Abstract
Chemokine receptors are critical regulators of cell migration in the context of immune surveillance, inflammation and development. The G protein-coupled chemokine receptor, CXCR4, is specifically implicated in cancer metastasis and HIV-1 infection. Here we report five independent crystal structures of CXCR4 bound to an antagonist small molecule IT1t and a cyclic peptide CVX15 at 2.5–3.2 Å resolution. All structures reveal a consistent homodimer with an interface involving helices V and VI that may be involved in regulating signaling. The location and shape of the ligand binding sites differ from other G protein-coupled receptors and are closer to the extracellular surface. These structures provide new clues about the interactions between CXCR4 and its natural ligand CXCL12 and with the HIV-1 glycoprotein gp120.
Introduction
Chemokine receptors are G protein-coupled receptors (GPCRs) that, together with their small protein ligands, regulate the migration of many different cell types, most notably leukocytes (1–3). CXCR4, one of 19 known human chemokine receptors, is activated exclusively by the chemokine CXCL12 (also known as Stromal Cell-Derived Factor-1, SDF-1) and couples primarily through Gi proteins. Targeted deletion of CXCR4 or CXCL12 in mice confers embryonic lethality and exhibits defects in vascular and CNS development, hematopoiesis, and cardiogenesis (4–5). CXCR4 has been associated with more than 23 types of cancers where it promotes metastasis, angiogenesis and growth/survival (6–10). Furthermore, T-tropic HIV-1 uses CXCR4 as a co-receptor for viral entry into host cells (11). Thus, the discovery that endogenous CXCL12 inhibits HIV-1 entry suggested the therapeutic potential of targeting CXCR4 to block viral infection (12–13). Despite a wealth of data related to CXCR4 and GPCRs in general, many aspects of ligand binding and signaling are poorly understood at the molecular level. For instance, CXCR4 has a propensity to form hetero- and homo-oligomers (14–15), and such oligomerization could play a role in the allosteric regulation of CXCR4 signaling (16). While structural understanding of GPCRs has benefited from a number of recent breakthroughs (17–20), coverage of the superfamily’s phylogenetic tree is incomplete, and a structure of a GPCR that is activated by a protein ligand has not been reported.
Protein engineering, ligand selection, and structure determination
Here we report the crystal structures of human CXCR4 in complex with a small molecule antagonist at 2.5 Å resolution and with a cyclic peptide inhibitor at 2.9 Å resolution. Three stabilized constructs (CXCR4-1, CXCR4-2 and CXCR4-3; Table S1) expressed in baculovirus-infected Spodoptera frugiperda (Sf9) insect cells were selected for structural studies based on thermal stability, monodispersity, and lipid matrix diffusion. Similar to the previously determined high-resolution structures of the β2-adrenergic receptor (β2AR) (17, 21) and A2A adenosine receptor (A2AAR) (18), the CXCR4 constructs contain a T4 lysozyme (T4L) fusion inserted between transmembrane (TM) helices V and VI at the cytoplasmic side of the receptor. In addition, all three constructs contain a thermostabilizing L1253.41W mutation (22–23). The constructs differ in the precise T4L junction site, the position of the C-terminal truncation, as well as a T2406.36P mutation in CXCR4-3, and required further stabilization with ligands to facilitate purification and crystallization. Two antagonists were selected for crystallization trials based on ligand solubility, binding affinity, and induced protein thermostability (Table S2, S3): a small, drug-like, isothiourea derivative (IT1t) (24) and CVX15, a 16-residue cyclic peptide analog of the horseshoe crab peptide polyphemusin, that was previously characterized as an HIV-inhibiting and anti-metastatic agent (25–27).
Prior to crystallization trials, the effects of the protein engineering on CXCR4 function were evaluated using radioligand binding and calcium flux assays. CXCR4-WT expressed in Sf9 cells binds a [3H]bis(imidazolylmethyl) amine analog (BIMA) with similar affinity as the same construct expressed in HEK293 cells (Kd 3.5 ± 1.5 and 3.7 ± 1.4 nM, respectively). All other constructs expressed in Sf9 cells also show similar binding affinity to BIMA and IT1t (Table S3). However, CXCR4-1 and CXCR4-2 display lower binding affinity for the CVX15 peptide compared to CXCR4-WT and CXCR4-3. Calcium flux assays demonstrated the expected result that these constructs do not activate G proteins (Fig. S1), due to the T4L insertion in the third intracellular loop, which is critical for G protein interactions. Assays with the same constructs lacking T4L confirmed that the stabilizing L1253.41W mutation, as well as the various C-terminal truncations, did not adversely affect calcium release, while the T2406.36P mutation, which is present only in the CXCR4-3 construct, abolished signaling.
After extensive optimization of crystallization conditions in lipidic mesophase, five distinct crystal forms were obtained (Table S4). CXCR4-1, CXCR4-2 and CXCR4-3 were co-crystallized with IT1t (two crystal forms for CXCR4-2), while crystals of CXCR4-3 were also obtained with CVX15. Data collection and refinement statistics for all five crystal forms are shown in Table S1 (28).
Overall architecture of CXCR4
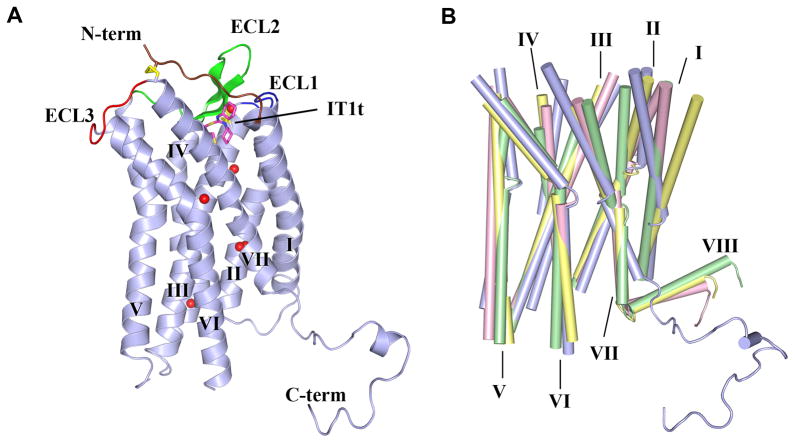
The overall structure of CXCR4 bound to the small molecule antagonist IT1t is conserved in all crystal forms with a Cα RMSD of 0.6 Å. Binding of the CVX15 cyclic antagonist peptide induced conformational differences relative to IT1t in the CXCR4-3/CVX15 structure (Cα RMSD 0.9 Å). For clarity, we focus on the highest resolution crystal form of CXCR4-2/IT1t (2.5 Å, monomer A) for discussion of the CXCR4 structural features and comparison with other GPCR structures. The final model includes 326 of the 352 residues of CXCR4 and residues 2–161 of T4L. The remaining N-terminal 26 residues did not have interpretable density and are presumed to be disordered. The main fold of CXCR4 consists of the canonical bundle of 7 TM α-helices (Fig. 1A), which shows about the same level of structural divergence from 7TM helical bundles of previously solved GPCR structures (Cα RMSDs ~2.0–2.2 Å) (Fig. 1B). The most striking differences in the disposition of the TM helices of CXCR4 are the following: i) The extracellular end of helix I is shifted towards the central axis of the receptor by 9 Å compared to β2AR and by more than 3 Å compared to A2AAR; ii) helix II makes a tighter helical turn at Pro922.58 resulting in ~120° rotation of its extracellular end compared to other GPCR structures (this rotation essentially introduces a one-residue gap in the sequence alignment that would result in wrong residues facing the ligand-binding pocket in a homology model that did not account for the rotation); iii) both intracellular and extracellular tips of helix IV in CXCR4 substantially deviate (~5 and ~3 Å, respectively) from their consensus positions in other GPCRs; iv) the extracellular end of helix V in CXCR4 is about one turn longer; v) helix VI has a similar shape in all structures and is characterized by a sharp kink at the highly conserved residue, Pro2546.50; however, its extracellular end is shifted by ~3 Å in CXCR4 relative to β2AR and A2AAR; and finally vi) the extracellular end of helix VII in CXCR4 is two helical turns longer than in other GPCR structures. These two extra turns place Cys2747.25 at the tip of helix VII in a strategic position to form a disulfide bond with Cys28 in the N-terminal region. Taken together, these multiple differences suggest that accurate homology modeling of even the CXCR4 TM bundle, let alone the entire structure, would be challenging.
Fig. 1.
Overall fold of the CXCR4/IT1t complex and comparison with other GPCR structures. (A) Overall fold of the CXCR4-2/IT1t. The receptor is colored blue. The N-terminus, ECL1, ECL2 and ECL3 are highlighted in brown, blue, green and red, respectively. The compound IT1t is shown in a magenta stick representation. The disulfide bonds are yellow. Conserved water molecules (67) are shown as red spheres. (B) Comparison of TM helices for CXCR4 (blue), β2AR (PDB ID: 2RH1; yellow), A2AAR (PDB ID: 3EML; green) and rhodopsin (PDB ID: 1U19; pink).
The extracellular interface of CXCR4 consists of 34 N-terminal residues, extracellular loop 1 (ECL1, residues 100–104) linking helices II and III, ECL2 (residues 174–192) linking helices IV and V, and ECL3 (residues 267–273) linking helices VI and VII (Fig. 1A). Clear density starts at Pro27, adjacent to Cys28, which pins the base of the N-terminal segment to Cys2747.25 at the tip of helix VII via a disulfide bond; these two cysteines are conserved in all chemokine receptors except CXCR5 and CXCR6 (Fig. S2). Another disulfide links Cys1093.25 with Cys186 of ECL2, which is the largest extracellular loop in CXCR4. While ECL2 length, sequence and secondary structure vary dramatically in GPCRs, the disulfide connecting ECL2 with the extracellular end of helix III is highly conserved in chemokine receptors and most other Class A GPCRs. Both disulfide bonds at the extracellular side of CXCR4 are critical for ligand binding (29), and the crystal structure shows that they function by constraining ECL2 and the N-terminal segment (residues 26–34), thereby shaping the entrance to the ligand binding pocket.
The intracellular side of CXCR4 contains intracellular loop 1 (ICL1, residues 65–71) linking helices I and II, ICL2 (residues 140–149) linking helices III and IV, and ICL3 (residues 225–230) linking helices V and VI, and the C-terminus. ICL3 also contains T4L inserted between Ser229 and Lys230 and flanked by short linkers (GS-T4L-GS). Structural alignment of CXCR4 with high resolution GPCR structures indicates that the intracellular half of the TM bundle is structurally more conserved (Cα RMSDs with β2AR, A2AAR and rhodopsin are 1.8, 1.9 and 1.4 Å, respectively) than the extracellular half (2.6, 2.2 and 2.2 Å, respectively). Therefore, it comes as a surprise that in all five CXCR4 structures, helix VII is about one turn shorter at the intracellular side, ending right after the GPCR-conserved NPxxY motif, and that all structures lack the short α-helix VIII (Fig. 1B). The C-terminal part of CXCR4 beyond Ala3037.54 adopts an extended conformation and participates in a number of crystal contacts with the extracellular side of a symmetry-related molecule in the highest resolution crystal form, CXCR4-2/IT1t, (Fig. S4A), and is not traceable in the other four CXCR4 structures. Due to its structural persistence and common α-helical sequence motif (F[RK]xx[FL]xxx[LF]), helix VIII was thought to be a regular structural element of all Class A GPCRs. However, CXCR4 contains only a partially conserved motif FKxxAxxxL, and while it may be capable of forming an α-helix under certain conditions, this helix would be less stable due to replacement of Phe/Leu with Ala. In addition, CXCR4 lacks a putative palmitoylation site at the end of helix VIII, which anchors to the lipid membrane in many GPCRs.
Construct CXCR4-3 contains a T2406.36P mutation near the intracellular side of helix VI, which results in retention of ligand binding affinity, but abolishes signaling (Table S3 and Fig. S1). Comparison of the CXCR4-3 structure with CXCR4-1 and CXCR4-2 reveals that the only effect of the T2406.36P mutation is the disruption of a short section of helix VI between Lys2346.30 and Pro2406.36. Since helix VI is thought to be one of the major players in the signaling mechanism (30–31), disruption of its structure would likely impact G protein binding and activation. Thus, T2406.36P represents a novel structure-based uncoupling mutation.
Molecular recognition of the small molecule IT1t and the cyclic CVX15 peptide by CXCR4
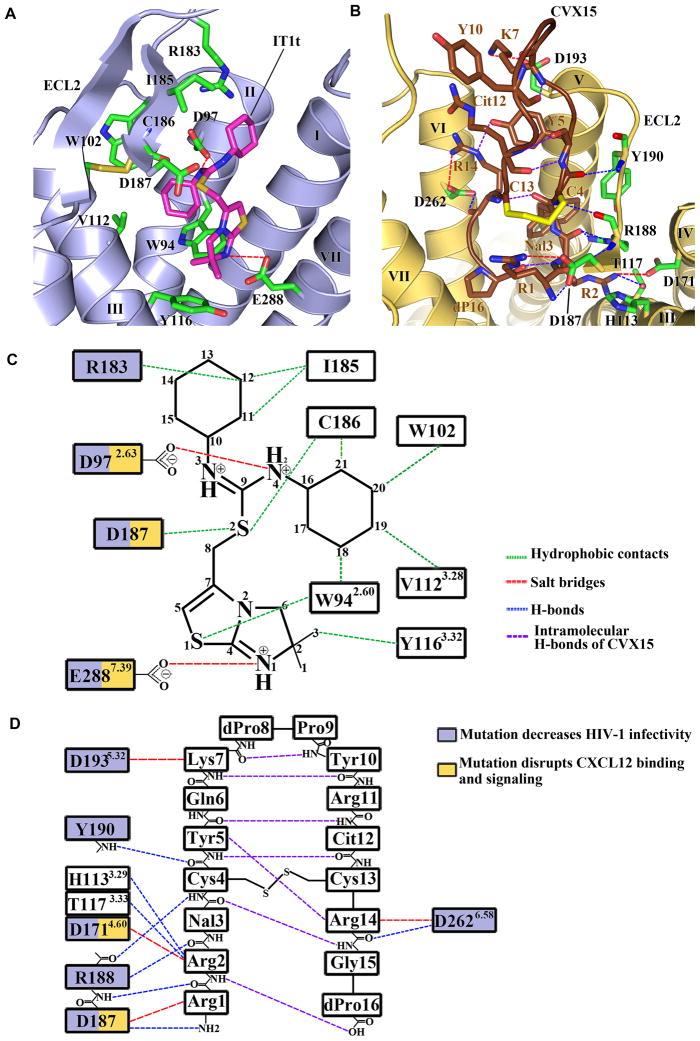
Strong electron density was observed for IT1t in the binding cavity of both subunits of the CXCR4 homodimer (Fig. S3A). Compared to previous GPCR structures, the cavity is larger, more open and is located closer to the extracellular surface (Fig. 2A, C, Fig. 3B & Table S5). The IT1t ligand occupies part of the pocket defined by side chains from helices I, II, III and VII, but makes no contact with helices IV, V and VI, in stark contrast to ligands in previous GPCR structures. The nitrogens of the symmetrical isothiourea group are both protonated with a net positive resonance charge, one of them (N4) forming a salt bridge (2.7 Å) with the Asp972.63 side chain. Note, that the electron density does not preclude the existence of a very similar ligand conformation with a flipped thiourea group, in which the N3 nitrogen forms a salt bridge to Asp972.63, and the N4 nitrogen makes a polar interaction with mainchain carbonyl of Cys186 in ECL2. The importance of both nitrogens is supported by a reduction in binding affinity of ~100-fold upon methylation of one of them (24). Both cyclohexane rings fit into small subpockets, making hydrophobic contacts with CXCR4. Connected by a short flexible linker, the imidazo-thiazole ring system is the only part of the ligand that contacts helix VII, in particular by making a salt bridge (2.8 Å) between the protonated imidazo-thiazole N1 and Glu2887.39 (32).
Fig. 2.
CXCR4 ligand-binding cavities for the small molecule IT1t and the cyclic peptide CVX15. (A) CXCR4 ligand-binding cavity for the small molecule IT1t. IT1t (magenta) and the residues of the receptor (green) involved in the ligand interactions are shown in stick representation. Nitrogen atoms are blue and sulfur atoms are yellow. Hydrophobic contacts are shown as green dashed lines, salt bridges are red, and hydrogen bonds are blue. Only the helices involved in the receptor-ligand interaction and part of ECL2 are shown. (B) CXCR4 ligand-binding cavity for the peptide CVX15. The residues of CVX15 (brown) and the residues of the receptor (green) involved in receptor-ligand polar interactions are shown in stick representation. The Cys4-Cys13 disulfide bridge in CVX15 is shown as a yellow stick. Intramolecular hydrogen bonds of CVX15 are shown as purple dashed lines. (C) Schematic representation of the interactions between CXCR4 and IT1t in the ligand binding pocket. Mutations reported to decrease HIV-1 infectivity and disrupt CXCL12 binding and signaling are indicated with blue and yellow squares, respectively (56, 68). (D) Schematic representation of the interactions between CXCR4 and CVX15 in the ligand binding pocket.
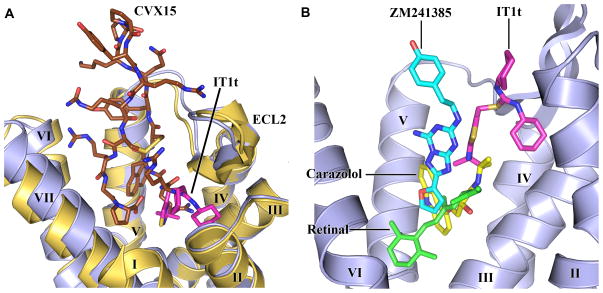
Fig. 3.
CXCR4 ligand-binding modes and comparison with other GPCR structures. (A) Comparison of the ligand-binding modes for IT1t and CVX15. CXCR4 molecules in the CXCR4- 2/IT1t and CXCR4-3/CVX15 complexes are colored blue and yellow, respectively. IT1t (magenta) and CVX15 (brown) are shown as sticks. (B) Comparison of the small molecule ligand-binding modes for CXCR4, β2AR (PDB ID: 2RH1), A2AAR (PDB ID: 3EML) and rhodopsin (PDB ID: 1U19). Only CXCR4 helices are shown (blue). The ligands IT1t (for CXCR4, magenta), carazolol (for β2AR, yellow), ZM241385 (for A2AAR, cyan) and retinal (for rhodopsin, green) are shown in stick representation.
In the CXCR4-3/CVX15 complex, the bulky 16-residue ligand fills most of the binding pocket volume (Fig. 2B, D, Fig. S3B & Table S5). The peptide forms a disulfide-stabilized (Cys4-Cys13) β-hairpin, with dPro8-Pro9 at the tip of the turn exposed to the extracellular milieu. The N-terminal part of the peptide backbone from Arg1 to Cys4 forms hydrogen bonds to CXCR4 backbone residues Asp187-Tyr190, adding a partial third strand to the ECL2 β-hairpin. The core specific interactions are formed by two arginines at the peptide N-terminus: Arg1 makes polar interactions with Asp187 (3.1 Å), while Arg2 interacts with Thr1173.33 (2.9 Å) and Asp1714.60 (3.0 Å) and may form an additional hydrogen bond with His1133.29 (2.9 Å) depending on its protonation state. The bulky naphthalene ring of Nal3 is anchored in a hydrophobic region bordered by helix V. Arg14 makes a salt bridge with Asp2626.58 (3.2 Å), and an intramolecular hydrogen bond to the Tyr5 side chain, which in turn makes hydrophobic contacts with helix V side chains. Finally, the C-terminal d-proline is buried in the pocket next to the N-terminus of the peptide, making a water-mediated interaction with Asp2887.39 side chain of CXCR4. The importance of the above interactions is supported by structure-activity relationship analyses of a series of CVX15 analogues (25).
The small molecule and peptide ligand binding sites significantly overlap (Fig. 3A). As CVX15 fills the entire pocket, some conformational variations between the two complexes are not surprising. CVX15 binding induces significant deviations in the base of the receptor N-terminus (residues 29–33), as well as a minor adjustment of extracellular tips of helices VI (~1Å inward), VII (~1Å tangential) and V (~0.3Å outward). Major differences, observed between binding of IT1t and CVX15 to CXCR4 compared to ligand binding modes in β2AR, A2AAR and rhodopsin (Fig. 3B), highlight the structural plasticity of GPCR binding sites.
Receptor dimerization
CXCR4 has been previously shown to homo- and hetero-dimerize, constitutively and upon ligand binding, by many different experimental methods (14–15, 33–39). While the functional importance of dimerization remains incompletely characterized, a significant body of data suggests that it has important in vivo pharmacological effects. For example, WHIM syndrome has been linked to mutations in the C-terminus of CXCR4 and results in truncated variants that exhibit enhanced signaling and fail to desensitize and internalize upon CXCL12 stimulation. As a primarily heterozygous disease in which truncated CXCR4 is co-expressed with WT receptor, dimerization has been proposed as the most likely mechanism to explain the dominance of mutant CXCR4 over the WT receptor (40–41). The structures presented here corroborate the concept of CXCR4 dimerization and define the dimer interface for a human GPCR with substantial buried surface area (850 Å2). A similar parallel, symmetric dimer of CXCR4 is observed in all five crystal forms (Fig. 4 & Fig. S4), suggesting that these contacts represent a biologically relevant homodimer interface.
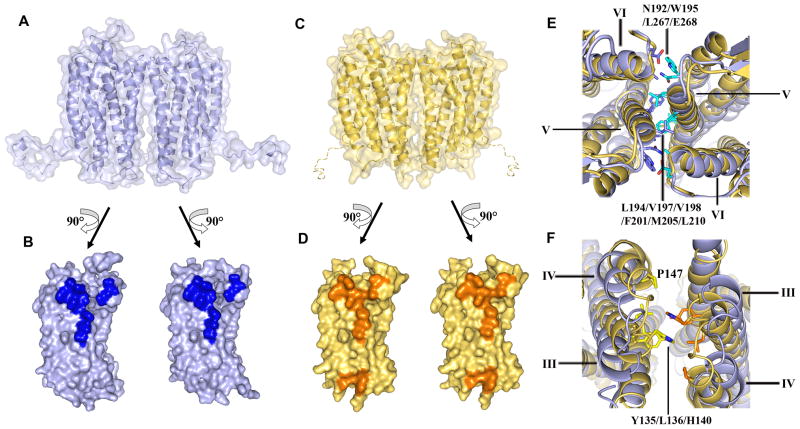
Fig. 4.
Dimer interactions in CXCR4-2/IT1t and CXCR4-3/CVX15. (A) Molecular surface representation of the CXCR4 dimer in CXCR4-2/IT1t (blue). (B) Dimer interface in CXCR4-2/IT1t. The surface involved in dimerization is highlighted in dark blue. (C) Molecular surface representation of the CXCR4 dimer in CXCR4-3/CVX15 (yellow). A hypothetical path of the C-terminus, which is not observed in the CXCR4-3/CVX15 structure, is shown as a dashed curve. (D) Dimer interface in CXCR4-3/CVX15. The surface involved in dimer interaction is highlighted in orange. (E) Top view of the extracellular side of the dimers. Two structures show similar interactions via helices V and VI. Residues of CXCR4-2/IT1t involved in the dimer interaction are shown in stick representation, and colored blue in molecule A, cyan in molecule B. (F) Bottom view of the intracellular side of the dimers. Contacts can only be observed at the intracellular tips of helices III and IV, and ICL2 in CXCR4-3/CVX15. The residues of CXCR4-3/CVX15 involved in the dimer interaction are shown in stick representation, and colored yellow and orange. These interactions are not present in the CXCR4/IT1t complex.
In dimers of CXCR4 bound to IT1t, the monomers interact only at the extracellular side of helices V and VI, leaving at least a 4 Å gap between the intracellular regions, which is presumably filled by lipids (Fig. 4A, B & Table S6). Dimer association is driven mostly by hydrophobic interactions involving Leu1945.33/Val1975.36/Val1985.37, as well as Phe2015.40-Phe2015.40, Met2055.44-Met2055.44, and Leu2105.49-Leu2105.49 contacts. A substantial role is also played by a Trp1955.34-Leu2676.63 contact, which includes both side-chain stacking and a hydrogen bond from Trp1955.34 (NE1) to the main chain carbonyl oxygen of Leu2676.63. Another specific polar interaction includes a hydrogen-bonding network between the side chains of Asn192 and Glu268 in opposing receptors, which also involves the main-chain carbonyl oxygens of Leu2666.62 and Trp1955.34. Pro191 in ECL2 likely plays a role in this network by stabilizing the Trp1955.34 side-chain conformation. As these contacts persist throughout all five crystal forms, they are likely genuine, rather than artifacts of crystallization (Fig. 4E).
In addition, dimers of CXCR4 bound to CVX15 are stabilized by interactions at the intracellular ends of helices III and IV, and ICL2, controlled largely by hydrophobic interactions of Tyr1353.51, Leu1363.52, His140 and Pro147 side chains (~400 Å2 buried) (Fig. 4C, D, F & Table S6). It appears that binding of the bulky CVX15 peptide induces a small tilt in the extracellular part of helix V, which brings the intracellular parts of opposing receptors into close contact. This type of ligand-induced conformational change could explain the cooperative binding observed with certain CXCR4 ligands, as well as the effects of allosteric modulators. Specifically, binding of a ligand to one receptor could induce a structural change in helix V of the second receptor, thereby modifying the ligand binding affinity to the second receptor, resulting in either negative or positive cooperativity. Extending this concept to chemokine receptor heterodimers, CXCR4 has been reported to dimerize with CCR2 and CCR5 and both complexes show negative binding cooperativity with their ligands not only in vitro but also in vivo (36, 38), an observation which may have significant implications for drug efficacy.
The CXCR4 dimer is strikingly different from previous models of GPCR dimerization, which suggested contacts through either helix I or helices IV/V (42–46) and implied contacts throughout the length of the TM bundle. It is also notable that with the exception of Trp1955.34 (conservation ~70%), little sequence conservation is found among chemokine receptors for the residues that constitute the dimerization site, even though many receptors have been shown to oligomerize (39). The specific nature of the interactions may facilitate the ability of CXCR4 to heterodimerize with other chemokine receptors (36, 38, 47) as well as GPCRs outside of the chemokine family (48), although one cannot discount the possibility that many modes of oligomerization may exist.
Implications for the two-site model of chemokine binding and complexes of CXCR4 with CXCL12 and gp120
The known structures of chemokines, including CXCL12, feature a disordered N-terminal domain that largely controls receptor signaling and is hypothesized to penetrate the receptor helical bundle (49–50). The chemokine N-terminus is followed by a core globular domain, which is thought to bind to the receptor N-terminus and ECLs, forming an interaction site that confers affinity and specificity (51). The separation of the binding and signaling functions has led to the so-called “two-site” model of receptor binding with the chemokine core domain being the “site one” docking domain and the chemokine N-terminus being the “site two” signaling trigger (49, 52–53). The NMR structure of CXCL12 complexed to a 38-residue, sulfotyrosine-containing peptide derived from the CXCR4 N-terminus has been determined (PDB ID: 2K05) (54). This structure is thought to represent at least part of the “site one” complex and reveals important interactions between CXCL12 and residues that are absent from the CXCR4 receptor structure, including three sulfated tyrosines.
The peptide and small molecule complexes of CXCR4 identify the likely “site two” of the chemokine signaling trigger. The IT1t compound and CVX15 peptide have both been characterized as competitive inhibitors of CXCL12, and many of the receptor-ligand contacts in the co-crystal structures presented are important for CXCL12 binding, including the acidic Asp187, Glu2887.39 and Asp972.63 (Fig. 2) (55–56). The CVX15 peptide, rich in basic residues, may trace to some extent the path of the N-terminal signaling peptide of CXCL12 (KPVSLSYR), and the binding site of IT1t may point to the major anchor region for this domain. Furthermore, our preliminary modeling studies suggest that Lys1, the most critical residue in CXCL12 for receptor activation, could reach into the CXCR4 pocket and interact with one of these acidic residues (Fig. 5). The extensive binding site mapped out by the CVX15 peptide also clarifies how progressive shortening of the CXCL12 N-terminus leads to a gradual loss of binding affinity (49). Taken together, these data suggest that the small molecule and cyclic peptide block ligand binding by acting as orthosteric competitors of the CXCL12 N-terminal signaling trigger, providing strong support for the two-site model of binding. Along these lines, a recent NMR study showed that the CXCR4 antagonist AMD3100 could displace the CXCL12 N-terminus from the receptor without displacing the chemokine core domain (57).
Fig. 5.
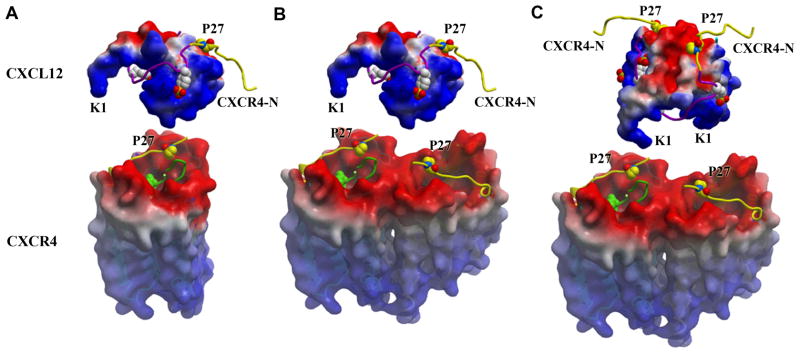
Stoichiometry of possible CXCR4/CXCL12 binding/signaling complexes. No information on the orientation of CXCL12 with respect to CXCR4 is implied from the models presented. (A) Monomeric CXCR4 binding monomeric CXCL12, (B) dimeric CXCR4 binding monomeric CXCL12, (C) dimeric CXCR4 binding dimeric CXCL12 at either one or both orthosteric sites on each protomer. Alternatively, the 2:2 complex could involve two CXCL12 monomers binding dimeric CXCR4 (not shown). Both CXCR4 and CXCL12 surfaces are colored according to their electrostatic potential from red (negative) to blue (positive), highlighting the charge complementarity of these proteins. The portion of the CXCR4 N-terminal domain (CXCR4-N) present in both the CXCL12 complex (PDB ID: 2K05) and crystal structures of this study is colored yellow, while the remainder is purple (Site 1). Pro27 and the three sulfotyrosines from the CXCR4 N-terminus are represented with space-filling models. The CVX15 peptide (green ribbon) is shown in one CXCR4 receptor per panel and suggests the binding site for Lys1 and the rest of the flexible N-terminal region of CXCL12, which is critical for receptor activation (Site 2). Figures were prepared using ICM software (www.Molsoft.com).
Chemokines are able to bind their receptors as monomers in order to activate cell migration (58). However, chemokine oligomers, including CXCL12, appear to be functional and induce alternative signaling responses such as cellular activation or signals to halt migration (54, 59–60) giving rise to the concept that these complexes dynamically change their stoichiometries and structures as part of their functional regulation. Given the oligomeric nature of CXCR4 and the complementary electrostatic surfaces of the ligand and receptor, one can envision CXCL12 binding the receptor as a 1:1, 1:2, or 2:2 ligand:receptor complex (Fig. 5). Additional experiments will be necessary to fully define the relevance and functional implications of different chemokine:receptor stoichiometries and structures. Nevertheless, the current CXCR4 structures are compatible with emerging concepts of signaling diversity induced by alternative binding modes of the ligands.
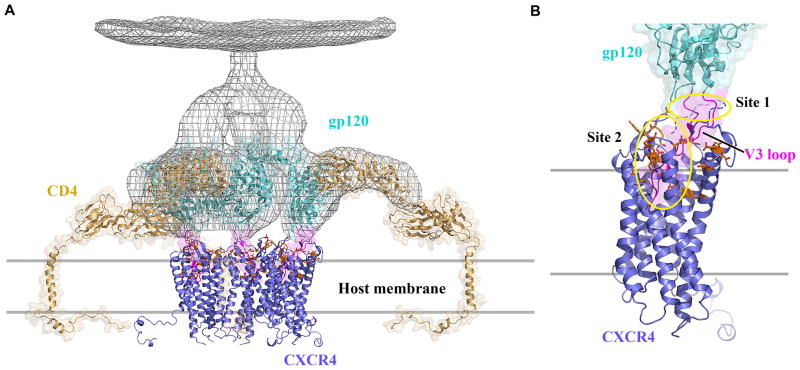
CXCR4 and the related CCR5 serve as co-receptors for HIV-1 viral particles, facilitating their entry into cells. Structures have been reported for the other key components of the entry complex, HIV-1 glycoproteins gp120 and gp41, and the host leukocyte glycoprotein receptor CD4 (61–63). The N-termini of CXCR4 and CCR5, including sulfated tyrosine residues, have been implicated in gp120 binding, analogous to CXCL12 recognition (64). Other structural features critical to the interaction involve the gp120 V3 loop, which becomes exposed on CD4 binding (65), and then interacts with CXCR4 ECL2 and ECL3. The basic character of the protruding V3 loop along with acidic residues in the CXCR4 binding pocket have been reported to be important for HIV-1 infectivity (Fig. 2C & D) (56, 66), suggesting that the loop could also penetrate the pocket (Fig. 6). Thus, the CXCR4 structures suggest testable hypotheses regarding interaction of CXCR4 with its natural ligand and with HIV-1 gp120. The real challenge will be in understanding the dynamic changes in these complexes that result in signal transduction and viral fusion. As further details of these interactions are resolved, new opportunities for drug discovery efforts targeting specific functional states of the receptor will emerge.
Fig. 6.
Model of early stages of the HIV-1 entry process. (A) Viral entry begins with binding of envelope spikes consisting of a heterotrimer (gp120)3 (gp41)3 (gray wire, EM databank ID: emd_5020 and emd_5023; PDB ID: 3DNO) to CD4 on the surface of host target cells. Glycoprotein gp120 (core structure, cyan, PDB ID: 2QAD) interacts with CD4 (tan, PDB ID: 1WIP and 2KLU). This interaction triggers conformational changes in gp120 that increase the exposure of the third variable loop V3 (magenta) and a region of gp120 between inner and outer domains. CCR5 or CXCR4 (blue) is then recruited as a co-receptor. The number of spikes involved in viral entry and the number of molecules of CD4 or CXCR4 binding to a single spike are unknown; here three CD4 molecules are represented, which results in the close approach of gp120 molecules to the host cell membrane where the interaction with three CXCR4 molecules is depicted. (B) By analogy to a two-site model based on CCR5 (64), the N-terminus of CXCR4 containing sulfotyrosines (site1, circled in yellow) binds first to the base of the V3 loop inducing further conformational changes in gp120 that enable V3 to bind to the extracellular side of CXCR4, primarily ECL2, ECL3 and the ligand-binding cavity (site 2, circled in yellow). CXCR4 residues previously shown to affect gp120 binding are shown as sticks with carbons colored in orange. A hypothetical path of the CXCR4 N-terminus, which is not observed in the current structure, is shown as a blue dashed curve. Only CXCR4 monomers are shown for clarity, although dimers are also possible. Fig. 1, 2, 3, 4 and 6 were prepared using PyMOL.
Supplementary Material
Acknowledgments
This work was supported in part by the Protein Structure Initiative grant U54 GM074961 for structure production, NIH Roadmap Initiative grant P50 GM073197 for technology development, NIH grants R21 RR02536 and R21 AI087189 to VC, and Pfizer. Additionally, TMH acknowledges support from National Institutes of Health R01 AI037113 and R01 GM081763 and DJH from F32 GM083463. The authors thank Jeffrey Velasquez for help on molecular biology, Tam Trinh and Kirk Allin for help on baculovirus expression, Ian Wilson and Dennis Burton for careful review and scientific feedback on the manuscript, Bill Schief for electron miscroscopy models of gp120/gp41/CD4 complex, Gye Won Han for evaluating the structure quality and preparation for PDB submission, and Angela Walker for assistance with manuscript preparation. The authors acknowledge Erik La Chapelle on chemistry tool compound synthesis, Anil Rane on radiolabelling of [3H]BIMA, and Mei Cui for help developing [3H]BIMA binding assay; Yuan Zheng, The Ohio State University, and Martin Caffrey, Trinity College (Dublin, Ireland), for the generous loan of the in meso robot (built with support from the National Institutes of Health [GM075915], the National Science Foundation [IIS0308078], and Science Foundation Ireland [02-IN1-B266]); and Janet Smith, Robert Fischetti and Nukri Sanishvili at the GM/CA-CAT beamline at the Advanced Photon Source, for assistance in development and use of the minibeam and beamtime. The GM/CA-CAT beamline (23-ID) is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Atomic coordinates and structure factors have been deposited in the Protein Data Bank with identification codes 3ODU (CXCR4-2/IT1t, P21), 3OE0 (CXCR4-3/CVX15, C2), 3OE8 (CXCR4-2/IT1t, P1), 3OE9 (CXCR4-3/IT1t, P1), 3OE6 (CXCR4-1/IT1t, I222).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Materials and Methods
Reference
References and Notes
- 1.Baggiolini M. Nature. 1998;392:565. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 2.Moser B, Wolf M, Walz A, Loetscher P. Trends Immunol. 2004;25:75. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Mackay CR. Nat Immunol. 2001;2:95. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- 4.Ma Q, et al. Proc Natl Acad Sci U S A. 1998;95:9448. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Nature. 1998;393:595. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 6.Muller A, et al. Nature. 2001;410:50. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 7.Balkwill F. Semin Cancer Biol. 2004;14:171. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Zlotnik A. J Pathol. 2008;215:211. doi: 10.1002/path.2350. [DOI] [PubMed] [Google Scholar]
- 9.Fulton AM. Curr Oncol Rep. 2009;11:125. doi: 10.1007/s11912-009-0019-1. [DOI] [PubMed] [Google Scholar]
- 10.Teicher BA, Fricker SP. Clin Cancer Res. 2010;16:2927. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Broder CC, Kennedy PE, Berger EA. Science. 1996;272:872. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 12.Bleul CC, et al. Nature. 1996;382:829. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 13.Oberlin E, et al. Nature. 1996;382:833. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 14.Babcock GJ, Farzan M, Sodroski J. J Biol Chem. 2003;278:3378. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- 15.Percherancier Y, et al. J Biol Chem. 2005;280:9895. doi: 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Norcross M. Drug Discov Today. 2008;13:625. doi: 10.1016/j.drudis.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Cherezov V, et al. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaakola VP, et al. Science. 2008;322:1211. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warne T, et al. Nature. 2008;454:486. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Nature. 2008;454:183. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 21.Hanson MA, et al. Structure. 2008;16:897. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth CB, Hanson MA, Stevens RC. J Mol Biol. 2008;376:1305. doi: 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.In Ballesteros-Weinstein numbering, a single most conserved residue among the class A GPCRs is designated x.50, where x is the transmembrane helix number. All other residues on that helix are numbered relative to this conserved position.
- 24.Thoma G, et al. J Med Chem. 2008;51:7915. doi: 10.1021/jm801065q. [DOI] [PubMed] [Google Scholar]
- 25.Tamamura H, et al. FEBS Lett. 2003;550:79. doi: 10.1016/s0014-5793(03)00824-x. [DOI] [PubMed] [Google Scholar]
- 26.DeMarco SJ, et al. Bioorg Med Chem. 2006;14:8396. doi: 10.1016/j.bmc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Moncunill G, et al. Mol Pharmacol. 2008;73:1264. doi: 10.1124/mol.107.042911. [DOI] [PubMed] [Google Scholar]
- 28.Materials and methods are available as supporting material on Science Online.
- 29.Zhou H, Tai HH. Arch Biochem Biophys. 2000;373:211. doi: 10.1006/abbi.1999.1555. [DOI] [PubMed] [Google Scholar]
- 30.Scheerer P, et al. Nature. 2008;455:497. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann KP, et al. Trends Biochem Sci. 2009;34:540. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Experimental measurement of pKa via Multiplexed Capillary Electrophoresis gave pKa1=5.39, pKa2=7.89 and pKa3=9.92, indicating that the conjugated acid of isothiourea N4 has a pKa of 5.4, which is also consistent with the predication (pKa=4.84±0.60) by pKa database from ACD Labs; and suggesting that N4 is unprotonated at the physiological condition of pH=7.4. Crystallization of the CXCR4-2/IT1t complex was performed at pH 5.5 and the acidic microenvironment of Glu2887.39 may further shift the equilibrium towards protonation.
- 33.Vila-Coro AJ, et al. FASEB J. 1999;13:1699. [PubMed] [Google Scholar]
- 34.Luker KE, Gupta M, Luker GD. FASEB J. 2009;23:823. doi: 10.1096/fj.08-116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. Blood. 2009;113:6085. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- 36.Sohy D, et al. J Biol Chem. 2009;284:31270. doi: 10.1074/jbc.M109.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, He L, Combs CA, Roderiquez G, Norcross MA. Mol Cancer Ther. 2006;5:2474. doi: 10.1158/1535-7163.MCT-05-0261. [DOI] [PubMed] [Google Scholar]
- 38.Sohy D, Parmentier M, Springael JY. J Biol Chem. 2007;282:30062. doi: 10.1074/jbc.M705302200. [DOI] [PubMed] [Google Scholar]
- 39.Salanga CL, O’Hayre M, Handel T. Cell Mol Life Sci. 2009;66:1370. doi: 10.1007/s00018-008-8666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balabanian K, et al. Blood. 2005;105:2449. doi: 10.1182/blood-2004-06-2289. [DOI] [PubMed] [Google Scholar]
- 41.Lagane B, et al. Blood. 2008;112:34. doi: 10.1182/blood-2007-07-102103. [DOI] [PubMed] [Google Scholar]
- 42.Fanelli F, De Benedetti PG. J Comput Aided Mol Des. 2006;20:449. doi: 10.1007/s10822-006-9064-0. [DOI] [PubMed] [Google Scholar]
- 43.Filizola M. Life Sci. 2010;86:590. doi: 10.1016/j.lfs.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemoto W, Toh H. Curr Protein Pept Sci. 2006;7:561. doi: 10.2174/138920306779025657. [DOI] [PubMed] [Google Scholar]
- 45.Reggio PH. AAPS J. 2006;8:E322. doi: 10.1007/BF02854903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vohra S, et al. Biochem Soc Trans. 2007;35:749. doi: 10.1042/BST0350749. [DOI] [PubMed] [Google Scholar]
- 47.Isik N, Hereld D, Jin T. PLoS One. 2008;3:e3424. doi: 10.1371/journal.pone.0003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oscar† MP, et al. Eur J Immunol. 2008;38:537. [Google Scholar]
- 49.Crump MP, et al. EMBO J. 1997;16:6996. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dealwis C, et al. Proc Natl Acad Sci U S A. 1998;95:6941. doi: 10.1073/pnas.95.12.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta SK, Pillarisetti K, Thomas RA, Aiyar N. Immunol Lett. 2001;78:29. doi: 10.1016/s0165-2478(01)00228-0. [DOI] [PubMed] [Google Scholar]
- 52.Clark-Lewis I, et al. J Leukoc Biol. 1995;57:703. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 53.Wells TN, et al. J Leukoc Biol. 1996;59:53. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 54.Veldkamp CT, et al. Sci Signal. 2008;1:ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brelot A, et al. J Virol. 1999;73:2576. doi: 10.1128/jvi.73.4.2576-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brelot A, Heveker N, Montes M, Alizon M. J Biol Chem. 2000;275:23736. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 57.Kofuku Y, et al. J Biol Chem. 2009;284:35240. doi: 10.1074/jbc.M109.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paavola CD, et al. J Biol Chem. 1998;273:33157. doi: 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- 59.Appay V, Brown A, Cribbes S, Randle E, Czaplewski LG. J Biol Chem. 1999;274:27505. doi: 10.1074/jbc.274.39.27505. [DOI] [PubMed] [Google Scholar]
- 60.Czaplewski LG, et al. J Biol Chem. 1999;274:16077. doi: 10.1074/jbc.274.23.16077. [DOI] [PubMed] [Google Scholar]
- 61.Diskin R, Marcovecchio PM, Bjorkman PJ. Nat Struct Mol Biol. 2010;17:608. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pancera M, et al. Proc Natl Acad Sci U S A. 2010;107:1166. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwong PD, et al. Nature. 1998;393:648. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang CC, et al. Science. 2007;317:1930. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang CC, et al. Science. 2005;310:1025. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doranz BJ, et al. J Virol. 1999;73:2752. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Angel TE, Chance MR, Palczewski K. Proc Natl Acad Sci U S A. 2009;106:8555. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian S, et al. J Virol. 2005;79:12667. doi: 10.1128/JVI.79.20.12667-12673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.