Abstract
The nuage is a germline-specific perinuclear structure that remains functionally elusive. Recently, the nuage in Drosophila was shown to contain two of the three PIWI proteins – Aubergine and Argonaute 3 (AGO3) – that are essential for germline development. The PIWI proteins bind to PIWI-interacting RNAs (piRNAs) and function in epigenetic regulation and transposon control. Here, we report a novel nuage component, PAPI (Partner of PIWIs), that contains a TUDOR domain and interacts with all three PIWI proteins via symmetrically dimethylated arginine residues in their N-terminal domain. In adult ovaries, PAPI is mainly cytoplasmic and enriched in the nuage, where it partially colocalizes with AGO3. The localization of PAPI to the nuage does not require the arginine methyltransferase dPRMT5 or AGO3. However, AGO3 is largely delocalized from the nuage and becomes destabilized in the absence of PAPI or dPRMT5, indicating that PAPI recruits PIWI proteins to the nuage to assemble piRNA pathway components. As expected, papi deficiency leads to transposon activation, phenocopying piRNA mutants. This further suggests that PAPI is involved in the piRNA pathway for transposon silencing. Moreover, AGO3 and PAPI associate with the P body component TRAL/ME31B complex in the nuage and transposon activation is observed in tral mutant ovaries. This suggests a physical and functional interaction in the nuage between the piRNA pathway components and the mRNA-degrading P-body components in transposon silencing. Overall, our study reveals a function of the nuage in safeguarding the germline genome against deleterious retrotransposition via the piRNA pathway.
Keywords: AGO3, TDRD, Partner of PIWI, Nuage, Transposon silencing, Drosophila
INTRODUCTION
The nuage has long been recognized as a perinuclear structure that is present in germline cells in diverse organisms; yet its function remains elusive. Recently, the nuage has been shown to contain two of the three Drosophila PIWI proteins, Aubergine (AUB) and Argonaute 3 (AGO3) (Brennecke et al., 2007; Harris and Macdonald, 2001; Li et al., 2009). Argonaute (AGO)/PIWI family proteins are essential for germline development, stem cell self-renewal, epigenetic regulation and transposon silencing (Malone and Hannon, 2009; Thomson and Lin, 2009). These proteins contain PAZ and PIWI domains, and are divided into AGO and PIWI subfamilies (Jinek and Doudna, 2009). AGO subfamily proteins bind microRNAs (miRNAs) or small interfering RNAs (siRNAs) that are ~21 nulceotides and form the core of RNA-induced silencing complex (RISC) in regulating the translation and degradation of mRNAs, respectively (Ghildiyal et al., 2010; Siomi and Siomi, 2009); however, PIWI subfamily proteins bind to PIWI-interacting RNAs (piRNAs) that are usually 24-33 nucleotides (Aravin et al., 2006; Aravin et al., 2001; Brennecke et al., 2007; Girard et al., 2006; Grivna et al., 2006; Houwing et al., 2007; Ruby et al., 2006; Saito et al., 2006; Vagin et al., 2006; Watanabe et al., 2006; Yin and Lin, 2007) and are apparently produced by a Dicer-independent mechanism (Vagin et al., 2006). AGO proteins are ubiquitously expressed in somatic and germline cells, whereas PIWI proteins are mostly restricted to the germline and are essential for germline development (Brennecke et al., 2007; Cox et al., 1998; Cox et al., 2000; Gunawardane et al., 2007; Harris and Macdonald, 2001; Megosh et al., 2006; Saito et al., 2006; Wiederhecker et al., 2009). In addition, AGO proteins accumulate in P-bodies that are presumed to be the sites for mRNA storage and degradation (Jagannath and Wood, 2009; Liu et al., 2005); PIWI proteins, however, when in the cytoplasm, are enriched in the germline-specific organelles such as polar granules in early embryos or the nuage in the adult germline, both of which are essential for germline development (Brennecke et al., 2007; Chen et al., 2009; Harris and Macdonald, 2001; Megosh et al., 2006).
Polar granules and the nuage are both electron-dense structures that are rich in protein and RNA (Saffman and Lasko, 1999). Many of the piRNA pathway components localize to the nuage, suggesting that the nuage may function as a cytoplasmic site where post-transcriptional transposon silencing occurs (Brennecke et al., 2007; Chen et al., 2009; Cook et al., 2004; Gunawardane et al., 2007; Harris and Macdonald, 2001; Lim and Kai, 2007; Malone et al., 2009; Pane et al., 2007; Patil and Kai, 2010; Vagin et al., 2006). Mutants of the nuage components, such as vasa, maelstrom, armitage, zucchini, squash, krimper and tejas, exhibit defects in piRNA production and de-repression of transposons (Cook et al., 2004; Lim and Kai, 2007; Pane et al., 2007; Patil and Kai, 2010), linking the nuage to the piRNA pathway. Despite this progress, the exact molecular function of the nuage remains elusive.
Recent studies have shown that PIWI, but not AGO, subfamily proteins contain clustered symmetrically dimethylated arginine residues (sDMAs) in their N termini (Kirino et al., 2009; Nishida et al., 2009; Vagin et al., 2009). In Drosophila, the sDMAs of three PIWI proteins (PIWI, AUB and AGO3) are specifically catalyzed by a protein methyltransferase (PRMT) encoded by the dPRMT5 (csul/dart5 – FlyBase) gene (Kirino et al., 2009). This modification is crucial for their interaction with TUDOR-domain-containing proteins (Chen et al., 2009; Kirino et al., 2009; Kirino et al., 2010; Nishida et al., 2009; Reuter et al., 2009; Shoji et al., 2009; Vagin et al., 2009; Vasileva et al., 2009; Wang et al., 2009). Thus, sDMAs may reflect a molecular motif that is specific to PIWI subfamily proteins and key to understanding the action mechanism of PIWI proteins.
Here, we report the discovery of a novel nuage component, herein named Partner of PIWIs (PAPI). PAPI is a novel TUDOR-domain-containing protein that interacts specifically with PIWI subfamily proteins, especially AGO3. Moreover, we show that PAPI interacts with AGO3 via sDMAs in its N-terminal domain. This interaction is essential for the recruitment of AGO3 to the nuage and for transposon silencing, thus revealing symmetric dimethylation as a mechanism that recruits PIWI proteins and their mediated transposon-silencing mechanism to the nuage. Furthermore, we describe the physical association between the AGO3-PAPI complex and P-body proteins in the nuage, which further link these two pathways in the post-transcriptional regulation of transposon silencing in the nuage.
MATERIALS AND METHODS
Drosophila strains
The w1118 strain was used as a wild-type strain. The mutant alleles used in this study were: dPRMT5 (dart5 or csule00797) (Gonsalvez et al., 2006), ago3t2/t2 (Thomson and Lin, 2009), tral1/2 (Wilhelm et al., 2005) and piwi2 (Cox et al., 1998). Full-length papi-coding sequence was amplified using papi-FL-F (5′-CACCATGTTGCGCAACACGCCTTTCGGTG-3′) and papi-FL-R (5′-CTAATGCGCGCTAGCACCATTTGTGGT-3′) with cDNA generated from ovarian RNA as template, cloned into pENTR/D-TOPO, and combined into pPFMW (The Drosophila Gateway Vector Collection) according to the manufacturer's protocol. The resulted plasmid was injected into w1118 embryos to generate transgenic flies. The expression of transgene was driven by Act5C-Gal4 driver. papi RNAi strain was obtained from VDRC Stock Center (transformant ID 2553). The expression of papi hairpin-RNA was induced by Act5C-Gal4 and nosVP16-Gal4 drivers.
Yeast two-hybrid screens and assays
PIWI yeast two-hybrid (H2Y) screens and specificity tests were performed as previously described (Brower-Toland et al., 2007). All bait and prey constructs for Y2H interaction analyses were generated by cloning PCR-amplified fragments into pEG202 (bait vector) or pJG4-5 (prey vector) using previously cloned cDNAs as template.
To construct PIWI deletion series for Y2H, deletion series fragments were amplified from PIWI cDNA and cloned into the BamHI and NotI sites of the pEG202 bait vector by introducing a BglII site into the 5′ PCR primer and a NotI site into the 3′ PCR primer. The following primers were used: 5′ PCR Primer PIWI-1, AAA-AAG-ATC-TTA-ATG-GCT-GAT-GAT-CAG-GGA-CGT; 5′ PCR Primer PIWI-2, AAA-AAG-ATC-TTA-AAA-GTT-ATG-CGC-ACC-GAG-ACG; 5′ PCR Primer PIWI-3, AAA-AAG-ATC-TTA-GAG-ATG-CGC-TCA-AAC-TTT-CAG-C; 3′ PCR Primer PIWI-4, AAA-AGC-GGC-CGC-TTA-TTT-GGG-ATC-TGA-GCG-CAC-ACA; 3′ PCR Primer PIWI-5, AAA-AGC-GGC-CGC-TTA-GGC-ATT-GAG-CCC-AGT-CAC-TCG; 3′ PCR Primer PIWI-6, AAA-AGC-GGC-CGC-TTA-GTG-AGT-TAT-TTC-GGT-GCC-CA. The bold nucleotides are translation start or stop codons; the underlined nucleotides are restriction sites.
The deletion clones used the following combinations of these primers: for PIWI residues 1-843, primers PIWI-1/PIWI-4; for PIWI residues 1-491, primers PIWI-1/PIWI-5; for PIWI residues 492-843, primers PIWI-1/PIWI-6; for PIWI residues 1-257, primers PIWI-2/ PIWI-4; for PIWI residues 258-491, primers PIWI-2/ PIWI-5.
To construct AGO3 deletion series, deletion series fragments were amplified from AGO3 cDNA generated from ovarian RNA and cloned into the BamHI and XhoI sites of the pEG202 bait vector by introducing a BglII site into the 5′ PCR primer and a XhoI site into the 3′ PCR primer. The following primers were used: forward primer AGO3-1, 5′-CCCCCCAGATCTCCATGTCTGGAAGAGGAAATTTG-3′; forward primer AGO3-2, AAA-AAG-ATC-TTA-AAA-GTT-ATG-CGC-ACC-GAG-ACG; forward primer AGO3-3, 5′-CCCCCCAGATCTCCCAAAAAACT-GTTCTAGAAATGC-3′; reverse primer AGO3-4, 5′-CCCCCCTCGAGTTAAAGATAAAATAGTTTTTCAGAAAGTG-3′; 3′ PCR Primer AGO3-5, AAA-AGC-GGC-CGC-TTA-GGC-ATT-GAG-CCC-AGT-CAC-TCG; 3′ PCR Primer AGO3-6, 5′-CCCCCTCGAGTTACAAAGTATACGATGCGAAACGTC-3′. The deletion clones used the following combinations of these primers: for AGO3 residues 1-867, primers AGO3-1/AGO3-4; for AGO3 residues 1-403, primers AGO3-1/AGO3-5; for AGO3 residues 404-867, primers AGO3-2/AGO3-4; for AGO3 residues 1-289, primers AGO3-1/AGO3-6; for AGO3 residues 290-403, primers AGO3-3/AGO3-5.
To construct PAPI deletion series, deletion series fragments were amplified from PAPI cDNA generated from ovarian RNA and cloned into the BamHI and NotI sites of the pJG4-5 prey vector by introducing a BglII site into the 5′ PCR primer and a NotI site into the 3′ PCR primer. The following primers were used: 5′ PCR Primer PAPI-1, AAA-AAG-ATC-TTA-ATG-GCT-GAT-GAT-CAG-GGA-CGT; 5′ PCR Primer PAPI-2, AAA-AAG-ATC-TTA-AAA-GTT-ATG-CGC-ACC-GAG-ACG; 5′ PCR Primer PAPI-3, AAA-AAG-ATC-TTA-GAG-ATG-CGC-TCA-AAC-TTT-CAG-C; 3′ PCR Primer PAPI-4, AAA-AGC-GGC-CGC-TTA-TTT-GGG-ATC-TGA-GCG-CAC-ACA; 3′ PCR Primer PAPI-5, AAA-AGC-GGC-CGC-TTA-GGC-ATT-GAG-CCC-AGT-CAC-TCG; 3′ PCR Primer PAPI-6, AAA-AGC-GGC-CGC-TTA-GTG-AGT-TAT-TTC-GGT-GCC-CA. The deletion clones used the following combinations of these primers: for PAPI residues 1-576, primers PAPI-1/PAPI-4; for PAPI residues 1-257, primers PAPI-1/PAPI-5; for PAPI residues 258-576, primers PAPI-1/PAPI-6; for PAPI residues 258-385, primers PAPI-2/PAPI-4; for PAPI residues 386-576, primers PAPI-2/ PAPI-5.
To generate mutant PIWI Y2H bait, arginine residues were changed to lysines (wild-type 6-GRGR was changed to mutant 6-GKGK) by site-directed mutagenesis (QuickChange XL, Stratagene) using the following primers: forward PCR primer, 5′-ATGGCTGATGATCAGGGAAAAGGAAAAAGGCGTCCACTTAACGAAGATGAT-3′; and reverse PCR primer 5′-GGAATCATCTTCGTTAAGTGGACGCCTTTTTCCTTTTCCCTGATCATCAGCCAT-3′. To generate mutant AGO3 Y2H bait, arginine residues were changed to lysines (wild-type 67-GRGRAR was changed to mutant 67-GKGKAK) by site-directed mutagenesis (QuickChange XL, Stratagene) using the following primers: forward PCR primer 5′-GTAAACATCTCGGTCGGCAAAGGAAAAGCTAAGCTTATAGACACATT-3′ and reverse PCR primer 5′-AATGTGTCTATAAGCTTAGCTTTTCCTTTGCCGACCGAGATGTTTAC-3′.
Antibody generation
Guinea pig anti-PAPI polyclonal antibody was generated against His-tagged full-length PAPI. Full-length PAPI coding sequence was amplified using papi-FL-F and papi-FL-R, cloned into pENTR/D-TOPO, and combined into pDEST17 (Invitrogen). His-tagged full-length PAPI protein was expressed and purified according to the manufacturer's protocol and injected into animals.
Immunostaining
Drosophila ovaries were dissected from adult flies and stained as described by Lin et al. (Lin et al., 1994). Embryos were collected, fixed and stained according to Patel et al. (Patel et al., 1989). For immunofluorescence staining the following antibodies were used: guinea pig anti-PAPI polyclonal antibody (1:500), mouse anti-AGO3 monoclonal antibody (1:500) (Gunawardane et al., 2007), mouse anti-PIWI monoclonal antibody (1:200) (Gunawardane et al., 2007), rabbit anti-Aub polyclonal antibody (1:1000) (Harris and Macdonald, 2001) and rabbit anti-Tral antibody (1:400) (Wilhelm et al., 2005). Alexa Fluor-488 or Alexa Fluor-568-conjugated goat anti-rabbit, anti-mouse or anti-guinea pig IgG secondary antibodies were purchased from Jackson ImmunoResearch Laboratory and were used at 1:500 dilution. Immunofluorescently labeled samples were also counterstained with DAPI as described previously (Lin et al., 1994). Images were taken using Leica TCS SP5 Spectral Confocal Microscope in the sequential scanning mode.
Immunoprecipitation
Drosophila ovaries or 0- to 12-hour embryos were homogenized in ice-cold IP buffer [100 mM potassium acetate, 0.1% Triton, 50 mM HEPES (pH 7.4), 2 mM magnesium acetate, 10% glycerol, 1 mM DTT, 20 U/ml RNase out (Invitrogen), 1× complete mini EDTA-free protease inhibitor cocktail (Roche)]. The immunoprecipitation was carried out with two negative controls: lysate plus beads and antibody plus beads. The extract was first precleared with equilibrated Protein A agarose sepharose (GE Healthcare) for 2 hours at 4°C. The equilibrated beads were bound to antibody for 2 hours at 4°C, washed with IP buffer and split into two fractions. One was saved as an antibody control and the other fraction was incubated with pre-cleared extract for 2 hours at 4°C. Meanwhile, the same amount of pre-cleared extract was added to bare beads as an extract plus beads control. The beads were washed with IP buffer five times, and proteins were eluted from the beads at 95°C in SDS sample buffer and analyzed by western blots. For RNase A treatment, RNase A (Sigma) was added to ovary extract to a final concentration of 20 U/ml. The extract was incubated for 2 hours at 4°C and 30 minutes at room temperature before being used for immunoprecipitation.
Western blot and quantification
Western blotting was performed with standard protocols with the following antibodies: mouse anti-HA monoclonal antibody (1:1000) (Santa Cruz Biotechnology), mouse anti-Myc 9B11 monoclonal antibody (Cell Signaling), rabbit anti-GAPDH polyclonal antibody (Sigma), rabbit anti-RNA polymerase II polyclonal antibody (Santa Cruz Biotechnology), guinea pig anti-PAPI polyclonal antibody (1:2000), mouse anti-AGO3 monoclonal antibody (1:2000) (Gunawardane et al., 2007), mouse anti-PIWI monoclonal antibody (1:1000) (Gunawardane et al., 2007), rabbit anti-Aub polyclonal antibody (1:5000) (Harris and Macdonald, 2001), rabbit anti-Tral antibody (1:5000) (Wilhelm et al., 2005), mouse anti-Me31b monoclonal antibody (1:5000) (Nakamura et al., 2001) and rabbit anti-TER94 polyclonal antibody (1:2000) (Nicchitta et al., 1991). HRP-conjugated goat anti-guinea pig, anti-rabbit or anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch Laboratory and were used at 1:10,000 dilution. All bands shown in quantification were exposed within the linear range. The band density was quantified with Kodak Molecular Imaging (MI) Software.
Nuclear/cytoplasmic fractionation
Drosophila ovaries were homogenized in cold IP buffer [100 mM potassium acetate, 0.1% Triton, 50 mM HEPES (pH 7.4), 2 mM magnesium acetate, 10% glycerol, 1 mM DTT, 20 U/ml RNase out (Invitrogen), 1× complete mini EDTA-free protease inhibitor cocktail (Roche)]. The crude nuclear pellet was collected at the speed of 1300 g for 10 minutes at 4°C. Supernatant was collected as cytoplasmic fraction. The above crude pellet was homogenized again, centrifuged at 1300 g for 10 minutes to collect the pellet as the nuclear pellet. Nuclear pellet was resuspended in the same volume of buffer used for the cytoplasmic fraction. Nuclear membranes were broken using sonication. The unbroken nuclei were pelleted at 1300 g for 10 minutes. The resulting supernatant is the nuclear fraction.
Sucrose gradient polysome fractionation
Drosophila ovaries or 0-12 hour embryos were homogenized in ice-cold IP buffer, and 0.5 ml of the extract was loaded onto continuous 15%-55% (w/w) linear sucrose gradient made by Density Gradient Fractionation System (Teledyne ISCO). The gradient was centrifuged at 150,000 g for 3 hours (Beckman, CA, USA). Fractions were collected and assayed by western blotting. For the EDTA treatment, the extract was treated with 20 mM EDTA before applying to the sucrose gradient (supplemented with 20 mM EDTA instead of magnesium acetate in all buffers).
Quantitative RT-PCR
RNA was isolated from Drosophila ovaries and quantification of the transposon transcripts by quantitative RT-PCR was performed as previously described (Li et al., 2009). The following primer pairs were used for quantitative RT-PCR: 412 forward (5′-CACCGGTTTGGTCGAAAG-3′) and reverse (5′-GGACATGCCTGGTATTTTGG-3′); Accord forward (5′-ACAATCCACCAACAGCAACA-3′) and reverse (5′-AAAAGCCAAAATGTCGGTTG-3′); Accord2 forward (5′-TTGCTTTCGGACTTCGTCTT-3′) and reverse (5′-TTCCACAACGAAAACAACCA-3′); Blood forward (5′-TGCCACAGTACCTGATTTCG-3′) and reverse (5′-GATTCGCCTTTTACGTTTGC-3′); Diver forward (5′-GGCACCACATAGACACATCG-3′) and reverse (5′-GTGGTTTGCATAGCCAGGAT-3′); Diver2 forward (5′-CTTCAGCCAGCAAGGAAAAC-3′) and reverse (5′-CTGGCAGTCGGGTGTAATTT-3′); gtwin forward (5′-TTCGCACAAGCGATGATAAG-3′) and reverse (5′-GATTGTTGTACGGCGACCTT-3′); gypsy forward (5′-GTTCATACCCTTGGTAGTAGC-3′) and reverse (5′-CAACTTACGCATATGTGAGT-3′); gypsy6 forward (5′-GACAAGGGCATAACCGATACTGTGGA-3′) and reverse (5′-AATGATTCTGTTCCGGACTTCCGTCT-3′); HeT-A forward (5′-CGCGCGGAACCCATCTTCAGA-3′) and reverse (5′-CGCCGCAGTCGTTTGGTGAGT-3′); Hopper forward (5′-GGCTGGCTTCAACAAAAGAA-3′) and reverse (5′-GGACTCCCGAAAACGTCATA-3′); I-element forward (5′-GACCAAATAAAAATAATACGACTTC-3′) and reverse (5′-AACTAATTGCTGGCTTGTTATG-3′); Invader1 forward (5′-GTACCGTTTTTGAGCCCGTA-3′) and reverse (5′-AACTACGTTGCCCATTCTGG-3′); Max forward (5′-TCTAGCCAGTCGAGGCGTAT-3′) and reverse (5′-TGGAAGAGTGTCGCTTTGTG-3′); mdg1 forward (5′-AACAGAAACGCCAGCAACAGC-3′) and reverse (5′-CGTTCCCATGTCCGTTGTGAT-3′); R1A1 forward (5′-AATTCCCGAGCTGTGCTAGA-3′) and reverse (5′-GTCTCAAGGCACCTTTCAGC-3′); rp49 forward (5′-CCGCTTCAAGGGACAGTATCTG-3′) and reverse (5′-ATCTCGCCGCAGTAAACGC-3′); Rt1a forward (5′-CCACACAGACTGAGGCAGAA-3′) and reverse (5′-ACGCATAACTTTCCGGTTTG-3′); ZAM forward (5′-ACTTGACCTGGATACACTCACAAC-3′) and reverse (5′-GAGTATTACGGCGACTAGGGATAC-3′); CG7082 (papi) forward (5′-TACAATCCAAAGGAGCAGG-3′) and reverse (5′-TGGCAGCAGCACTTACAG-3′); CG31935 forward (5′-AGTTCCCAGATCGAGAAGCTTC-3′) and reverse (5′-TCGTGCAACAGTTGTCATTGCG-3′); CG11160 forward (5′-GTAATGGTTTTGGCTCAGTCG-3′) and reverse (5′-CGTAGATCCTTCGGGTTCCA-3′).
RESULTS
PAPI Interacts with PIWI proteins
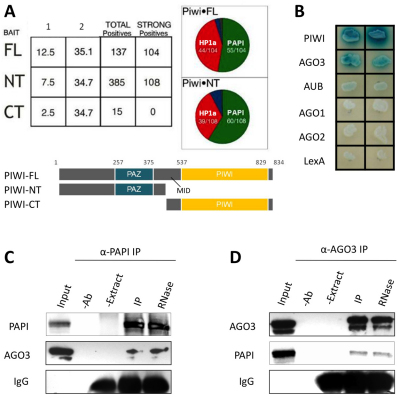
To discover new nuage components, we searched for proteins that interact with PIWI proteins in the nuage. To identify PIWI-interacting proteins, we performed yeast-two hybrid (Y2H) screens of a high-complexity ovarian cDNA library using three PIWI baits: full-length (PIWI-FL), N-terminal (PIWI-NT) and C-terminal (PIWI-CT) (Fig. 1A) (Brower-Toland et al., 2007). We screened 12.5, 7.5 and 2.5 million primary transformants with the three baits, and recovered 104, 108 and 0 strong positives, respectively. Fifty-five of 104 positives for PIWI-FL and 60 of 108 positives for PIWI-NT encoded cDNAs for the Drosophila CG7082 gene, herein named partner of piwis (papi; Fig. 1A). We tested the specificity of the PAPI interaction with proteins of the AGO/PIWI family by Y2H, and found that PAPI interacts with all three PIWI subfamily proteins: most strongly with PIWI, strongly with AGO3 and weakly with AUB, but fails to interact with AGO subfamily proteins AGO1 and AGO2 (Fig. 1B).
Fig. 1.
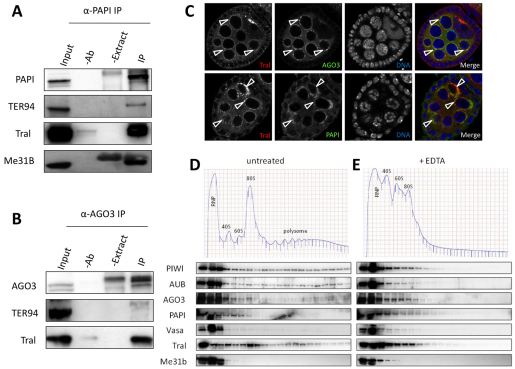
PAPI interacts with PIWI proteins, especially AGO3. (A) PAPI is a strong PIWI-interacting protein in a Y2H screen. Three baits were used in the Y2H screen: PIWI-FL, residues 1-843; PIWI-NT, residues 1-491; and PIWI-CT, residues 492-843. PAPI interacts strongly with the PIWI-FL and the PIWI-NT baits, but not with the PIWI-CT bait. (B) Y2H assay showing that PAPI interacts with PIWI proteins, especially AGO3, but not with AGO proteins or the LexA DNA-binding moiety of the bait plasmid pEG202. (C,D) AGO3 interacts with PAPI in vivo. Endogenous PAPI (C) and AGO3 (D) were immunoprecipitated from ovarian extract treated with or without RNase A using anti-PAPI (C) and anti-AGO3 (D) antibodies, respectively. Extract plus beads (−Ab) and antibody plus beads (−Extract) were used as negative controls for IP. Both immunoblots were probed for PAPI and AGO3. IgG bands were probed as a loading control. The interaction between PAPI and AGO3 is RNA independent.
To test whether these interactions occur in vivo, we immunoprecipitated endogenous PAPI from 0-12 hours embryonic extracts with a guinea pig polyclonal antibody that specifically recognizes PAPI (see Fig. S1A,B in the supplementary material). We probed a western blot of the PAPI co-immunoprecipitates with anti-PIWI, AUB and AGO3 antibodies, respectively. AGO3 associates with PAPI most strongly in vivo, and weaker association of PIWI and AUB with PAPI was also observed (see Fig. S2 in the supplementary material). The PAPI-AGO3 interaction was further confirmed by reciprocal co-immunoprecipitation, in which PAPI was present in AGO3 immunoprecipitates but not in negative controls (Fig. 1D). To examine whether the interaction between PAPI and AGO3 is RNA dependent, we treated the embryonic extract with RNase A prior to co-immunoprecipitation. RNase A treatment did not affect the association between PAPI and AGO3 (Fig. 1C,D), suggesting that PAPI and AGO3 form a complex in vivo in an RNA-independent manner.
PAPI binds to symmetrically dimethylated arginine residues in the N terminus of AGO3 via its TUDOR domain
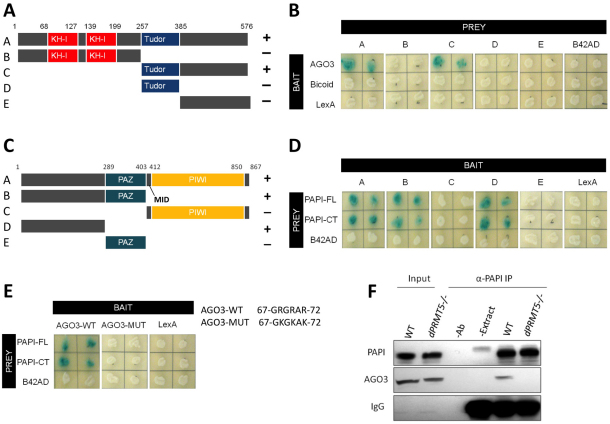
Having demonstrated an interaction between PAPI and PIWI proteins, we proceeded to map the domains of PAPI and AGO3 responsible for their interaction by Y2H. PIWI proteins contain an N-terminal domain, a MID domain and highly conserved PAZ and PIWI domains (Fig. 1A, Fig. 2C); however, PAPI contains two KH-I domains, a TUDOR domain and a C-terminal domain (Fig. 2A). We tested the interaction between a series of deletional variants of PAPI and AGO3, which revealed that the C-terminal half of PAPI that contains a TUDOR domain and the C-terminal domain was sufficient to interact with AGO3; however, the N-terminal half of PAPI that contains two KH-I domains did not contribute to the interaction (Fig. 2B). The TUDOR domain alone could not mediate the interaction, suggesting that the C-terminal domain is also necessary for binding to AGO3 (Fig. 2B). In the reciprocal experiment, we found that the N-terminal domain of AGO3 alone was sufficient to interact with both full-length PAPI (PAPI-FL) and the C-terminal half of PAPI (PAPI-CT). Neither the PAZ domain nor the PIWI domain of AGO3 can bind PAPI (Fig. 2D). Likewise, The PAPI-PIWI interaction was also mapped to the N-terminal domain of PIWI and C-terminal domain of PAPI (see Fig. S3B,D in the supplementary material). Collectively, these results suggest that the N-terminal domain of PIWI proteins interact with the C-terminal half of PAPI.
Fig. 2.
PAPI binds to the sDMAs in the N terminus of AGO3 via its TUDOR domain. (A) PAPI deletions used to map the AGO3-binding region. The positions of amino acid residues at various domain junctions are indicated. (B) Y2H assay indicating that the C-terminal domain of PAPI interacts with AGO3. Drosophila Bicoid protein and LexA were used as negative controls. (C) AGO3 deletions used to map the PAPI-binding region. The positions of amino acid residues at various domain junctions are indicated. (D) Y2H assay indicating that the N-terminal domain of AGO3 interacts with PAPI-FL and PAPI-CT. The B42AD moiety of the prey plasmid pJG4-5 was used as a negative control. (E) The sDMAs in the N terminus of AGO3 are required for the PAPI-AGO3 interaction. Triple sDMA mutant AGO3 (AGO3-MUT) does not interact with PAPI. Arginine to lysine mutations are indicated. (F) PAPI immunoprecipitates from wild-type (WT) and dPRMT5 mutant ovaries were subjected to western blotting analysis and probed with anti-PAPI and anti-AGO3 antibodies. AGO3 is detected only in the PAPI immunoprecipitates from wild-type ovaries. Extract plus beads (−Ab) and antibody plus beads (−Extract) were used as negative controls for IP. IgG was probed as a loading control.
TUDOR-domain-containing proteins are known to bind symmetrically dimethylated arginines (sDMAs) on their target proteins (Cote and Richard, 2005). It has been reported that the Drosophila dPRMT5 gene encodes a protein arginine methyltransferase (PRMT) that specifically catalyzes symmetric dimethylation of arginine residues of PIWI proteins (Kirino et al., 2009). Mass spectrometry analyses have revealed that PIWI, but not AGO, subfamily proteins are arginine methylated at their N termini (Chen et al., 2009; Nishida et al., 2009; Vagin et al., 2009), which is important for the recognition of specific TUDOR family proteins (Chen et al., 2009; Kirino et al., 2010; Nishida et al., 2009; Reuter et al., 2009; Shoji et al., 2009; Vagin et al., 2009; Wang et al., 2009). For example, TUDOR, one of the Drosophila TUDOR-domain-containing proteins, associates with AUB and AGO3 in an sDMA-dependent manner (Kirino et al., 2010; Nishida et al., 2009). The mouse TUDOR domain-containing proteins TDRD1, TDRD2 and TDRD9 also require the sDMA modification of the mouse PIWI proteins for their association (Chen et al., 2009; Reuter et al., 2009; Shoji et al., 2009; Vagin et al., 2009; Wang et al., 2009). We investigated the interaction between PAPI and AGO3 in the absence of dPRMT5 by co-immunoprecipitation of PAPI from dPRMT5 mutant ovaries. AGO3 was present in PAPI immunoprecipitates from wild-type but not dPRMT5 mutant ovaries (Fig. 2F), suggesting that loss of arginine symmetric di-methylation abolished AGO3-PAPI interaction.
To confirm that the loss of PAPI-AGO3 interaction in dPRMT5 mutant ovaries is indeed caused by the lack of sDMAs in the N-terminal domain of AGO3, we generated a mutant form of AGO3 by mutating the three clustered arginine residues previously identified to be symmetrically dimethylated (Kirino et al., 2009; Nishida et al., 2009) into lysine residues, and tested the interaction between PAPI and the mutant form of AGO3 by Y2H. Although the wild-type AGO3 interacted with both PAPI-FL and PAPI-CT, the mutated AGO3 failed to interact with either of them (Fig. 2E). We also mutated the clustered arginine residues in the N terminus of PIWI, and found that the mutant PIWI no longer interacts with PAPI (see Fig. S3E in the supplementary material). These results together indicate that PAPI interacts with PIWI proteins through the binding of its TUDOR domain to the clustered sDMAs in the N-terminal domain of PIWI proteins.
PAPI and AGO3 interact in the nuage
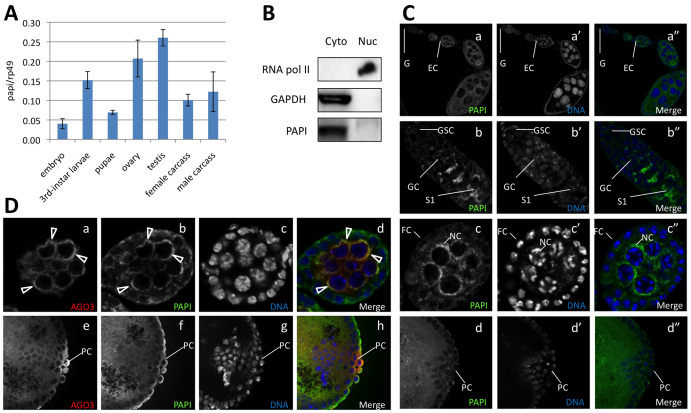
To investigate the expression profile of papi, we performed real-time PCR of papi at major developmental stages and in different tissues. The expression of papi was detected at all the developmental stages examined, including embryos, 3rd-instar larvae, pupae and adults (Fig. 3A). In adult flies, papi is most prominently expressed in ovaries and testes, and is also clearly detected in extra-gonadal somatic tissues (Fig. 3A).
Fig. 3.
PAPI and AGO3 colocalize in the nuage. (A) Quantitative RT-PCR analysis showing papi mRNA expression in embryos, 3rd instar larvae, pupae, ovaries, testes, adult female carcasses and adult male carcasses. (B) PAPI is primarily expressed in the cytoplasm. Purity of the nuclear and cytoplasmic fractions was confirmed using RNA PolII as a nuclear marker and GAPDH as a cytoplasmic marker. (C) Immunostaining of PAPI (green) in ovaries (a-c″) and embryos (d-d″). In adult fly ovaries, PAPI is localized to the cytoplasm, and is enriched in the germarium and the nuage. PAPI is cytoplasmic in embryos. No specific localization or strong deposition of PAPI was detected in early embryos. (D) PAPI partially colocalizes with AGO3 in the nuage (indicated by arrowheads) in ovaries and embryos. G, germarium; GSC, germline stem cells; GC, germline cyst; EC, egg chamber; FC, follicle cell; NC, nurse cells; S1, stage 1 egg chamber; PC, pole cells.
We then investigated whether PAPI colocalizes with PIWI proteins in the nuage by immunofluorescence microscopy. We first studied the cellular and subcellular localization of the PAPI protein during oogenesis and embryogenesis. In adult ovaries, PAPI is predominantly present in the cytoplasm of both germline cells and somatic follicle cells (Fig. 3C). Previous microarray analysis identified papi as one of the genes highly expressed in germline stem cells (Kai et al., 2005). We found that PAPI protein is expressed not only in germline stem cells, but also in germline cyst cells, nurse cells and oocytes, with a strong accumulation in region IIb germline cysts in the germarium (Fig. 3C, part b). As expected, in post-germarial egg chambers, PAPI accumulates in the perinuclear loci that appear to be the nuage (Fig. 3C, part c). In early embryos, PAPI is in the cytoplasm throughout the whole embryo, including the pole cells (Fig. 3C, part d). No specific localization or strong accumulation was detected shortly after egg laying or at later embryonic stages (not shown). To further confirm the cytoplasmic localization of PAPI, we performed a cytoplasm-nuclear fractionation of fly ovary extract. The purity of each fraction was assessed by western blot using GAPDH as a cytoplasmic marker and RNA polymerase II as a nuclear marker (Fig. 3B). Western blot analysis of PAPI in these fractions revealed that PAPI is predominantly cytoplasmic (Fig. 3B).
To verify PAPI as a component of the nuage, we further examined the expression pattern of a Flag-Myc-tagged PAPI transgene. The expression level and localization of the protein encoded by the transgene is very similar to that of the endogenous PAPI (see Fig. S4A,B in the supplementary material). In particular, Flag-Myc-tagged PAPI also accumulates in the perinuclear nuage region (see Fig. S4B in the supplementary material), where it colocalizes with endogenous PAPI (see Fig. S4C in the supplementary material). Therefore, PAPI is a novel component of the nuage.
Earlier studies have demonstrated that, similar to PAPI, AGO3 is localized in the cytoplasm, particularly enriched in the nuage in developing egg chambers (Aravin et al., 2007; Gunawardane et al., 2007; Li et al., 2009), and is ubiquitously expressed in embryos (Brennecke et al., 2008). We analyzed where the PAPI-AGO3 interaction occurs by examining the colocalization of PAPI and AGO3 using immunofluorescence microscopy. Co-immunostaining of PAPI and AGO3 showed that they share a very similar localization pattern. In adult female ovaries, their expression overlaps in the cytoplasm of germline stem cells, differentiating germline cysts and postgermarial egg chambers (data not shown). The colocalization is especially pronounced in the nuage of nurse cells (Fig. 3D, parts a-d). In early embryos, PAPI and AGO3 also colocalize in the cytoplasm (Fig. 3D, parts e-h). The colocalization pattern of PAPI and AGO3 indicates that they might interact physically and functionally in the cytoplasm both in early embryos and adult germline.
As PAPI also has weaker association with PIWI and AUB, we examined the potential colocalization of PAPI with PIWI and AUB. In adult fly ovaries, PIWI is predominantly nuclear, whereas PAPI is mainly in the cytoplasm. No obvious PAPI-PIWI colocalization was observed in ovaries (see Fig. S5A in the supplementary material), despite the interaction observed in Y2H and co-immunoprecipitation experiments. A possible explanation is that PAPI transiently interacts with PIWI in the nuage when PIWI enters the nucleus from the cytoplasmic sites where it is initially synthesized. Different from PIWI but similar to AGO3, AUB partially colocalizes with PAPI in the nuage in egg chambers (see Fig. S5B in the supplementary material).
PAPI and dPRMT5 are required for the localization of AGO3 to the nuage
To analyze the function of papi in the nuage, we knocked down the expression of papi using interfering RNA (RNAi) induced by the GAL4/UAS system. To induce RNAi expression, flies carrying papi shRNA were crossed to Act5C-GAL4 driver flies, leading to strong and nearly ubiquitous expression of shRNA in all tissues and developmental stages. Quantitative RT-PCR analysis revealed that the relative amounts of papi mRNA was reduced to less than 20% of the wild-type level (see Fig. S6A in the supplementary material). Western blot analysis of PAPI showed a ~80% drop in the protein level (see Fig. S6B in the supplementary material). The specificity of papi RNAi knockdown was examined by RT-PCR of the potential off targets. We first searched for all genes in the genome that are most homologous to the 293 bp papi-RNAi sequence. Two genes produced mRNAs that are most homologous to the papi-RNAi sequence – CG31935 and CG11160, each with an 18 bp complete match (see Fig. S7A,B in the supplementary material). In addition, two intergenic sequences also show an 18 bp complete match (see Fig. S7A,B in the supplementary material). Because the intergenic sequences do not encode genes or expressed RNAs, we examined whether the mRNA levels of CG31935 and CG11160 are affected by papi-RNAi. The papi RNAi specifically knocked down the expression of papi but not other two top score protein coding genes (see Fig. S7C in the supplementary material).
The papi-deficient flies are viable; however, female flies lay 30% fewer eggs from day 0 to day 5 compared with the control flies (see Fig. S6C in the supplementary material), indicating that loss of papi affects female fertility. Previous studies have suggested that mutations in piRNA pathway components, such as aub, ago3, vasa, armitage, spindleE, krimper and zucchini, disrupt the establishment of embryonic axis specification (Tushir et al., 2009). We observed that ~20% of eggs laid by papi-deficient mother displayed dorsal-ventral polarity defects. The weak phenotype is probably due to the incomplete knockdown of PAPI.
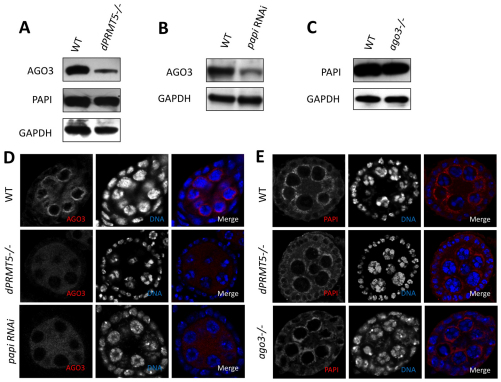
We then asked whether the expression and localization of AGO3 are affected in the absence of the AGO3-PAPI interaction in dPRMT5 mutant and papi-deficient ovaries. Western blot of ovary extract from dPRMT5 mutant ovaries showed a marked reduction in AGO3 protein level (Fig. 4A). Confocal microscopy further revealed that AGO3 localization was greatly reduced from the nuage and become evenly distributed in the cytoplasm of the germline cells (Fig. 4D). Similarly, partial knockdown of PAPI also leads to a reduction in AGO3 protein levels (Fig. 4B) and delocalization of AGO3 from the nuage (Fig. 4D). Unlike AGO3, both PIWI and AUB and properly localized in papi RNAi ovaries (see Fig. S8A,B in the supplementary material). To further determine whether the delocalization of AGO3 from the nuage in papi-deficient ovaries is due to the disruption of the general assembly and/or stability of the nuage or the failure of PAPI in recruiting AGO3 to the nuage, we examined the localization of VASA. VASA staining displays nuage morphology in papi-deficient ovaries (see Fig. S8C in the supplementary material), indicating that PAPI is not required for the general assembly/stability of the nuage. Collectively, these data suggest that the interaction with PAPI via the sDMAs of AGO3 is required for AGO3 to localize to the nuage.
Fig. 4.
The PAPI-AGO3 interaction is required for the localization of AGO3, but not PAPI, to the nuage. (A) Western blots of ovarian lysates from wild-type (WT) and dPRMT5 mutant (−/−) flies with AGO3 and PAPI antibodies. GAPDH was used as a loading control. dPRMT5 is required for the stability of AGO3 but not PAPI. (B) Western blots of ovary lysates from wild-type and papi RNAi flies with anti-AGO3 antibody. PAPI knockdown affects the level of AGO3 protein. (C) Western blots of ovary lysates from WT and ago3 mutant with anti-PAPI antibody. AGO3 mutation does not affect the level of PAPI protein. (D) Localization of AGO3 in wild-type, dPRMT5 mutant and papi RNAi egg chambers. AGO3 is largely delocalized from the nuage and more evenly distributed in the cytoplasm in dPRMT5 mutant and papi RNAi egg chambers. (E) Localization of PAPI in wild-type, dPRMT5 and ago3 mutant egg chambers. dPRMT5 and AGO3 are not required for the localization of PAPI to the nuage.
To analyze whether the mislocalization of AGO3 is affected by loss of germline or somatic PAPI, we knocked down PAPI in the germline tissues by expressing papi RNAi form a germline-specific GAL4 driver nosVP16-GAL4. Immunostaining of PAPI showed that PAPI was specifically knocked down in the germline cells but not the somatic follicle cells (see Fig. S9A in the supplementary material). Loss of germline PAPI also lead to a great delocalization of AGO3 from the nuage (see Fig. S9B in the supplementary material), suggesting that germline PAPI is responsible for the recruitment of AGO3 to the nuage.
dPRMT5 and AGO3 are not required for the localization of PAPI to the nuage
We then examined the expression and distribution of PAPI in dPRMT5 and ago3 mutant ovaries. Western blot of ovarian extracts from dPRMT5 and ago3 mutant ovaries showed a similar level of PAPI protein when compared with wild type (Fig. 4A,C). Furthermore, the localization of PAPI was not affected by dPRMT5 or ago3 mutations (Fig. 4D,E), as PAPI is still highly enriched in the nuage. Therefore, the localization of PAPI to the nuage is not dependent on the PAPI-AGO3 interaction or any other interaction that is mediated by sDMAs.
PAPI and AGO3 associate with the TRAL/ME31B complex in the nuage
Previous work has demonstrated that the piRNA pathway is closely related to processing (P) bodies (Lim et al., 2009). For example, AGO3 and AUB colocalize in germline cells with Decapping protein 1/2 (DCP1/2) and the Maternal expression at 31B (ME31B) protein, which are known components of P bodies (Lim et al., 2009). In addition, ME31B, Trailer hitch (TRAL) and Transitional endoplasmic reticulum 94 (TER94) proteins are present in the same RNA-protein complex (Wilhelm et al., 2005) and are associated with AUB and TUDOR in polar granules – the germline specific organelle essential for germline establishment in Drosophila (Thomson et al., 2008). Moreover, tral mutant female flies are sterile and lay eggs with dorsoventral patterning defects (Wilhelm et al., 2005), which is very similar to the phenotype of ago3 and other piRNA pathway mutants (Cook et al., 2004; Gonzalez-Reyes et al., 1997; Li et al., 2009; Lim and Kai, 2007; Pane et al., 2007; Wilson et al., 1996). To investigate whether the TRAL/ME31B complex physically and functionally interacts with the AGO3/PAPI complex, we co-immunoprecipitated PAPI from ovarian extracts and immunoblotted for TRAL, ME31B and TER94. We found that TRAL, ME31B and TER94 all specifically co-immunoprecipitated with PAPI (Fig. 5A), which indicates a physical association of PAPI with the TRAL/ME31B complex. Similar co-immunoprecipitation was performed with anti-AGO3 antibody. TRAL and TER94 were also found to associate with AGO3 (Fig. 5B). As both anti-AGO3 and anti-ME31B antibodies are mouse monoclonal, and the molecular weight of ME31B is very close the IgG heavy chain, we were not able to visualize ME31B in AGO3 co-immunoprecipitates.
Fig. 5.
AGO3 and PAPI associate with TRAL/ME31B complex in nuage. (A) PAPI immunoprecipitates from ovary lysates were probed with the indicated antibodies. Extract plus beads (−Ab) and antibody plus beads (−Extract) were used as negative controls for immunoprecipitation. (B) AGO3 immunoprecipitates from ovary lysates were probed with indicated antibodies. (C) TRAL colocalizes with PAPI and AGO3 in the nuage (arrowheads). (D) TRAL/ME31B and PAPI co-fractionated with AGO3 in the non-ribosomal fractions. Polysome fractionation of 0-12 hour embryo extract was performed in a 15%-50% (w/w) sucrose gradient. The fractions were probed with the antibodies indicated. (E) Embryonic extract was treated with EDTA prior to sucrose gradient fractionation. The fractions were probed with the antibodies indicated.
We next further examined whether TRAL, PAPI and AGO3 colocalize in the nuage by confocal microscopy. We observed that TRAL is particularly enriched in the nuage, where it colocalizes with PAPI and AGO3 (Fig. 4C), in addition to its previously reported accumulation in distinct foci in nurse cells (Wilhelm et al., 2005). Taken together, these results suggest that AGO3 and PAPI associate with the TRAL/ME31B complex in the nuage where they may function together in the piRNA pathway.
PAPI and the TRAL/ME31B complex co-fractionate with PIWI proteins in the non-ribosomal RNP pool
Recent studies have suggested that mRNAs are actively selected for piRNA production for regulatory purposes (Robine et al., 2009; Saito et al., 2009) and that P-body proteins negatively regulate translation by mRNA degradation (Eulalio et al., 2007a; Parker and Sheth, 2007), even though the formation of P-bodies has recently been suggested as a consequence of silencing (Eulalio et al., 2007b). Given the similarities between P bodies in somatic cells and the nuage in the germline, we wondered whether the PIWI-PAPI interaction in the nuage is related to translational regulation. We performed a polysome fractionation analysis using a sucrose gradient, and analyzed the distribution of the PIWI proteins, PAPI, the nuage marker VASA and the P-body proteins TRAL and ME31B in these fractions by western blotting analysis. All three PIWI proteins are present in RNP fractions as well as in fractions containing ribosomal subunits, monosomes and polysomes. TRAL is also present in these fractions. However, PAPI, VASA and ME31B are only present in the RNP fractions but excluded from the polysome fractions (Fig. 5D), indicating that the nuage and P body components are represented by the RNP fractions. To test whether the co-fractionation of PIWI proteins with ribosomes truly reflects their association, we treated the lysate with EDTA to dissociate polysomes and monosomes to large and small ribosomal subunits. Upon EDTA treatment, PIWI proteins and TRAL are eliminated from the polysome fractions, and shifted to the RNP fraction (Fig. 5E), supporting the notion that they are associated with polysomes. Our results suggest that there are two pools of Drosophila PIWI proteins: one is in the active translating pool, where they are associated with polysomes and might be involved in translational regulation; the other one is in the ribosome-free RNP fractions, which probably contain the nuage or polar granules components. The interaction of PIWI proteins with PAPI and P-body components occurs in the RNP fractions, which is excluded from the active translation pool.
A subset of transposons are de-silenced in papi and tral mutant ovaries
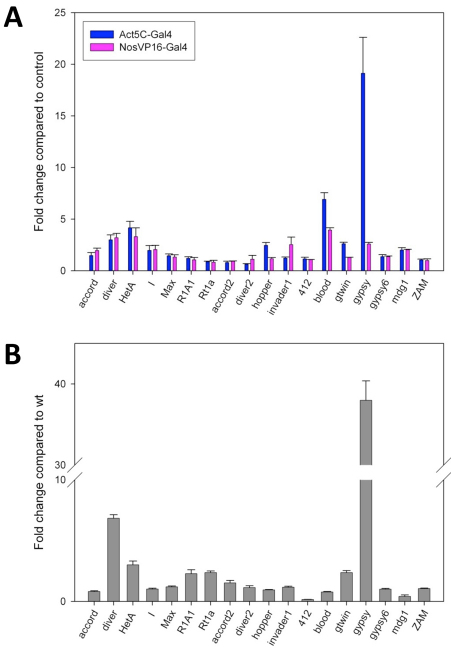
Because PAPI recruits PIWI proteins to the nuage and PIWI proteins are known to repress transposition, we explored whether PAPI and the nuage has a role in silencing transposition. Previous tilling array analysis showed that in the absence of ago3, 32 out of 64 group I transposons and 14 out of 26 group III transposons showed increased expression, and group II transposons were not significantly altered (Li et al., 2009). Given that PAPI binds to the sDMAs in the N terminus of AGO3, which is required for the localization of AGO3 to the nuage, we asked whether PAPI in the nuage is involved in transposon silencing. We examined the expression of three groups of transposons in papi-deficient ovaries by quantitative RT-PCR. We observed a trend of transposon expression in papi-deficient ovaries similar to that of the ago3 mutant. Four out of six group I transposons were de-silenced, including accord, diver, HetA and I-element (Fig. 6A). There is no significant change in the expression of four group II transposons except that the hopper transcript is upregulated by 2.5-fold (Fig. 6A). The expression of four out of seven group III transposons was significantly increased, including blood, gtwin, gypsy and mdg1, among which gypsy mRNA was drastically increased by 18-fold when compared with the wild-type level (Fig. 6A). To determine whether the increased transposon activity is due to the reduction of PAPI in the soma or germline, we examined the transposon activity in nosVP16-Gal4/papi-RNAi ovaries. The three group III transposons (blood, gtwin and gypsy) showed much lower levels of upregulation in nosVP16-Gal4/papi-RNAi ovaries when compared with Act5C-Gal4/papi-RNAi ovaries (Fig. 6A), suggesting that group III transposons are mostly regulated by PAPI in somatic cells. The de-repression of transposons in papi-deficient ovaries suggests that, like PIWI proteins, PAPI is also involved transposon silencing in both somatic and germline cells, and that in germline the nuage is a main, if not exclusive, site for transposon silencing.
Fig. 6.
PAPI and TRAL are required for the silencing of a subset of transposons. (A) Quantitative RT-PCR was performed to determine the expression of group I, group II and group III transposons in Act5C-Gal4/+, Act5C-Gal4/papi-RNAi, nosVP16-Gal4/+ and nosVP16-Gal4/papi-RNAi flies relative to that of rp49. The ratio of the relative transcript levels of Act5C-Gal4/papi-RNAi ovaries versus that of Act5C-Gal4/+ ovaries represents the fold change of transcript expression in ovaries with papi expression reduced in both somatic and germline cells. However, the ratio of the relative transcript levels of Act5C-Gal4/papi-RNAi ovaries versus that of nosVP16-Gal4/+ ovaries represents the fold change of transcript expression in ovaries with papi expression reduced only in germline cells. (B) Quantitative RT-PCR was performed to determine the expression of group I, group II and group III transposons, relative to rp49, in wild-type (WT) and tral mutant ovaries. The ratio of the relative transcript levels in tral mutant ovaries versus that of wild-type ovaries represents the fold change of transcript expression in tral mutant ovaries.
We next investigated whether the P-body components that associated with AGO3 and PAPI are also involved in regulating transposon activity by examining the transposon expression in tral mutant ovaries. In the absence of TRAL, three of six group I transposons were upregulated (Fig. 6B). No significant change was observed in the expression of four group II transposons (Fig. 6B). The expression of three out of six group III transposons was increased. Among these three transposons, gypsy mRNA was increased 38-fold compared with wild-type, whereas gtwin and gypsy6 mRNAs were only increased by a few folds (Fig. 6B). It has been shown that P-body components TWIN, DCP1 and SKI3, which are involved in mRNA degradation, are also involved in repressing HetA mRNA expression (Lim et al., 2009). Our results provide further evidence for the involvement of the P-body components in the nuage in transposon regulation.
DISCUSSION
Although the nuage has long been discovered in the germline of diverse organisms, little is known about its function. In this study, we identified and molecularly characterized a novel nuage component, PAPI. PAPI is a TUDOR-domain-containing protein that recruits PIWI proteins, especially AGO3, to the nuage and stabilizes them. The interaction between PAPI and AGO3 in the nuage is mediated by sDMAs in the N-terminal domain of AGO3 but is RNA independent. Previous studies have suggested the nuage as the cytoplasmic loci where post-transcriptional silencing of transposons occurs (Brennecke et al., 2007; Cook et al., 2004; Gunawardane et al., 2007; Harris and Macdonald, 2001; Li et al., 2009; Lim and Kai, 2007; Lim et al., 2009; Malone et al., 2009; Pane et al., 2007; Patil and Kai, 2010; Vagin et al., 2006). In addition, loss of Drosophila TUDOR protein has been shown to affect the localization of AUB to the nuage and to alter the piRNA profile (Nishida et al., 2009). Our new findings, together with these observations, indicate that TUDOR-domain-containing proteins might serve as a platform for the recruitment of PIWI proteins to the nuage and for the assembly of piRNA pathway components. A subset of transposons are de-repressed in papi deficient ovaries, suggesting that PAPI is involved in transposon silencing in the nuage, just like other piRNA pathway components. Our study thus reveals a function of the nuage in safeguarding the germline genome against deleterious retrotransposition via the piRNA pathway.
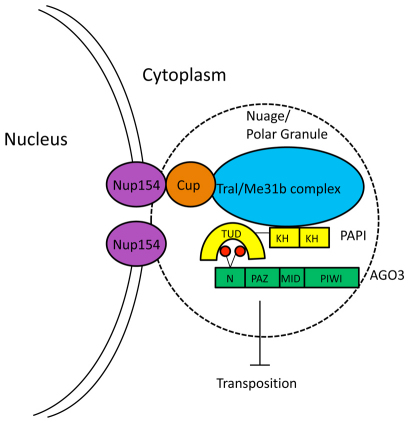
Furthermore, we have identified a physical association of PAPI and AGO3 with the TRAL/ME31B complex and their colocalization in the nuage, and the role of these P body proteins in silencing the expression of some transposons. The TRAL/ME31B complex has been shown to interact with CUP (Wilhelm et al., 2005), which also associates with the nuclear pore complex component NUP154 (Grimaldi et al., 2007). Our findings reveal an exciting physical and functional link between the piRNA machinery and the P body components in the nuage and a mechanism for nuage localization to the nuclear periphery. The P body proteins are well known for their function in mRNA processing and degradation, yet the piRNA machinery regulates transposon silencing by reducing the level of their mRNAs. The physical interaction between these two machineries, with the functional relationship among known components of these two machineries in the nuage illustrated in Fig. 7, raises the intriguing possibility that these two pathways work together in the nuage as a post-transcriptional mechanism to degrade transposon mRNAs, leading to transposon silencing. In addition, these data implicate the interaction of between the TRAL/ME31B complex and NUP154 via CUP as a mechanism of nuage localization to the nuclear periphery.
Fig. 7.
A working model. A working model summarizing the current knowledge on the interaction of PAPI (yellow), AGO3 (green) and TRAL/ME31B complex (blue) and their localization in the nuage via CUP (tan) anchoring to the nuclear pore complex component Nup154 (purple) in silencing transposition.
Supplementary Material
Acknowledgements
We thank Dr Seth Findley for the initial identification of PAPI as a PIWI interactor via Y2H screen; Dr Haruhiko Siomi for anti-PIWI and anti-AGO3 antibodies; Dr Phillip Zamore for ago3t2 and ago3t3 mutants; Dr James Wilhelm for tral1 and tral2 mutants, and anti-Tral antibody; and Dr Paul Lasko for anti-AUB antibody. We are grateful to Celina Juliano, Vamsi Gangaraju, Jae Eun Kwak, Toshiaki Watanabe, Jacob Gonzales and Vladimir Shteyn for their critical reading of the manuscript. This work was supported by NIH grants R01-HD33760 and DP1-OD006825 as well as the G. Harold and Leila Y. Mathers Charitable Foundation to H.L. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.059287/-/DC1
References
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M. J., Kuramochi-Miyagawa S., Nakano T., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203-207 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Naumova N. M., Tulin A. V., Vagin V. V., Rozovsky Y. M., Gvozdev V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017-1027 [DOI] [PubMed] [Google Scholar]
- Aravin A. A., Hannon G. J., Brennecke J. (2007). The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761-764 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G. J. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089-1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Malone C. D., Aravin A. A., Sachidanandam R., Stark A., Hannon G. J. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B., Findley S. D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S. C., Lin H. (2007). Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21, 2300-2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Jin J., James D. A., Adams-Cioaba M. A., Park J. G., Guo Y., Tenaglia E., Xu C., Gish G., Min J., et al. (2009). Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl. Acad. Sci. USA 106, 20336-20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook H. A., Koppetsch B. S., Wu J., Theurkauf W. E. (2004). The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 116, 817-829 [DOI] [PubMed] [Google Scholar]
- Cote J., Richard S. (2005). Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280, 28476-28483 [DOI] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Baker J., Chang L., Qiao D., Lin H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715-3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. N., Chao A., Lin H. (2000). piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503-514 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. (2007a). P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8, 9-22 [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. (2007b). P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 27, 3970-3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Xu J., Seitz H., Weng Z., Zamore P. D. (2010). Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16, 43-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G. J., Carmell M. A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199-202 [DOI] [PubMed] [Google Scholar]
- Gonsalvez G. B., Rajendra T. K., Tian L., Matera A. G. (2006). The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 16, 1077-1089 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., Elliott H., St Johnston D. (1997). Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development 124, 4927-4937 [DOI] [PubMed] [Google Scholar]
- Grimaldi M. R., Cozzolino L., Malva C., Graziani F., Gigliotti S. (2007). nup154 genetically interacts with cup and plays a cell-type-specific function during Drosophila melanogaster egg-chamber development. Genetics 175, 1751-1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna S. T., Pyhtila B., Lin H. (2006). MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 103, 13415-13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L. S., Saito K., Nishida K. M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M. C. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587-1590 [DOI] [PubMed] [Google Scholar]
- Harris A. N., Macdonald P. M. (2001). Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128, 2823-2832 [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L. M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D. V., Blaser H., Raz E., Moens C. B., et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69-82 [DOI] [PubMed] [Google Scholar]
- Jagannath A., Wood M. J. (2009). Localization of double-stranded small interfering RNA to cytoplasmic processing bodies is Ago2 dependent and results in up-regulation of GW182 and Argonaute-2. Mol. Biol. Cell 20, 521-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Doudna J. A. (2009). A three-dimensional view of the molecular machinery of RNA interference. Nature 457, 405-412 [DOI] [PubMed] [Google Scholar]
- Kai T., Williams D., Spradling A. C. (2005). The expression profile of purified Drosophila germline stem cells. Dev. Biol. 283, 486-502 [DOI] [PubMed] [Google Scholar]
- Kirino Y., Kim N., de Planell-Saguer M., Khandros E., Chiorean S., Klein P. S., Rigoutsos I., Jongens T. A., Mourelatos Z. (2009). Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 11, 652-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y., Vourekas A., Sayed N., de Lima Alves F., Thomson T., Lasko P., Rappsilber J., Jongens T. A., Mourelatos Z. (2010). Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16, 70-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Vagin V. V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M. D., Syrzycka M., Honda B. M., et al. (2009). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. K., Kai T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104, 6714-6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. K., Tao L., Kai T. (2009). piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186, 333-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Yue L., Spradling A. C. (1994). The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120, 947-956 [DOI] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. (2005). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7, 719-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Hannon G. J. (2009). Molecular evolution of piRNA and transposon control pathways in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 74, 225-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C. D., Brennecke J., Dus M., Stark A., McCombie W. R., Sachidanandam R., Hannon G. J. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh H. B., Cox D. N., Campbell C., Lin H. (2006). The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 16, 1884-1894 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Amikura R., Hanyu K., Kobayashi S. (2001). Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233-3242 [DOI] [PubMed] [Google Scholar]
- Nicchitta C. V., Migliaccio G., Blobel G. (1991). Biochemical fractionation and assembly of the membrane components that mediate nascent chain targeting and translocation. Cell 65, 587-598 [DOI] [PubMed] [Google Scholar]
- Nishida K. M., Okada T. N., Kawamura T., Mituyama T., Kawamura Y., Inagaki S., Huang H., Chen D., Kodama T., Siomi H., et al. (2009). Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28, 3820-3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A., Wehr K., Schupbach T. (2007). zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12, 851-862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635-646 [DOI] [PubMed] [Google Scholar]
- Patel N. H., Martin-Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S. (1989). Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, 955-968 [DOI] [PubMed] [Google Scholar]
- Patil V. S., Kai T. (2010). Repression of retroelements in Drosophila germline via piRNA pathway by the tudor domain protein Tejas. Curr. Biol. 20, 724-730 [DOI] [PubMed] [Google Scholar]
- Reuter M., Chuma S., Tanaka T., Franz T., Stark A., Pillai R. S. (2009). Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 16, 639-646 [DOI] [PubMed] [Google Scholar]
- Robine N., Lau N. C., Balla S., Jin Z., Okamura K., Kuramochi-Miyagawa S., Blower M. D., Lai E. C. (2009). A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr. Biol. 19, 2066-2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Jan C., Player C., Axtell M. J., Lee W., Nusbaum C., Ge H., Bartel D. P. (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193-1207 [DOI] [PubMed] [Google Scholar]
- Saffman E. E., Lasko P. (1999). Germline development in vertebrates and invertebrates. Cell. Mol. Life Sci. 55, 1141-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Nishida K. M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M. C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214-2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M. C. (2009). A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296-1299 [DOI] [PubMed] [Google Scholar]
- Shoji M., Tanaka T., Hosokawa M., Reuter M., Stark A., Kato Y., Kondoh G., Okawa K., Chujo T., Suzuki T., et al. (2009). The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell 17, 775-787 [DOI] [PubMed] [Google Scholar]
- Siomi H., Siomi M. C. (2009). On the road to reading the RNA-interference code. Nature 457, 396-404 [DOI] [PubMed] [Google Scholar]
- Thomson T., Lin H. (2009). The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25, 355-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T., Liu N., Arkov A., Lehmann R., Lasko P. (2008). Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 125, 865-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushir J. S., Zamore P. D., Zhang Z. (2009). SnapShot: Fly piRNAs, PIWI proteins, and the ping-pong cycle. Cell 139, 634 [DOI] [PubMed] [Google Scholar]
- Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320-324 [DOI] [PubMed] [Google Scholar]
- Vagin V. V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G. J., Aravin A. A. (2009). Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749-1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A., Tiedau D., Firooznia A., Muller-Reichert T., Jessberger R. (2009). Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 19, 630-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Saxe J. P., Tanaka T., Chuma S., Lin H. (2009). Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 19, 640-644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H. (2006). Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732-1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhecker G. S., Chen L., Gondarenko A., Lipson M. (2009). Controlling photonic structures using optical forces. Nature 462, 633-636 [DOI] [PubMed] [Google Scholar]
- Wilhelm J. E., Buszczak M., Sayles S. (2005). Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9, 675-685 [DOI] [PubMed] [Google Scholar]
- Wilson J. E., Connell J. E., Macdonald P. M. (1996). aubergine enhances oskar translation in the Drosophila ovary. Development 122, 1631-1639 [DOI] [PubMed] [Google Scholar]
- Yin H., Lin H. (2007). An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450, 304-308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.