Abstract
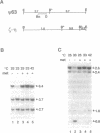
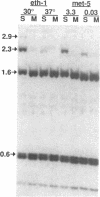
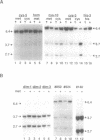
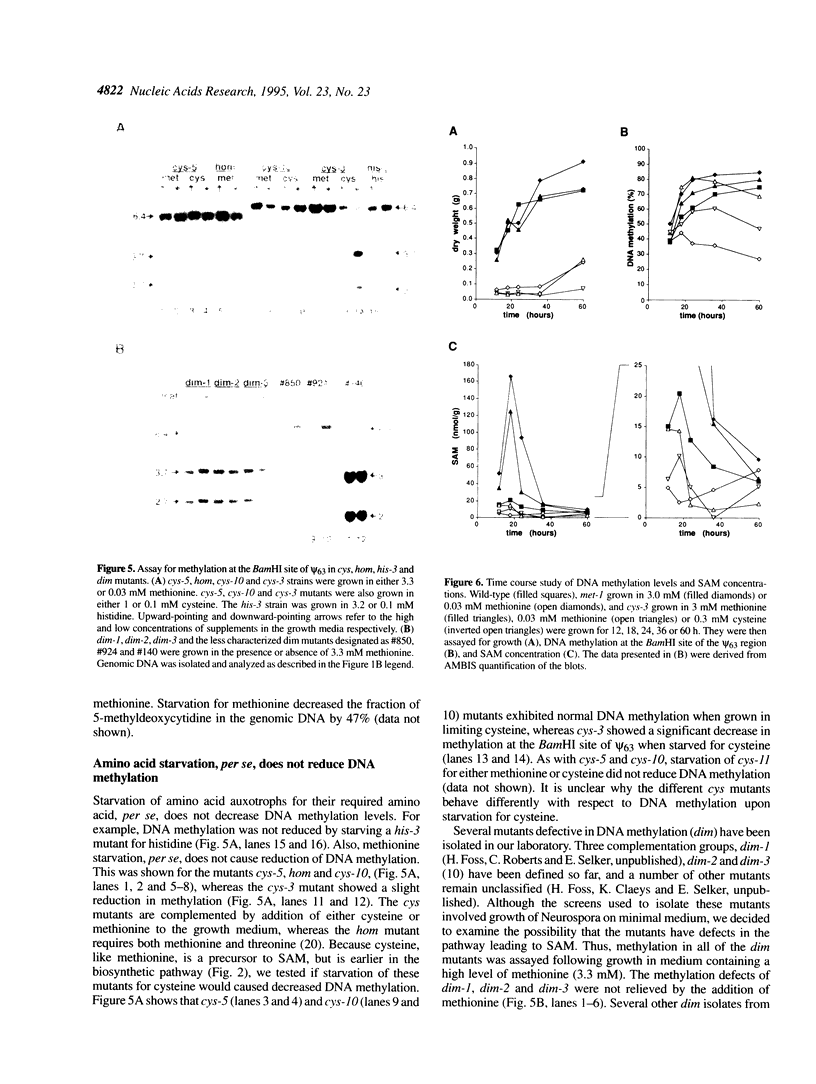
A temperature-sensitive methionine auxotroph of Neurospora crassa was found in a collection of conditional mutants and shown to be deficient in DNA methylation when grown under semipermissive conditions. The defective gene was identified as met-3, which encodes cystathionine-gamma-synthase. We explored the possibility that the methylation defect results from deficiency of S-adenosylmethionine (SAM), the presumptive methyl group donor. Methionine starvation of mutants from each of nine complementation groups in the methionine (met) pathway (met-1, met-2, met-3, met-5, met-6, met-8, met-9, met-10 and for) resulted in decreased DNA methylation while amino acid starvation, per se, did not. In most of the strains, including wild-type, intracellular SAM peaked during rapid growth (12-18 h after inoculation), whereas DNA methylation continued to increase. In met mutants starved for methionine, SAM levels were most reduced (3-11-fold) during rapid growth while the greatest reduction in DNA methylation levels occurred later. Addition of 3 mM methionine to cultures of met or cysteine-requiring (cys) mutants resulted in 5-28-fold increases in SAM, compared with wild-type, at a time when DNA methylation was reduced approximately 40%, suggesting that the decreased methylation during rapid growth in Neurospora is not due to limiting SAM. DNA methylation continued to increase in a cys-3 mutant that had stopped growing due to methionine starvation, suggesting that methylation is not obligatorily coupled to DNA replication in Neurospora.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barry C., Faugeron G., Rossignol J. L. Methylation induced premeiotically in Ascobolus: coextension with DNA repeat lengths and effect on transcript elongation. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4557–4561. doi: 10.1073/pnas.90.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Burton E. G., Metzenberg R. L. Regulation of methionine biosythesis in Neurospora crassa. Arch Biochem Biophys. 1975 May;168(1):219–229. doi: 10.1016/0003-9861(75)90244-1. [DOI] [PubMed] [Google Scholar]
- Culp L. A., Black P. H. DNA synthesis in normal and virus-transformed mammalian cells after methionine deprivation. Biochim Biophys Acta. 1971 Oct 14;247(2):220–232. doi: 10.1016/0005-2787(71)90672-1. [DOI] [PubMed] [Google Scholar]
- Eden S., Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994 Apr;4(2):255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- Evans H. H., Evans T. E., Littman S. Methylation of parental and progeny DNA strands in Physarum polycephalum. J Mol Biol. 1973 Mar 15;74(4):563–572. doi: 10.1016/0022-2836(73)90047-8. [DOI] [PubMed] [Google Scholar]
- Evans H. H., Evans T. E. Methylation of the deoxyribonucleic acid of Physarum polycephalum at various periods during the mitotic cycle. J Biol Chem. 1970 Dec 10;245(23):6436–6441. [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Giordano M., Mattachini M. E., Cella R., Pedrali-Noy G. Purification and properties of a novel DNA methyltransferase from cultured rice cells. Biochem Biophys Res Commun. 1991 Jun 14;177(2):711–719. doi: 10.1016/0006-291x(91)91846-5. [DOI] [PubMed] [Google Scholar]
- Greene R. C., Hunter J. S., Coch E. H. Properties of metK mutants of Escherichia coli K-12. J Bacteriol. 1973 Jul;115(1):57–67. doi: 10.1128/jb.115.1.57-67.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: two DNA methyltransferases with BsuRI specificity. II. Catalytic properties, substrate specificity, and mode of action. J Biol Chem. 1981 Sep 10;256(17):9346–9351. [PubMed] [Google Scholar]
- HAROLD F. M. Accumulation of inorganic polyphosphate in mutants of Neurospora crassa. Biochim Biophys Acta. 1960 Dec 4;45:172–188. doi: 10.1016/0006-3002(60)91438-4. [DOI] [PubMed] [Google Scholar]
- Hoffman D., Kumar A. M., Spitzer A., Gupta R. K. NMR measurement of intracellular water volume in rat kidney proximal tubules. Biochim Biophys Acta. 1986 Dec 19;889(3):355–360. doi: 10.1016/0167-4889(86)90198-9. [DOI] [PubMed] [Google Scholar]
- Hoffman R. M. Unbalanced transmethylation and the perturbation of the differentiated state leading to cancer. Bioessays. 1990 Apr;12(4):163–166. doi: 10.1002/bies.950120404. [DOI] [PubMed] [Google Scholar]
- Jacobson E. S., Chen G. S., Metzenberg R. L. Unstable S-Adenosylmethionine synthetase in an ethionine-resistant strain of Neurospora crassa. J Bacteriol. 1977 Nov;132(2):747–748. doi: 10.1128/jb.132.2.747-748.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J. A., Richards E. J. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 1995 Jan 11;23(1):130–137. doi: 10.1093/nar/23.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr D. S., Flavin M. The regulation of methionine synthesis and the nature of cystathionine gamma-synthase in Neurospora. J Biol Chem. 1970 Apr 10;245(7):1842–1855. [PubMed] [Google Scholar]
- Koujima I., Hayashi H., Tomochika K., Okabe A., Kanemasa Y. Adaptational change in proline and water content of Staphylococcus aureus after alteration of environmental salt concentration. Appl Environ Microbiol. 1978 Mar;35(3):467–470. doi: 10.1128/aem.35.3.467-470.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Metzenberg R. L., Stevens J. N., Selker E. U., Morzycka-Wroblewska E. Identification and chromosomal distribution of 5S rRNA genes in Neurospora crassa. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2067–2071. doi: 10.1073/pnas.82.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Raschke E., McClelland M. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1993 Jul 1;21(13):3139–3154. doi: 10.1093/nar/21.13.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., Metzenberg R. L., Raju N. B., Selker E. U., Barry E. G. Reversal of a Neurospora translocation by crossing over involving displaced rDNA, and methylation of the rDNA segments that result from recombination. Genetics. 1986 Nov;114(3):791–817. doi: 10.1093/genetics/114.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., Newmeyer D., Björkman M. Chromosomal loci of Neurospora crassa. Microbiol Rev. 1982 Dec;46(4):426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977 Oct 25;252(20):7265–7272. [PubMed] [Google Scholar]
- Russell P. J., Rodland K. D., Rachlin E. M., McCloskey J. A. Differential DNA methylation during the vegetative life cycle of Neurospora crassa. J Bacteriol. 1987 Jun;169(6):2902–2905. doi: 10.1128/jb.169.6.2902-2905.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwencke J., De Robichon-Szulmajster H. The transport of S-adenosyl-L-methionine in isolated yeast vacuoles and spheroplasts. Eur J Biochem. 1976 May 17;65(1):49–60. doi: 10.1111/j.1432-1033.1976.tb10388.x. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Fritz D. Y., Singer M. J. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993 Dec 10;262(5140):1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Garrett P. W. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Jensen B. C., Richardson G. A. A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science. 1987 Oct 2;238(4823):48–53. doi: 10.1126/science.2958937. [DOI] [PubMed] [Google Scholar]
- Selker E. U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Stevens J. N. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Stevens J. N. Signal for DNA methylation associated with tandem duplication in Neurospora crassa. Mol Cell Biol. 1987 Mar;7(3):1032–1038. doi: 10.1128/mcb.7.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J. C., Rideout W. M., 3rd, Jones P. A. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992 Dec 24;71(7):1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- Theiss G., Schleicher R., Schimpff-Weiland G., Follmann H. DNA methylation in wheat. Purification and properties of DNA methyltransferase. Eur J Biochem. 1987 Aug 17;167(1):89–96. doi: 10.1111/j.1432-1033.1987.tb13307.x. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R. A., Richards E. J. Arabidopsis thaliana DNA methylation mutants. Science. 1993 Jun 25;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wainfan E., Dizik M., Stender M., Christman J. K. Rapid appearance of hypomethylated DNA in livers of rats fed cancer-promoting, methyl-deficient diets. Cancer Res. 1989 Aug 1;49(15):4094–4097. [PubMed] [Google Scholar]
- Wheatley D. N., Inglis M. S., Foster M. A., Rimington J. E. Hydration, volume changes and nuclear magnetic resonance proton relaxation times of HeLa S-3 cells in M-phase and the subsequent cell cycle. J Cell Sci. 1987 Aug;88(Pt 1):13–23. doi: 10.1242/jcs.88.1.13. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Wyszynski M., Gabbara S., Bhagwat A. S. Cytosine deaminations catalyzed by DNA cytosine methyltransferases are unlikely to be the major cause of mutational hot spots at sites of cytosine methylation in Escherichia coli. Proc Natl Acad Sci U S A. 1994 Feb 15;91(4):1574–1578. doi: 10.1073/pnas.91.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P. H. Determination of tissue S-adenosylmethionine by radioenzymatic assay. Methods Enzymol. 1983;94:66–69. doi: 10.1016/s0076-6879(83)94011-9. [DOI] [PubMed] [Google Scholar]
- Yu P. H. Radioenzymatic estimation of S-adenosylmethionine in rat brain regions and subcellular fractions. Anal Biochem. 1978 Jun 1;86(2):498–504. doi: 10.1016/0003-2697(78)90774-1. [DOI] [PubMed] [Google Scholar]