcript from Thermus thermophilus interacting with the cognate synthetase were determined by footprinting. Backbone bond protection against cleavage by iodine of the phosphorothioate-containing transcripts was found in the anticodon stem-loop, the D stem-loop and the acceptor stem and weak protection was also seen in the variable loop. Most of the protected phosphates correspond to regions around known identity elements of tRNA(Phe). Enhancement of cleavage at certain positions indicates bending of tRNAPhe upon binding to the enzyme. When applied to the three-dimensional model of tRNA(Phe) from yeast the majority of the protections occur on the D loop side of the molecule, revealing that phenylalanyl-tRNA synthetase has a rather complex and novel pattern of interaction with tRNAPhe, differing from that of other known class II aminoacyl-tRNA synthetases.
Full text
PDF
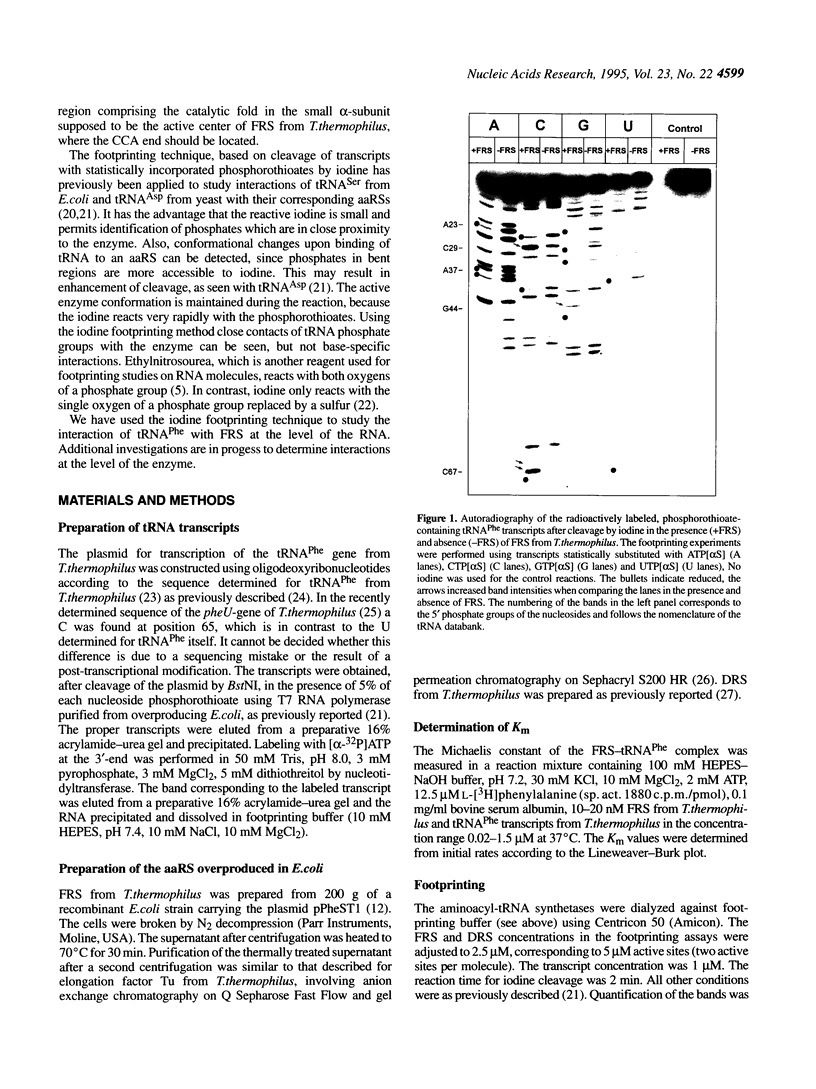
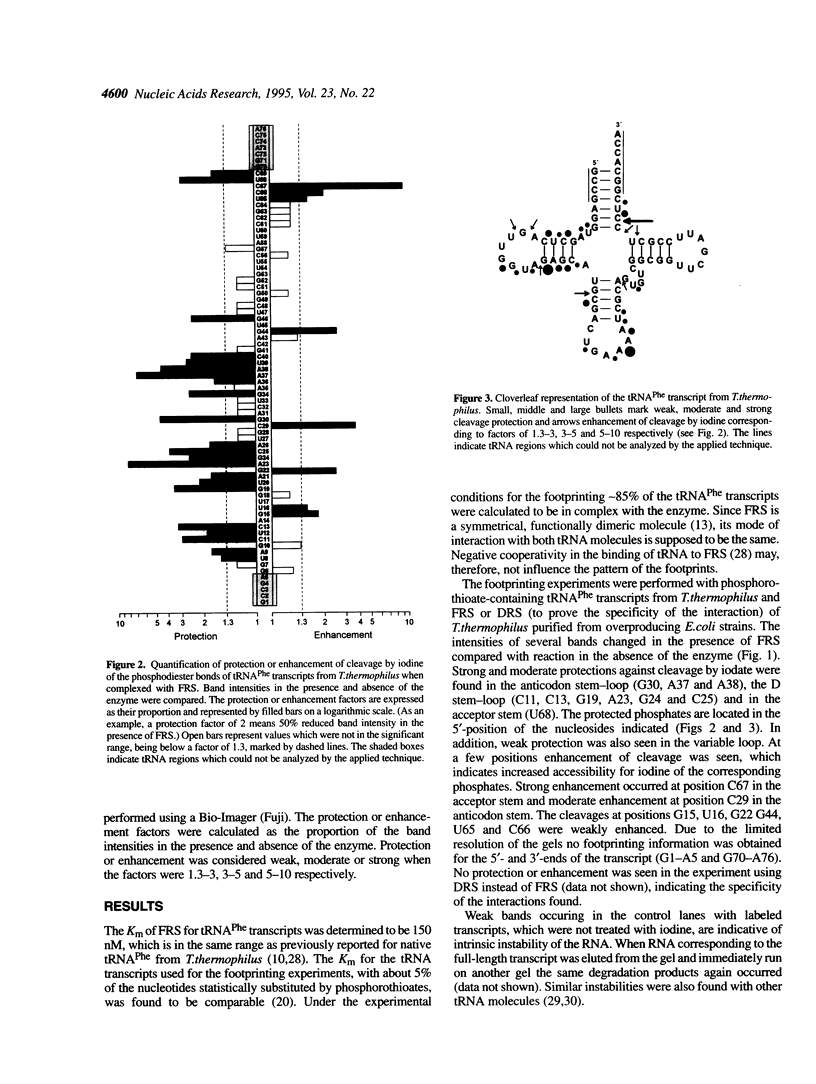
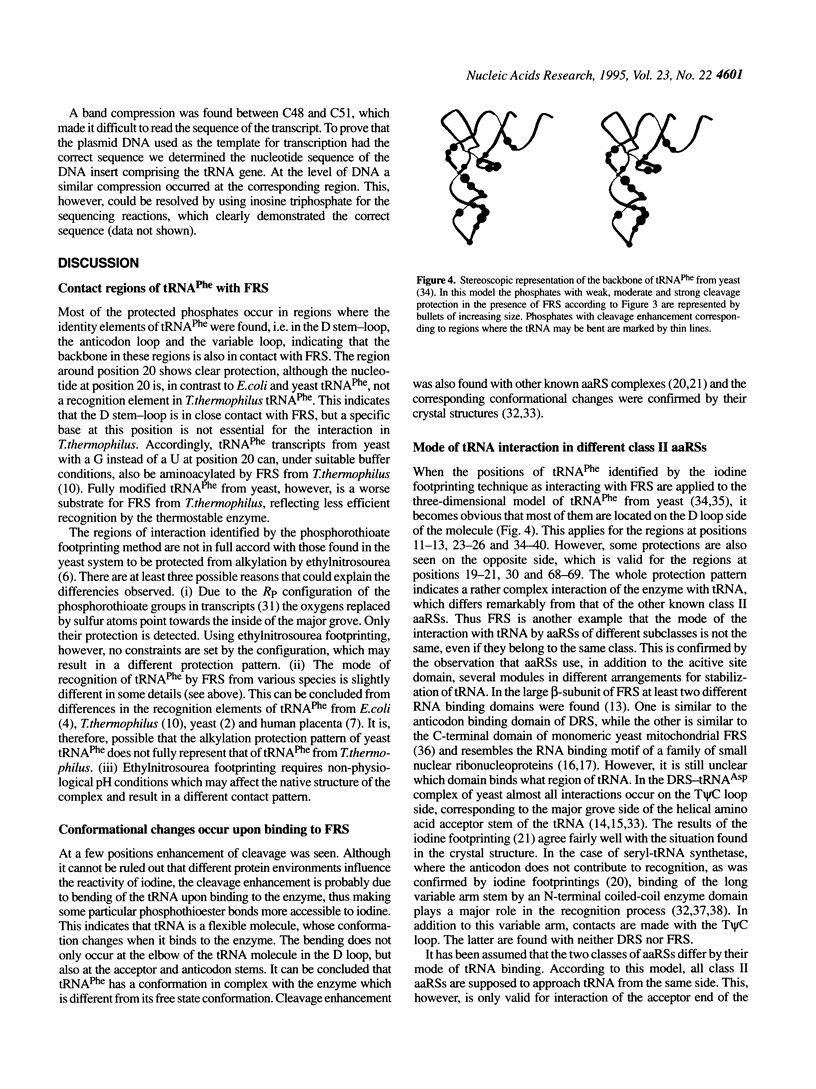

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankilova V. N., Vlassov V. V., Knorre D. G., Melamed N. V., Nuzdihna N. A. Involvement of the D-stem of tRNAPhe (E. coli) in interaction with phenylalanyl-tRNA synthetase as shown by chemical modification. FEBS Lett. 1975 Dec 1;60(1):168–171. doi: 10.1016/0014-5793(75)80444-3. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994 Mar 11;263(5152):1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Blank J., Grillenbeck N. W., Kreutzer R., Sprinzl M. Overexpression and purification of Thermus thermophilus elongation factors G, Tu, and Ts from Escherichia coli. Protein Expr Purif. 1995 Oct;6(5):637–645. doi: 10.1006/prep.1995.1084. [DOI] [PubMed] [Google Scholar]
- Borel F., Vincent C., Leberman R., Härtlein M. Seryl-tRNA synthetase from Escherichia coli: implication of its N-terminal domain in aminoacylation activity and specificity. Nucleic Acids Res. 1994 Aug 11;22(15):2963–2969. doi: 10.1093/nar/22.15.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Eriani G., Rees B., Ruff M., Boeglin M., Mitschler A., Martin F., Gangloff J., Thierry J. C., Moras D. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994 Jan 15;13(2):327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Thierry J. C., Moras D. Yeast aspartyl-tRNA synthetase: a structural view of the aminoacylation reaction. Biochimie. 1993;75(12):1117–1123. doi: 10.1016/0300-9084(93)90011-g. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Moras D. Conformational changes and dynamics of tRNAs: evidence from hydrolysis patterns. Cold Spring Harb Symp Quant Biol. 1987;52:113–121. doi: 10.1101/sqb.1987.052.01.016. [DOI] [PubMed] [Google Scholar]
- Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Frugier M., Florentz C., Giegé R. Efficient aminoacylation of resected RNA helices by class II aspartyl-tRNA synthetase dependent on a single nucleotide. EMBO J. 1994 May 1;13(9):2218–2226. doi: 10.1002/j.1460-2075.1994.tb06499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Puglisi J. D., Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Grawunder U., Schön A., Sprinzl M. Sequence and base modifications of two phenylalanine-tRNAs from Thermus thermophilus HB8. Nucleic Acids Res. 1992 Jan 11;20(1):137–137. doi: 10.1093/nar/20.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A. D., Potter B. V., Eperon I. C. Stereospecificity of nucleases towards phosphorothioate-substituted RNA: stereochemistry of transcription by T7 RNA polymerase. Nucleic Acids Res. 1987 May 26;15(10):4145–4162. doi: 10.1093/nar/15.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hountondji C., Schmitter J. M., Beauvallet C., Blanquet S. Affinity labeling of Escherichia coli phenylalanyl-tRNA synthetase at the binding site for tRNAPhe. Biochemistry. 1987 Aug 25;26(17):5433–5439. doi: 10.1021/bi00391a033. [DOI] [PubMed] [Google Scholar]
- Keller B., Hennecke H. Cloning and sequencing of the pheU gene for tRNA(Phe) of Thermus thermophilus HB8, and genomic mapping of the pheU and pheST genes. FEMS Microbiol Lett. 1994 Nov 1;123(3):275–279. doi: 10.1111/j.1574-6968.1994.tb07236.x. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Kreutzer R., Kruft V., Bobkova E. V., Lavrik O. I., Sprinzl M. Structure of the phenylalanyl-tRNA synthetase genes from Thermus thermophilus HB8 and their expression in Escherichia coli. Nucleic Acids Res. 1992 Aug 25;20(16):4173–4178. doi: 10.1093/nar/20.16.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Nucleotides that contribute to the identity of Escherichia coli tRNA(Phe). J Mol Biol. 1988 Aug 20;202(4):697–709. doi: 10.1016/0022-2836(88)90551-7. [DOI] [PubMed] [Google Scholar]
- Moor N., Nazarenko I., Ankilova V., Khodyreva S., Lavrik O. Determination of tRNA(Phe) recognition nucleotides for phenylalanyl-tRNA synthetase from Thermus thermophilus. Biochimie. 1992 Apr;74(4):353–356. doi: 10.1016/0300-9084(92)90112-r. [DOI] [PubMed] [Google Scholar]
- Mosyak L., Reshetnikova L., Goldgur Y., Delarue M., Safro M. G. Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus. Nat Struct Biol. 1995 Jul;2(7):537–547. doi: 10.1038/nsb0795-537. [DOI] [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990 Dec 6;348(6301):515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Nazarenko I. A., Peterson E. T., Zakharova O. D., Lavrik O. I., Uhlenbeck O. C. Recognition nucleotides for human phenylalanyl-tRNA synthetase. Nucleic Acids Res. 1992 Feb 11;20(3):475–478. doi: 10.1093/nar/20.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E. T., Uhlenbeck O. C. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1992 Oct 27;31(42):10380–10389. doi: 10.1021/bi00157a028. [DOI] [PubMed] [Google Scholar]
- Poterszman A., Plateau P., Moras D., Blanquet S., Mazauric M. H., Kreutzer R., Kern D. Sequence, overproduction and crystallization of aspartyl-tRNA synthetase from Thermus thermophilus. Implications for the structure of prokaryotic aspartyl-tRNA synthetases. FEBS Lett. 1993 Jul 5;325(3):183–186. doi: 10.1016/0014-5793(93)81069-c. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Reshetnikova L., Khodyreva S., Lavrik O., Ankilova V., Frolow F., Safro M. Crystals of the phenylalanyl-tRNA synthetase from Thermus thermophilus HB8 complexed with tRNA(Phe). J Mol Biol. 1993 Jun 5;231(3):927–929. doi: 10.1006/jmbi.1993.1338. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Rudinger J., Puglisi J. D., Pütz J., Schatz D., Eckstein F., Florentz C., Giegé R. Determinant nucleotides of yeast tRNA(Asp) interact directly with aspartyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5882–5886. doi: 10.1073/pnas.89.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry. 1990 Mar 13;29(10):2523–2532. doi: 10.1021/bi00462a014. [DOI] [PubMed] [Google Scholar]
- Sanni A., Hountondji C., Blanquet S., Ebel J. P., Boulanger Y., Fasiolo F. Interaction of the tRNA(Phe) acceptor end with the synthetase involves a sequence common to yeast and Escherichia coli phenylalanyl-tRNA synthetases. Biochemistry. 1991 Mar 5;30(9):2448–2453. doi: 10.1021/bi00223a022. [DOI] [PubMed] [Google Scholar]
- Sanni A., Walter P., Boulanger Y., Ebel J. P., Fasiolo F. Evolution of aminoacyl-tRNA synthetase quaternary structure and activity: Saccharomyces cerevisiae mitochondrial phenylalanyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8387–8391. doi: 10.1073/pnas.88.19.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P., Giegé R., Moras D., Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov V. V., Kern D., Romby P., Giegé R., Ebel J. P. Interaction of tRNAPhe and tRNAVal with aminoacyl-tRNA synthetases. A chemical modification study. Eur J Biochem. 1983 May 16;132(3):537–544. doi: 10.1111/j.1432-1033.1983.tb07395.x. [DOI] [PubMed] [Google Scholar]