Abstract
Major challenges of glycomics are to characterize a glycome and identify functional glycans as ligands for glycan-binding proteins (GBPs). To address these issues we have developed a general strategy termed shotgun glycomics. We focus on glycosphingolipids (GSLs), a challenging class of glycoconjugates recognized by toxins, antibodies, and GBPs. We derivatized GSLs extracted from cells with a heterobifunctional fluorescent tag suitable for covalent immobilization. Fluorescent GSLs were separated by multidimensional chromatography, quantified, and coupled to glass slides to create GSL shotgun microarrays. The microarrays were interrogated with cholera toxin, antibodies, and sera from patients with Lyme disease to identify biologically relevant GSLs that were subsequently characterized by mass spectrometry. Shotgun glycomics incorporating GSLs and potentially glycoprotein-derived glycans provides an approach to accessing the complex glycomes of animal cells and offers a strategy for focusing structural analyses on functionally significant glycans.
Keywords: glycosphingolipids, glycan array, fluorescent labeling, immobilization, functional glycomics
Introduction
Biologically important glycans found in glycosphingolipids and glycoproteins1–4 comprise the complex glycomes of cells and tissues. They are recognized by glycan-binding proteins (GBPs) expressed by human and animal cells, pathogens, and antibodies developed against host and foreign glycans5–7. Despite enormous progress in high performance liquid chromatography (HPLC), lectin affinity chromatography, mass spectrometry (MS), and glycan microarrays8, 9, chemically defining a glycome, the complete list of glycan structures that occur in a cell, tissue, or organism, remains elusive.
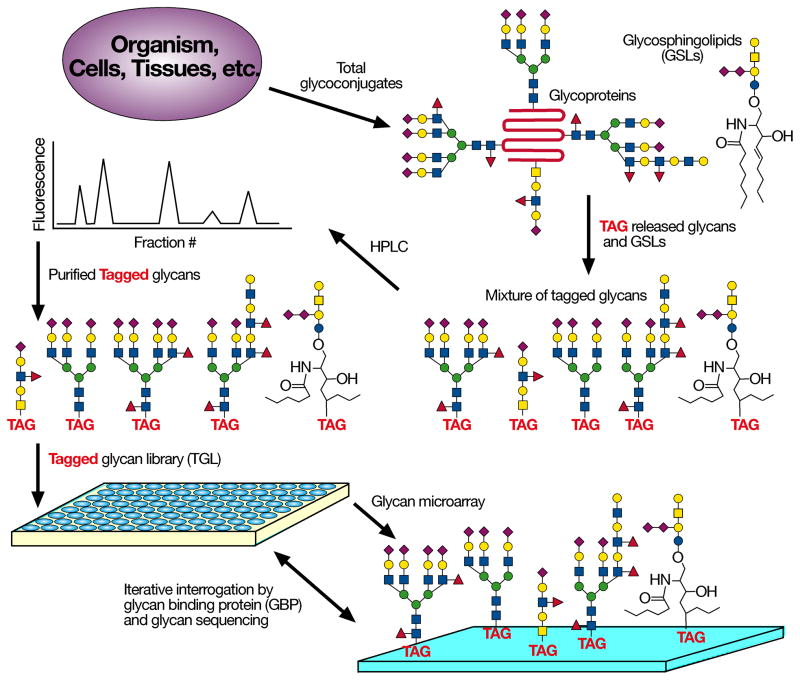
Because glycan structural analyses are difficult and require specialized technologies, a means of focusing analyses on biologically relevant glycans is desired. Since practical exploration of a cellular glycome should also incorporate quantitative and functional information about glycans, we devised the strategy of Shotgun Glycomics. We derivatize glycans from cellular glycosphingolipids (GSLs) and glycoproteins to generate fluorescently labeled glycans that can be separated, quantified, and covalently printed on glass slides or other surfaces for interrogation by GBPs and antibodies (Fig. 1). This approach is comparable in some ways to shotgun genomics, since the ultimate goal is the elucidation of structures (sequence) and functions of the glycome or genome.
Figure 1.
Schematic for Shotgun glycomics Glycans are released chemically or enzymatically from glycoproteins, GSLs are modified directly; the fluorescently labeled products are separated, quantified, and printed to create microarrays available for interrogation with GBPs.
We have developed a fluorescent tag N-(aminoethyl)-2-amino benzamide (AEAB) with an available aryl amine for conjugation to free glycans and an alkyl amine for efficient conjugation to reactive surfaces10. However, a major hurdle to shotgun glycomics and functional glycomics in general is the challenge of derivatizing GSLs. Glycans of GSLs are linked to a sphingosine moiety11, and while enzymatic release of the glycans from GSLs is feasible12, the complete loss of the aglycone may compromise GBP recognition. Therefore, we developed an approach for fluorescently labeling GSLs that permits easy derivatization, quantification, and separation by HPLC, and immobilization to glass slides to generate GSL shotgun microarrays. These microarrays allow interrogation by GBPs and antibodies, permitting structural analyses of GSLs recognized by GBPs.
Results
Ozonolysis, fluorescent conjugation, printing of GSLs
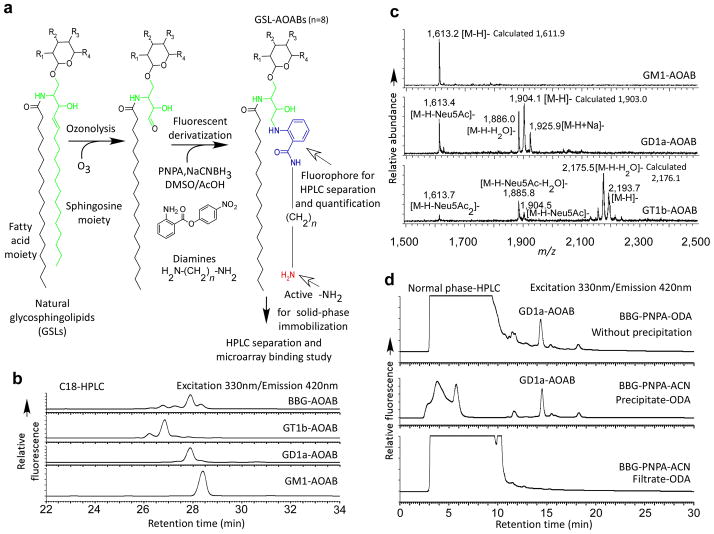
Ozonolysis of the common sphingosine moiety in GSLs generates a free aldehyde, readily reactive with the heterobifunctional p-nitrophenyl anthranilate (PNPA)13 through reductive amination to form a GSL-PNPA derivative, bearing a p-nitrophenyl ester as an excellent leaving group (Fig. 2a). The derivative precipitates from the product mixture with acetonitrile, and upon reaction with diamines, transforms to a fluorescent labeled GSL derivative, which retains an alkyl amine that can be covalently coupled to appropriate surfaces.
Figure 2.
Fluorescent derivatization of GSLs for shotgun glycomics: (a) The derivatization of GSLs with a bifunctional linker; (b) The C18-HPLC profiles of AOAB derivatization of GM1, GD1a, GT1b and BBG mixture detected by fluorescence; (c) The MALDI-TOF spectra of GM1-AOAB, GD1a-AOAB and GT1b-AOAB purified by HPLC as shown in (b). The spectra were acquired in the reflective negative mode; (d) The normal phase HPLC profiles of crude ODA treatment of BBG-PNPA conjugates without precipitation, the precipitate and the filtrate of BBG-PNPA mixture after addition of acetonitrile.
The approach was evaluated and optimized using the monosialyl ganglioside GM1 (Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ1-ceramide) (Supplementary Fig. 1). We chose NHS-activated slides for further studies, due to their generally lower background, and the ozone, PNPA, and octane-1,8-diamine (ODA) derivatization with its C8 extension, due to its relatively higher sensitivity of detection on microarray. We further evaluated this procedure with other GSLs, including GD1a, GT1b, and a mixture of bovine brain gangliosides (BBG) (Fig. 2b-d). C18-HPLC profiles (Fig. 2b) of the products from GM1, GD1a, and GT1b each show a dominant fluorescent peak, suggesting that the reactions are highly specific. No substantial desialylation occurred for individual gangliosides or the BBG mixture based on HPLC. We purified all products by HPLC and characterized them by MALDI-TOF (Fig. 2c) and confirmed all expected masses. We observed partial losses of sialic acids in the MALDI process, especially for multi-sialylated gangliosides.
To ensure that precipitation of GSL-PNPA in acetonitrile after reductive amination was quantitative, we treated the filtrate (Fig. 2d, bottom profile), precipitate (Fig. 2d, middle profile), and crude reductive amination product without precipitation (Fig. 2d, top profile) with a large excess of ODA. Comparison of HPLC profiles of the products indicated that the precipitation is complete. We then compared the underivatized GSLs and GSL-AOAB products using TLC. Both underivatized GSLs and the GSL-AOAB products were detected with orcinol staining (Supplementary Fig. 2a), while only the AOAB derivatives showed fluorescence under UV light (Supplementary Fig. 2b). The orcinol staining suggested the nearly quantitative AOAB-derivatization, since the TLC profiles after derivatization did not show more complexity. The fluorescent nature of the derivatives greatly facilitates preparative TLC and TLC protein overlay without destructive chemical staining.
Shotgun GSL microarray generated from BBG
We treated BBG with ozone, PNPA, and ODA, and separated the AOAB-labeled mixture of fluorescent GSLs from excess reagents by semi-preparative C18 HPLC and further fractionated it by 2D-HPLC (Supplementary Fig. 3a-c). The normal phase columns retain GSL-AEABs via their glycan moieties and provide good separations. The normal phase fractions are further resolved in a 2nd dimension by reverse phase HPLC, which simultaneously desalts each of the normal phase fractions. The resolved GSL-AOAB derivatives comprise the Tagged Glycolipid Library (TGL) and possess a free alkyl-amine function for printing as a GSL microarray and subsequent MS characterization. We quantified, characterized, and printed equimolar concentrations (10 μM) of the recovered peaks on NHS-slides. This BBG microarray contains 40 GSL-AOAB fractions plus controls. The compositional information of these fractions from MALDI-TOF/TOF was generated in an automatic fashion (Supplementary Table 1). We interrogated this GSL shotgun microarray with biologically-relevant GBPs: cholera toxin subunit B (CTSB) (Fig. 3a), and anti-GD1a antibody (Fig. 3b). These GBPs primarily bound GSLs in fractions #9 and #24, respectively. We characterized the glycan structures in these fractions by MS and MS/MS (Fig. 3c,d). The parent ions matched the expected compositions, which were subjected to collision-induced dissociation MS/MS analysis for more detailed structural analysis. The loss of labile Neu5Ac (−291) gave abundant fragment ions. Furthermore, the sequential loss of Hex (−162), HexNAc (−203), Hex (−162), Hex (−162) in the MS/MS spectra of fractions #9 and #24 is consistent with the structure asialoGM1, Galβ1-3GalNAcβ1-4Galβ1-4Glcβ1-R, confirming the general ganglioside core structure. Further, the fragment ion at 1248 (1247 for fraction #24) identifies Neu5Ac-Gal-Glc-ceramide-AOAB, and excludes the possibility of other isomers, such as GM1b and GD1b. The results allow the predictions that fraction #9 is GM1-AOAB, whereas fraction #24 is GD1a-AOAB (Neu5Acα2-3Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ1-R). The anti-GD1a antibody also exhibited cross-reactivity to other fractions (#8, #22, #26, #30). This result highlights the value of the GSL shotgun microarray to explore the specificities of monoclonal antibodies.
Figure 3.
Binding assay on the BBG GSL-AOAB microarray prepared from 2D HPLC separation: (a) cholera toxin subunit B (CTSB) at 0.1 μg ml−1 and (b) anti-GD1a antibody at 1:20 dilution of ascites fluid on the BBG-AOAB microarray. Average RFU of 4 replicates is reported, error bars = standard deviation. Structural characterization of bound fraction (c) #9 and (d) #24 by MS and MS/MS.
Serum antibody binding to Shotgun GSL microarray
To further evaluate the general utility of this approach using the BBG microarray, we screened for anti-GSL antibodies in serum from patients diagnosed with Lyme disease (Fig. 4a,b and Supplementary Table 2). Studies show a variety of peripheral neuropathies14, including those linked with Lyme disease15, are associated with anti-ganglioside antibodies. While both control and Lyme sera showed comparable weak binding to several gangliosides, only one BBG fraction (#12) showed statistically significant recognition by patient sera (P < 0.05) compared to control sera (Fig. 4b). Out of 10 patients, 5 showed a relatively high IgG response (>100 normalized relative fluorescence units, RFU) and 2 showed medium IgG response (50–100 normalized RFU) against fraction #12. Only 1 of 8 control sera showed a high IgG response and 1 showed a medium level IgG response against fraction #12 (Supplementary Table 2). We analyzed the MS and MS/MS data of fraction #12 (Fig. 4c), which suggested a composition of (Hex)3(HexNAc)1(Neu5Ac)2-H2O. MS/MS confirmed the composition with a clear Hex-Hex-HexNAc-Hex pattern, consistent with a ganglioside tetrasaccharide. The neutral loss of H2O may have occurred during ionization; however considerably shorter retention time (34.76 min) of this derivative compared to standard GD1a-AOAB’s (40.46 min) on normal phase HPLC suggests lower hydrophilicity, which might result from dehydration within the molecule. Although fraction #12 is a disialyl ganglioside, its MS/MS pattern is dramatically different from that of GD1a-AOAB, for example (Fig. 3d). There is an abundant fragment ion at 1321.4 from loss of two Neu5Acs (Neu5Ac2-H2O), but absolutely no fragment ion was observed due to loss of one Neu5Ac, suggesting another linkage between the two Neu5Ac moieties besides the common α2,8 glycosidic bond, possibly through formation of an internal ester or anhydro ether bond. Furthermore, the fragment ion at 1521.9, due to loss of HexNAc-Hex without loss of Neu5Ac, indicates no terminal Neu5Ac attached to the far most Gal at the non-reducing end and suggests a structure closely related to GD1b. Further studies including comparison with GD1b-AOAB prepared from standard, neuraminidase resistance, and formation of an amide with ethylenediamine strongly support the prediction that fraction #12 is GD1b-lactone (Supplementary Fig. 4). GD1b-lactone has been identified in brain tissues and melanoma cells16, 17. It can also be generated under acidic conditions in vitro18. Further study is now warranted with a larger set of serum samples from patients and controls. These results demonstrate the power of shotgun glycomics in identifying a relevant, but unusual and relatively low abundant GSL, within the mixture of starting material.
Figure 4.
The binding of Lyme disease and control sera on the BBG microarray: (a) comparison of IgG binding of sera from Lyme disease patients and control sera (tested at 1:100 dilution). The average RFUs are normalized in each serum sample by setting the binding of fraction 33 in control and patient serum to 100. (b) Distribution of binding by patient and control sera over six selected GSL-AOAB fractions (#12, 17, 26, 33, 39, and 40). Blue squares = individual control sera values, Red circles = individual patient sera values. P values, calculated with Student’s t-test, are given for the comparison of control to patient for the selected 6 glycans. * = P < 0.05. (c) Proposed structural characterization of bound fraction #12 by MS and MS/MS.
Shotgun Glycomics of GSLs from erythrocytes and PC3 cells
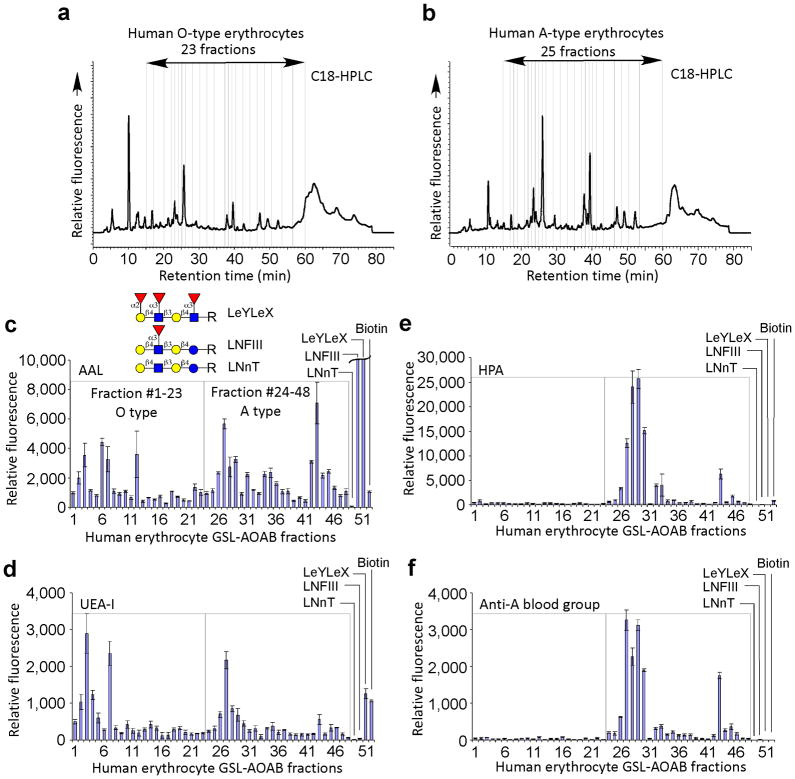
To further explore the general applicability of this method using whole cells, we prepared GSL-AOABs from human erythrocytes of blood types O and A. Human erythrocytes contain minute amounts of GSLs expressing blood group antigens, as most blood group antigens are found in glycoproteins19. We extracted GSLs from erythrocyte ghosts and subjected them to AOAB derivatization. The C18-HPLC profiles of O- and A-erythrocyte GSL-AOAB are similar (Fig. 5a,b). The TGL of O-erythrocyte GSL-AOAB and A-erythrocyte GSL-AOAB were comprised of 23 and 25 fractions, respectively. After separation and quantification, we printed and interrogated the TGL shotgun arrays with several GBPs. Binding by AAL, specific for α-linked fucose, suggested the general occurrence of fucose (Fig. 5c), while binding of several fractions by UEA-1, specific for α1–2 fucose, in both O-erythrocytes and A-erythrocytes (Fig. 5d), indicated the occurrence of H-antigen in both blood types. Interestingly, HPA, specific for terminal α-GalNAc, and anti-blood group A antibody showed binding only to several GSL-AOAB fractions prepared from A-erythrocytes with no cross reactivity to O-erythrocytes GSL-AOAB fractions (Fig. 5e,f). In future studies we will explore the blood group structures of such GSLs recognized by these specific antibodies. The sensitive and specific detection of these extremely scarce GSL structures through shotgun glycomics demonstrated the success of this strategy.
Figure 5.
The GSL microarray from human erythrocytes and its interrogation with lectins and antibodies: (a) C18-HPLC profiles of O-blood type erythrocyte GSL-AOAB, 23 fractions; (b) C18-HPLC profiles of A-blood type erythrocyte GSL-AOAB, 25 fractions; The binding of plant lectins (c) AAL (1 μg ml−1), (d) UEA-1 (10 μg ml−1), and (e) HPA (10μg ml−1), and (f) anti-blood group A antibody (10 μg ml−1) where GSL-AOAB #1-23 were from human blood group O erythrocytes, #24-48 were from human blood group A erythrocytes, and #49–52 were controls, including the AEAB derivatives of LNnT, LNFIII, LeyLex and biotin. Average RFU of 4 replicates is reported, error bars = standard deviation.
Many antibodies and other GBPs recognize tumor cells and may be useful in diagnostics and therapeutics. Therefore, we tested the shotgun glycomics approach with the cultured prostate cancer cell line PC3, which has been used for immunizations to develop antibodies that may have therapeutic potential and recognize undefined glycolipid antigens20. Using 1×107 PC3 cells, we prepared the mixture of GSL-AOAB derivatives as described above from GSLs extracted from the whole cell pellet. The C18-HPLC profile (Supplementary Fig. 5a) is quite different from that of erythrocytes (Fig. 5a,b). Thirty-three fractions were collected, printed on a microarray, and assayed with several plant lectins (Supplementary Fig. 6), showing differential binding to different fractions and controls. Ten major fractions were characterized by MALDI-TOF/TOF (Supplementary Table 3). Overall, the results predict that sulfated globosides (HexNAc-Hex-Hex-Hex-ceramide) are the major structures in PC3 GSL-AOABs, along with sialylated globosides (Supplementary Table 3). To illustrate the sulfated globoside structure of the PC3 GSLs, we show the analysis of one fraction (#16) (Supplementary Fig. 5b). The MS and MS/MS analyses using both positive and negative modes suggest a globo-series GSL structure with a sulfated pentasaccharide glycan. The GSL-AOAB fractions can be processed in a second dimension to generate individual glycans, whose composition can be determined by MS and MS/MS and printed as a microarray to further explore the specificities of antibodies against PC3 cells, such as the F77 antibody20 that is presumably directed against glycolipid epitopes.
Discussion
The Shotgun Glycomics approach complements our recently developed strategies for derivatizing N- and O-glycans released from glycoproteins using either non-reductive or reductive labeling with the fluorescent bifunctional AEAB reagent for separating and attaching glycans10, 21. Thus, glycans releasable from glycoproteins together with cellular GSLs can now be analyzed in a shotgun format for studies on recognition by GBPs, including antibodies and toxins. Importantly, the GSLs and the N-and O-glycans of cells can all be derivatized with similar 2-aminobenzamide-based fluorescent tags, which also permits the purification and covalent coupling through similar chemistries for all classes of cellular glycans. Thus, the cellular glycome can be functionally analyzed prior to focused sequencing approaches on relevant glycans. Shotgun microarrays prepared from natural sources complement and enlarge existing defined glycan microarrays, which are mainly comprised of chemo/enzymatically prepared glycans22. The TGLs prepared using the shotgun approach represent a permanent repository of a particular glycome and are available for detailed structural analysis or may be used for printing additional glycan arrays or coupling the glycans to other solid surfaces to assess interactions with novel GBPs. As the structures in a TGL are defined, so is the glycome of the cells or tissues used for its preparation, providing a strategy for expanding the number of structures on defined glycan arrays.
In this study we focused on GSLs, which are challenging due to their amphipathic nature and lack of sensitive chromophores for detection. In most cases, detection and quantification of GSLs employ destructive chemical sprays on TLC or relatively insensitive absorbance approaches23. In previous approaches to overcome the poor detection sensitivity during HPLC separation of GSLs, the component glycans were released from lipid and analyzed as fluorescent glycans by HPLC24, 25, but this provided no capability to explore their recognition by GBPs. Historically, studies on recognition of GSLs by various GBPs have been complex and often based on separation of GSLs by TLC and overlay with GBPs or ELISA-type formats26, 27. Although unrelated to natural GSLs as we have described, some prior approaches have involved generating neo-glycolipids from free glycans, and analyzing their recognition by GBPs using microarray28.
Ozonolysis, previously used to release oligosaccharides or conjugate sugars to reactive surfaces29, is specific to unsaturated bonds, as typically occur in sphingosine30, and permits easy fluorescent derivatization by PNPA and subsequently diamines. We have demonstrated the utility of bifunctional fluorescent tags to label free reducing glycans with diaminopyridine31, 32, AEAB10, 21, and PNPA13, to generate glycoconjugates with a functional amino group for subsequent immobilization on reactive surfaces. By exploiting the alkene present in the sphingosine moiety of most GSLs, we modified and adapted the PNPA-diamine labeling procedure for glycomic analysis of GSLs. In shotgun glycomics, the fractions, which are printed at equimolar concentrations on a microarray, are interrogated with biologically-relevant GBPs, so that structural analyses will be focused on only those glycans recognized by the GBP. Thus, with only nmol levels of GSL-AOAB purified from natural sources, it is possible to study the binding properties of proteins or microorganisms to different GSL-AOABs by microarray technology.
Our studies indicate that sera from patients with Lyme disease (borreliosis) express anti-glycolipid antibodies. Lyme disease is initiated from a bacterial infection with Borrelia burgdorferi, but the pathogenesis is believed to be related to auto-antibodies towards glycolipids15, 33. The observed IgG response to a disialylated ganglioside GD1b-lactone in BBG presumably arises from cross reactivity of the IgG generated against the bacterial infection, since glycolipid antigens are known to be present in B. burgdorferi33, 34. The characterization of GD1b-lactone provides a structural hint for future studies on the pathogenesis of Lyme disease, and more detailed analyses of such epitopes may lead to the development of diagnostic methods and therapeutic reagents. Similarly, GSL microarrays and TGLs prepared from human erythrocytes and PC3 cells indicate that our approach may be especially useful in identifying anti-carbohydrate antibodies20 to human tumors and could be important in biomarker discoveries for diagnostics and treatments.
Online Methods
Ganglioside standards were purchased (Calbiochem) and stored at −20°C until use. All chemicals were purchased (Sigma-Aldrich) and used without further purification. HPLC solvents were purchased (Fisher Scientific). An Ultraflex-II TOF/TOF system (Bruker Daltonics) was used for MALDI-TOF mass spectrometry analysis. Both reflective positive and reflective negative modes were used as indicated in the figures. 2,5-dihydroxybenzoic acid (5 mg ml−1 in 50% acetonitrile, 0.1% TFA) was freshly prepared as the matrix. 0.5 μl matrix solution was spotted on an Anchorchip target plate (200 μm or 400 μm) and air-dried. Then 0.5 μl solution was applied and air-dried. An Ozone Generator (Model OZV-8, Ozone Solutions) was used to generate ozone from high purity oxygen. Serum samples with a positive diagnosis for Lyme disease and control sera were obtained from Emory University Clinical Laboratory (IRB#00001614). PC3 cells (ATCC #CRL-1435) were grown in T-medium supplemented with 10% fetal bovine serum and 5% PenStrep (Invitrogen). Confluent PC3 cells were trypsinized, pelleted, and stored at −80°C until use.
Extraction of GSLs from cells
The extraction and desalting of GSLs from cells essentially followed protocols described previously35. Erythrocyte ghosts were prepared from ~300 ml each of A-type blood and O-type blood. Whole PC3 cell pellet (~0.5 ml, cell count ~5×107) was directly processed as described below. The wet human erythrocyte ghost pellet (or PC3 whole cell pellet) was homogenized with 3 volumes of water using a tip sonicator. The homogenate was added to 10.7 volumes of methanol slowly with stirring followed by 5.3 volumes of chloroform so that the final solvent ratio is 4:8:3 (C:M:W). The mixture was stirred for 30 min and centrifuged at 8,000×g for 30 min. The supernatant was poured into a separation funnel and 0.173 volume (relative to the supernatant) of water was slowly added. After gentle mixing, the upper phase was isolated. The upper phase was desalted with C18 Sep-pak, evaporated and redissolved in chloroform/methanol = 2/1 (v/v) for AOAB labeling.
Ozonolysis and fluorescent labeling of GSLs
GSLs (0.1–10 mg) were dissolved in 1–2 ml chloroform/methanol = 2/1 (v/v) and chilled in a dry ice/acetone bath. Ozone freshly generated by the ozone generator was passed through the solution for 1 min, while the blue color persisted. Nitrogen was bubbled through the solution for 1 min to remove the excess ozone. Methyl sulfide (Me2S), 100 μl, was added to destroy residual ozone; and after standing for 1 h at room temperature, the solution was dried under a stream of nitrogen. The ozonolysis products were labeled with PNPA as described previously for free glycans13. Briefly, 0.35 M PNPA and 1 M NaCNBH3 in DMSO/AcOH (7:3 v/v) were freshly prepared, and an equal amount (20 to 200 μl) of each solution was added to the dried residue and heated at 65°C for 2 h. Acetonitrile (10 volumes) was added and the mixture was cooled at −20°C for 2 h. The mixture was centrifuged and the supernatant was removed. To the precipitated GSL-PNPA derivatives, ODA (10% in DMSO, 20 to 200 μl) was added and the suspension was mixed on a vortex mixer for 30 minutes followed by the addition of 20 to 200 μl of 10% acetic acid. The mixture was centrifuged and the product was obtained from the supernatant.
High performance liquid chromatography (HPLC)
An HPLC CBM-20A system (Shimadzu), coupled with a UV detector (SPD-20A) and a fluorescence detector (RF-10Axl), was used for HPLC analysis and separation of GSL-AOABs. UV absorption at 330 nm or fluorescence at 330 nm excitation and 420 nm emission was used to detect GSL-AOABs in HPLC analyses and separations. Both UV absorption and fluorescence intensity were used for the quantification of GSLs with LNnT-AEAB as a standard.
For normal phase HPLC separation, a Zorbax NH2 column (250 mm × 4.6 mm) was used for analysis and a semi-preparative Zorbax NH2 column (250 mm × 9.2 mm) was used for preparative separations. The mobile phase used was acetonitrile, water, and 250 mM ammonium acetate (pH 4.5). In the analytical run, the concentration of water increased from 20% to 50% and the concentration of ammonium acetate buffer increased from 0 mM to 50 mM linearly over 25 min. In the preparative run, the concentration of water increased from 10% to 90% and the concentration of ammonium acetate buffer increased from 0 mM to 100 mM linearly over 120 min. For reverse phase HPLC with C18 column, a Vydac C18 column (250 mm × 4.6 mm) was used. The mobile phase was acetonitrile and water with 0.1% trifluoroacetic acid (TFA). The concentration of acetonitrile increased from 15% to 90% linearly over either 37.5 or 75 min.
Printing, binding assay, and scanning
NHS-activated slides were purchased (Schott). Epoxy slides were purchased (Corning). Non-contact printing was performed using a Piezoarray printer (Perkin Elmer). The average spot volume was within 10% variation of 1/3 nl. All samples were printed in phosphate buffer (300 mM sodium phosphates, pH 8.5). The processing for NHS and epoxy slides is described below. After printing, we boxed the slides loosely and put them in a high moisture chamber at 50°C and incubated for 1 h. We washed the slides and blocked with 50 mM ethanolamine in 0.1 M Tris buffer (pH 9.0) for 1 h. The slides were dried by centrifugation and stored desiccated at −20°C for future use. Before assay, we rehydrated the slides for 5 min in TSM buffer [20 mM Tris-HCl, 150 mM sodium chloride (NaCl), 0.2 mM calcium chloride (CaCl2), and 0.2 mM magnesium chloride (MgCl2)]. Biotin-hydrazine was printed as a positive control and also used for grid localization.
Biotinylated lectins (Vector Labs) and CTSB (Sigma) were used in the binding assay and the bound lectins were detected by a secondary incubation with cyanine 5-streptavidin (5μg ml−1, Invitrogen). Antibodies (anti-GD1a: Millipore, and anti-Blood group A: Santa Cruz Biotechnology) were detected by Alexa488-labeled corresponding secondary antibodies (5μg ml−1, Invitrogen). For multi-panel experiments on a single slide, we designed the array layout using Piezoarray software according to the dimension of a standard 16-chamber adaptor. We applied the adaptor to the slide to separate a single slide into 16 chambers sealed from each other during the assay.
The slides were scanned with a ProScanarray microarray scanner (Perkin Elmer) equipped with 4 lasers covering an excitation range from 488 nm to 637 nm. The scanned images were analyzed with ScanArray Express software. For cyanine 5 fluorescence, 649 nm (Ex) and 670 nm (Em) were used. For Alexa488 fluorescence, 495 nm (Ex) and 519 nm (Em) were used. All images obtained from the scanner were in grayscale and colored for easy discrimination.
Statistical analyses
For P value determination, patient (n = 10) and control (n = 8) sera were subjected to a two-tailed, unpaired Student’s t-test (unequal variance) and P < 0.05 was considered significant (alpha level 0.05).
Supplementary Material
Supplementary Figure 1 The bifunctional fluorescent derivatization of glycosphingolipids and the validation of microarray printing.
Supplementary Figure 2 Thin-layer chromatography (TLC) image of GM1, GD1a, GT1b, BBG, and their corresponding AOAB conjugates.
Supplementary Figure 3 The 2D HPLC separation of BBG GSL-AOABs.
Supplementary Figure 4 The elucidation of the structure of fraction 12.
Supplementary Figure 5 Fluorescent AOAB derivatization of PC3 cells GSLs.
Supplementary Figure 6 The lectin binding to PC3 cell glycosphingolipids microarray.
Supplementary Table 1 HPLC and Mass spectrometry characterization of BBG 2-dimensional fractions.
Supplementary Table 2 Raw data of antibody binding from control and Lyme disease patient sera.
Supplementary Table 3 MALDI-TOF MS and MS/MS data of the 10 peaks collected from C18 HPLC separation of PC3 cell GSL-AOABs.
Acknowledgments
This work was supported by in part by a Bridge Grant to R.D.C from the Consortium for Functional Glycomics from the National Institute of General Medical Sciences (Grant GM62116) and a EUREKA Grant (GM08544) to D.F.S. from the National Institute of General Medical Sciences. The authors have no financial interest to declare. We thank T. Burgess for helpful discussions.
Abbreviations
- GSL
glycosphingolipid
- AEAB
N-(2-aminoethyl)-2-amino- benzamide
- GSL-AEAB
Glycosphingolipid-AEAB conjugate
- ABAB
N-(aminobutyl)-2-amino benzamide
- AOAB
N-(aminooctyl)-2-amino benzamide
- TGL
tagged GSL/glycolipid library or tagged glycan library
- ConA
Concanavalin A
- CTSB
Cholera toxin subunit B
- HPLC
high performance liquid chromatography
- MAA
Maackia amurensis agglutinin
- NHS
N-hydroxysuccinimide
- RFU
relative fluorescence unit
- TLC
thin layer chromatography
- AAL
Aleuria aurantia agglutinin
- UEA-I
Ulex europaeus agglutinin I
- HPA
Helix, pomatia agglutinin
- BBG
Bovine brain gangliosides
- LNnT
Galβ1-4GlcNAcβ1-3Galβ1-4Glc
- LNFIII
Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4Glc
- LeyLex
Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAc
- Man5
Manα1-6(Manα1-3)Manα1-6(Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc
- NA2
Galβ1-4GlcNAcβ1-2Manα1-6(Galβ1-4GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc.
Footnotes
Author Contributions
X.S., R.D.C., and D.F.S. planned the project, and X.S., Y.L., B.X., H.J., C.Z., J.M.R., and R.J.M. carried out the experiments and supplied critical reagents. X.S., Y.L., J.H.M., R.D.C., and D.F.S. analyzed the data and wrote the manuscript, which was edited and commented on by the other authors.
References
- 1.Varki A, et al. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2009. [PubMed] [Google Scholar]
- 2.Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 3.Freeze HH, Aebi M. Altered glycan structures: the molecular basis of congenital disorders of glycosylation. Curr Opin Struct Biol. 2005;15:490–498. doi: 10.1016/j.sbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 5.Stowell SR, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamori M. A new turning point in glycosphingolipid research. Hum Cell. 2005;18:117–133. doi: 10.1111/j.1749-0774.2005.tb00002.x. [DOI] [PubMed] [Google Scholar]
- 7.Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol. 2010;28:107–130. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- 8.Comelli EM, et al. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology. 2006;16:117–131. doi: 10.1093/glycob/cwj048. [DOI] [PubMed] [Google Scholar]
- 9.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 10.Song X, et al. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 2009;16:36–47. doi: 10.1016/j.chembiol.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macher BA, Sweeley CC. Glycosphingolipids: structure, biological source, and properties. Methods Enzymol. 1978;50:236–251. doi: 10.1016/0076-6879(78)50026-8. [DOI] [PubMed] [Google Scholar]
- 12.Li YT, et al. Preparation of homogenous oligosaccharide chains from glycosphingolipids. Glycoconj J. 2009;26:929–933. doi: 10.1007/s10719-008-9125-9. [DOI] [PubMed] [Google Scholar]
- 13.Luyai A, Lasanajak Y, Smith DF, Cummings RD, Song X. Facile Preparation of Fluorescent Neoglycoproteins Using p-Nitrophenyl Anthranilate as a Heterobifunctional Linker. Bioconjug Chem. 2009 doi: 10.1021/bc900189h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willison HJ. Gangliosides as targets for autoimmune injury to the nervous system. J Neurochem. 2007;103 (Suppl 1):143–149. doi: 10.1111/j.1471-4159.2007.04718.x. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Monco JC, Seidman RJ, Benach JL. Experimental immunization with Borrelia burgdorferi induces development of antibodies to gangliosides. Infect Immun. 1995;63:4130–4137. doi: 10.1128/iai.63.10.4130-4137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terabayashi T, Kawanishi Y. Naturally occurring ganglioside lactones in Minke whale brain. Carbohydr Res. 1998;307:281–290. doi: 10.1016/s0008-6215(98)00018-4. [DOI] [PubMed] [Google Scholar]
- 17.Riboni L, et al. Natural occurrence of ganglioside lactones. Isolation and characterization of GD1b inner ester from adult human brain. J Biol Chem. 1986;261:8514–8519. [PubMed] [Google Scholar]
- 18.Bassi R, Riboni L, Sonnino S, Tettamanti G. Lactonization of GD1b ganglioside under acidic conditions. Carbohydr Res. 1989;193:141–146. doi: 10.1016/0008-6215(89)85113-4. [DOI] [PubMed] [Google Scholar]
- 19.Anstee DJ. The nature and abundance of human red cell surface glycoproteins. J Immunogenet. 1990;17:219–225. doi: 10.1111/j.1744-313x.1990.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G, et al. Suppression of human prostate tumor growth by a unique prostate-specific monoclonal antibody F77 targeting a glycolipid marker. Proc Natl Acad Sci U S A. 2010;107:732–737. doi: 10.1073/pnas.0911397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song X, Lasanajak Y, Xia B, Smith DF, Cummings RD. Fluorescent glycosylamides produced by microscale derivatization of free glycans for natural glycan microarrays. ACS Chem Biol. 2009;4:741–750. doi: 10.1021/cb900067h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muthing J. Analyses of glycosphingolipids by high-performance liquid chromatography. Methods Enzymol. 2000;312:45–64. doi: 10.1016/s0076-6879(00)12899-x. [DOI] [PubMed] [Google Scholar]
- 24.Ohara K, Sano M, Kondo A, Kato I. Two-dimensional mapping by high-performance liquid chromatography of pyridylamino oligosaccharides from various glycosphingolipids. J Chromatogr. 1991;586:35–41. doi: 10.1016/0021-9673(91)80022-9. [DOI] [PubMed] [Google Scholar]
- 25.Wing DR, et al. High-performance liquid chromatography analysis of ganglioside carbohydrates at the picomole level after ceramide glycanase digestion and fluorescent labeling with 2-aminobenzamide. Anal Biochem. 2001;298:207–217. doi: 10.1006/abio.2001.5393. [DOI] [PubMed] [Google Scholar]
- 26.Lopez PH, Schnaar RL. Determination of glycolipid-protein interaction specificity. Methods Enzymol. 2006;417:205–220. doi: 10.1016/S0076-6879(06)17015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magnani JL, et al. A monosialoganglioside is a monoclonal antibody-defined antigen of colon carcinoma. Science. 1981;212:55–56. doi: 10.1126/science.7209516. [DOI] [PubMed] [Google Scholar]
- 28.Stoll MS, et al. Fluorescent neoglycolipids. Improved probes for oligosaccharide ligand discovery. Eur J Biochem. 2000;267:1795–1804. doi: 10.1046/j.1432-1327.2000.01178.x. [DOI] [PubMed] [Google Scholar]
- 29.Laine RA, Yoggeswaran G, Hakomori S. Glycosphingolipids covalently linked to agarose gel or glass beads. Use of the compounds for purification of antibodies directed against globoside and hematoside. J Biol Chem. 1974;249:4460–4466. [PubMed] [Google Scholar]
- 30.Hirabayashi Y, Hamaoka A, Matsumoto M, Nishimura K. An improved method for the separation of molecular species of cerebrosides. Lipids. 1986;21:710–714. doi: 10.1007/BF02537245. [DOI] [PubMed] [Google Scholar]
- 31.Song X, Xia B, Lasanajak Y, Smith DF, Cummings RD. Quantifiable fluorescent glycan microarrays. Glycoconj J. 2008;25:15–25. doi: 10.1007/s10719-007-9066-8. [DOI] [PubMed] [Google Scholar]
- 32.Xia B, et al. Versatile fluorescent derivatization of glycans for glycomic analysis. Nature methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 33.Jones KL, et al. Strong IgG antibody responses to Borrelia burgdorferi glycolipids in patients with Lyme arthritis, a late manifestation of the infection. Clin Immunol. 2009;132:93–102. doi: 10.1016/j.clim.2009.03.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 35.Schnaar RL. Isolation of glycosphingolipids. Methods Enzymol. 1994;230:348–370. doi: 10.1016/0076-6879(94)30024-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 The bifunctional fluorescent derivatization of glycosphingolipids and the validation of microarray printing.
Supplementary Figure 2 Thin-layer chromatography (TLC) image of GM1, GD1a, GT1b, BBG, and their corresponding AOAB conjugates.
Supplementary Figure 3 The 2D HPLC separation of BBG GSL-AOABs.
Supplementary Figure 4 The elucidation of the structure of fraction 12.
Supplementary Figure 5 Fluorescent AOAB derivatization of PC3 cells GSLs.
Supplementary Figure 6 The lectin binding to PC3 cell glycosphingolipids microarray.
Supplementary Table 1 HPLC and Mass spectrometry characterization of BBG 2-dimensional fractions.
Supplementary Table 2 Raw data of antibody binding from control and Lyme disease patient sera.
Supplementary Table 3 MALDI-TOF MS and MS/MS data of the 10 peaks collected from C18 HPLC separation of PC3 cell GSL-AOABs.