Abstract
Steroid hormone receptors are members of a family of ligand inducible transcription factors, and regulate the transcriptional activation of target genes by recruiting coregulatory proteins to the pre-initiation machinery. The binding of these coregulatory proteins to the steroid hormone receptors is often mediated through their two activation functional domains, AF1, which resides in the N-terminal domain, and the ligand-dependent AF2, which is localized in the C-terminal ligand binding domain. Compared to other important functional domains of the steroid hormone receptors, our understanding of the mechanisms of action of the AF1 are incomplete, in part, due to the fact that, in solution, AF1 is intrinsically disordered (ID). However, recent studies have shown that AF1 must adopt a functionally active and folded conformation for its optimal activity under physiological conditions. In this review, we summarize and discuss current knowledge regarding the molecular mechanisms of AF1-mediated gene activation, focusing on AF1 conformation and coactivator binding. We further propose models for the binding/folding of the AF1 domains of the steroid hormone receptors and their protein-protein interactions. The population of ID AF1 can be visualized as a collection of many different conformations, some of which may be assuming the proper functional folding for other critical target binding partners that result in ultimate assembly of AF1:coactivator complexes and subsequent gene regulation. Knowledge of the mechanisms involved therein will significantly help in understanding how signals from a steroid to a specific target gene are conveyed.
Keywords: steroid hormone receptors, intrinsically disordered protein, activation function, protein:protein interactions, protein folding
Introduction
Intracellular transcription factors, such as the nuclear hormone receptors (NHRs) play many important biological roles in eukaryotic development, differentiation, reproduction and metabolic homeostasis among others (1–5). Proteins of the NHR super-family possess a modular structure with three major functional domains: i) a variable amino-terminal domain (NTD), ii) a highly conserved DNA-binding domain (DBD), and iii) a less conserved carboxyl-terminal ligand binding domain (LBD) (6–8). The NHR superfamily consists of receptors for steroids (glucocorticoid, mineralocorticoid, progesterone, estrogen, and androgen), thyroid/retinoid/vitamin D/peroxisome proliferator-activated receptor (PPAR), and the orphan receptors (for which ligands have not been identified) families (9,10). The classic mechanism of steroid action, based on the domain model, is that the ligand bound-receptor enters the nucleus where it binds to its response element DNA site that allows for the formation of a transcription initiation complex either directly or indirectly through subsequent binding to co-regulatory proteins (11–13). To a large extent, the composition of the receptor:coregulator complex assembly determines the final outcome of the target gene regulation by the receptor (14,15). Depending upon the interaction with coactivators or corepressors, NHRs up- or down-regulate the expression of specific gene(s) (14,15). Though the structural organization of the NHRs into NTD, DBD, and LBD regions is well characterized (Figure1), the regulation of transcription is poorly understood. This is due largely to the lack of detailed information concerning the function of their transcription activating domains (16–20). Two major transactivation domains of the NHRs are AF1 in the NTD, which acts in a constitutive manner, and a small but important AF2 sub-domain towards the C-terminal end, which functions in a ligand-dependent manner (16,17,21,22).
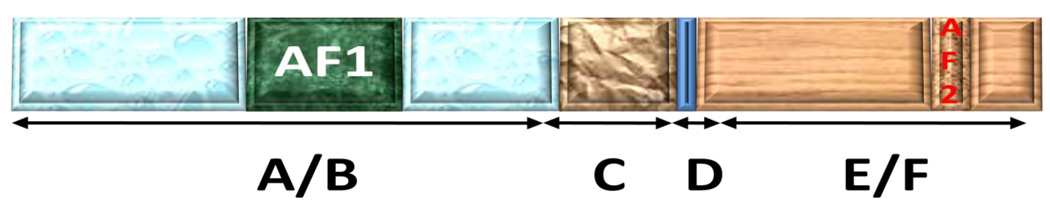
Figure 1. A general schematic representation of a SR showing functional domain arrangements.
A/B = N-terminal domain (NTD); C = DNA-binding domain (DBD); D = hinge region (in some cases also known as Tau2); E/F = ligand binding domain (LBD). Not all SRs contain F domain, and this is not required for hormone binding. Precise location and size of each domain may differ in each SR. These differences are most pronounced in the A/B region. A typical SR is composed of several functional domains. The variable N-terminal region (A/B) contains the ligand-independent AF1 transactivation domain, which can constitutively activate transcriptional activity of the SR lacking the LBD. The precise location and size of AF1 is different in each SR, and is shown in Table 1. The conserved DBD is responsible for the recognition and binding of site-specific DNA sequences. A variable linker region D connects the DBD to the conserved LBD as well as the dimerization surface. LBD is also the site for the interaction of various heat shock and other chaperone proteins for the un-liganded receptor. The ligand-independent transcriptional activation AF1 domain is contained within the A/B region, and the ligand-dependent AF2 core transactivation sub-domain is within the C-terminal portion of the receptor. Most of the known coregulatory proteins interact with these two AF domains. Conformational alterations in these regions play an important role in the recruitment of proper and efficient binding partners under physiological conditions. Except for the NTD, the three-dimensional structure of both DBD and LBD is known for several SRs.
While the two AFs can clearly act autonomously, there is increasing evidence that full receptor activity requires synergistic, perhaps even physical interactions between AF1 and AF2 (17,18,22–24). In many cases, synergistic effects between AF1 and AF2 are mediated by other coregulatory proteins that may directly interact with both AF1 and AF2, albeit with different regions of the binding partner protein (23,24). Usually AF2 interacts with the LXXLL motif of the binding protein whereas no consensus motif has been identified for AF1 (17,25,26). Though, it is believed that the AF1 interacting sites are outside the LXXLL motifs, it is still not known whether there is a consensus motif for AF1 interacting proteins, like those for AF2. To a large extent, the transactivation activities of these AFs have differential cell type- and promoter-specific effects, suggesting the possibility of selective interactions with differentially expressed coregulatory proteins (26–28). Due to the growing number of proteins identified that interact with NHRs, much attention has been focused on the role of protein:protein interactions in gene regulation by NHRs. In this context AF1 and AF2 domains, which physically and functionally interact with certain specific coregulators, are of immense importance. Although a systematic analysis of all possible NHR:cofactor interactions is still lacking, it is evident already that each NHR can interact with several different coregulatory proteins (29,30). Moreover, due to varied expression patterns, many cell types contain an assortment of different factors that can interact with a single receptor (31,32). It seems likely that the precise cofactor assembly, based on relative affinities and concentrations, and the NHR:cofactor ratios largely define the final transcriptional potential of each receptor in a given cell (25,33–36).
Three-dimensional structures of the DBD and LBD of several NHRs are known (37–48). The DBD solution structure shows a sequence of helices and loops folded to an overall globular shape (44–48).The crystal structure of NHR’s LBD consists of three sets of anti-parallel helices that form the sides and top of the globule, making a central pocket, the ligand-binding site (37–43). Mutational analyses have shown that the AF2 sub-domain is defined by amino acids within the “so called helix-12” (37). This sub-domain is believed to change its position upon ligand binding (37). For example, the crystal structure of the estrogen receptor LBD revealed that agonist-bound receptor changes the position of H12 so that the surface formed thereby allows interaction with coactivators, whereas antagonist-bound LBD fails to do so (37). This structural elucidation of LBD availed a great deal of information concerning how AF2 functions, and in explaining how signals are passed from a ligand to the specific target gene, but it fails to explain certain critical issues such as the importance of the AF1 domain in the process. The recently published crystal structure of the full length PPAR threw some light on the structural elements of the holo-receptor, however, the NTD structure was not clearly resolved and it appeared to be largely unstructured or intrinsically disordered (49). In spite of the discovery that AF1 is a major transactivation domain for several steroid receptors (SRs), not much is known about its structure and functions. A comprehensive understanding of AF1 actions is therefore required that can provide the structure:function relationship of the AF1, vital information to understanding how GR transmits transcriptional signals to regulate expression of a specific gene(s). Because AF1 exists in an ID state, the understanding of its structure and functions has languished (50–58). In terms of sequence homology and size, the NHRs’ NTD is the region least conserved among the family members, yet it possess powerful transactivation activity in those members with a long NTD sequence. This is particularly true for several steroid receptors (SRs) (5). It is known that AF1 interacts with other transcription factors, and the available data strongly suggest that conditional folding of AF1 is the key for these interactions and subsequent transcriptional activity (25,52,53,58–60). How and what kind of functional folded conformation AF1 adopts is an open question. In this review article, we have made an attempt to describe updated knowledge of our understanding regarding the SRs’ AF1s interaction with various coregulatory proteins, and the structural and functional consequences of such protein:protein interactions in regulating the transactivation activity of the AF1.
Steroid receptors contain intrinsically disordered (ID) regions in their N-terminal/AF1 domains
In recent years, it has become evident that eukaryotic genomes are highly enriched in ID proteins compared to prokaryotes, and seem to promote molecular recognition through their ability to form surfaces capable of binding specific macromolecular binding partners (61–67). Dunker and colleagues developed a method “predictor of natural disorder regions (PONDR)” to detect ID regions/domains within proteins and reported that extended regions of ID sequences are prevalent in a majority of transcription factor proteins, including several NHRs (68). In many transcription factor proteins, these ID regions are located in their activation domains, e.g. SRs’ AF1, suggesting that these activation domains may need to be highly flexible in order to efficiently carry out their functions (69–80). Their analyses established that the combination of low mean hydrophobicity and relatively high net charge, leading to ID protein/peptide, represents an important prerequisite for the lack of well defined compact structure in proteins or protein regions/domains under physiological conditions (69–72). Based on these physico-chemical analyses, several NHRs’ N-terminal activation domains may qualify to be ID regions. Characterization of the conformational propensities and function of such non-globular protein sequences, including those in SRs, represents a major challenge. The primary amino acid sequences of the NTDs of the SRs, which contain AF1, are much less conserved than are DBD and LBD regions (5,25). Though, the isolated NTDs of the SRs have poor sequence homology, in solution they share the characteristic of disordered structure, and possess a strong transactivation domain, AF1 (25). Such transcription factors generally work in conjunction with other proteins, and by multiple mechanisms (81–85). The SR AF1 is no exception to this phenomenon (25).
We have summarized the secondary structural elements of SRs AF1 domain in Table 1. It is evident from Table 1 that in spite of having variable size and locations, all the members of SR family have an AF1 domain, which consists of more than 70% amino acid sequences that are random coil in nature. In fact, studies from several laboratories including ours have confirmed the ID nature of several SRs’ NTD/AF1 using physico-chemical methods (25,54,55,58,60). Because the AF1 domain acts by interacting with specific coregulatory proteins, it is likely that its flexible and dynamic structure creates a favorable surface for AF1’s efficient interactions with partner binding proteins (25). This raises the question: what is the structural basis of the functional activity of the ID AF1 domain in the full length receptor? AF1 appears to be in an ID conformation in the full length PPAR (49), and our studies with the glucocorticoid receptor suggest that in a two-domain (NTD+DBD) preparation, AF1 also is not structured (51). Other studies have shown similar findings with the progesterone receptor (56). We propose that the ID nature of AF1 allows this region of the receptor to rapidly and reversibly adopt various configurations controlled by allosteric cooperativity between different domains, the selective responses to cell conditions, and heterologous protein partners control the regulation of target gene(s) (25). It is a well accepted fact now that ID regions (such as AF1) can be thought of as a large collection of rapidly inter-converting conformers (Figure 2). The number of such conformations may vary depending upon the cellular environment, and could exist as a collection of large, highly dynamic, and rapidly inter-converting structures (81–85). Some of these conformations could even exist in folded conformations; however, their equilibrium always will be shifted towards unstructured conformations at a given time and depends upon cellular crowding (such as presence of small cellular solutes) under physiological conditions (25,81–85).
Table 1.
A Summary of predicted intrinsically disordered structural elements within the AF1 domain of the steroid receptors (as calculated)*
| Receptor | Location of AF1 | Number of ID amino acids | % Random coil |
|---|---|---|---|
| GR | 77–262 | 129 | 69.35 |
| ERα | 56–127 | 55 | 75.35 |
| ERβ | 1–62 | 51 | 83.61 |
| PR | 456–546 | 63 | 69.23 |
| AR | 101–370 | 191 | 70.74 |
| MR | 170–433 | 229 | 87.07 |
Combet C., Blanchet C., Geourjon C. and Deléage G. NPS@: Network Protein Sequence Analysis. TIBS 2000 March Vol. 25, No 3 [291]:147–150
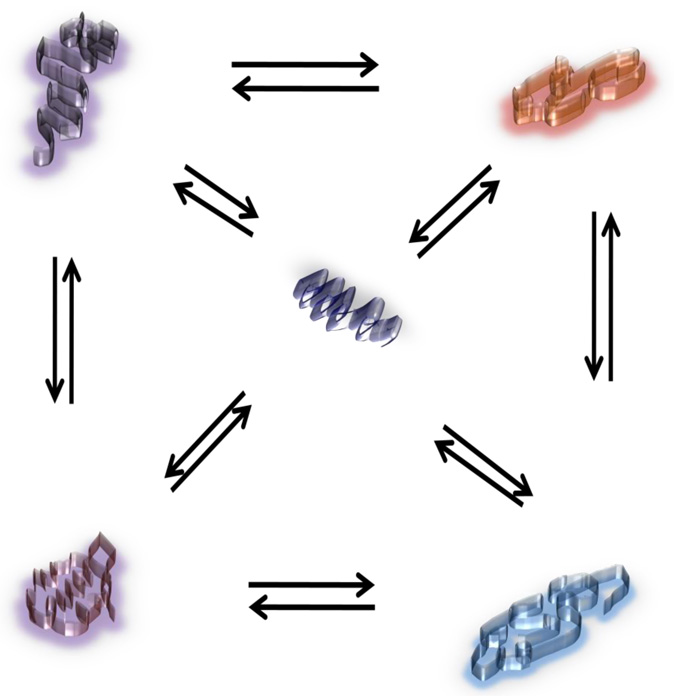
Figure 2. A schematic diagram showing various conformational states of an ID protein.
Shown is a hypothetical ID protein that can exist in a number of probable conformations. A few such possible conformations are illustrated in this figure (not real conformations). These conformations can be highly dynamic and can adopt various shapes in a very rapid and inter-convertible fashion. In other words, these conformations are reversible in nature. Under physiological conditions, depending upon the cellular environment, some of these conformers could well have limited residual structure. Depending on the relative probabilities of each state prior to stabilizing conditions (such as interaction of target molecules), the change in free energy for one or more of the stable conformation(s) may vary. In the case of ID AF1 domain of the SRs, disorder-to-order transition should provide access to the interaction energies between different cooperative elements to identify the most efficient and stable interactions in the final form of the complex. Further, the disorder-to-order transition that occurs upon complex formation (between AF1 and other binding partner proteins) can be localized to binding interfaces.
In the case of the ID AF1 domain of the SRs, we have found that the presence of various naturally occurring osmolytes can bring many (if not all) of these unstructured states into well folded globular structures with significant amounts of secondary and tertiary structures (53). Another interesting feature of these folding patterns is their cooperative nature indicating that AF1 can adopt a unique conformation that is functionally active as assessed by the ability of folded AF1 to interact with various coregulatory proteins and generate AF1-mediated transcriptional activity (53). It is striking that a high proportion of transcription factors contain ID regions that coincide with their activation domains (68). We propose that AF1’s ID nature allows it to rapidly “sample” the cellular environment until partners of appropriate concentration and affinity are found within a specific cell type. Once found, either by induced fit or selective binding of a particular conformer, a high-affinity AF1:binding partner interaction occurs. We further hypothesize that several factors can influence AF1 tertiary structure formation: binding of the DBD to DNA, ligand occupancy of the LBD and, in some circumstances, the type and concentrations of intracellular small solute molecules such as chemical chaperones.
Steroid receptors’ AF1 domains function in a cell type- and promoter-specific manner
As discussed above, each SR contains in its NTD an AF1, defined by molecular mapping that is responsible for a significant proportion of the transcription-activating potency of the SR. PRB (the larger form of the progesterone receptor) has two such regions, with the additional one, AF3, located in its N-terminal region, whereas PRA (the shorter form of the PR) has only AF1 just like other SRs (55,56,86). The evidence for transcription-controlling activity of the AF1s is compelling because mutations or deletions in AF1 lower the activity of the holo-receptor when an agonist ligand for the LBD is provided (5,25,87). In most cases AF1 is capable of providing constitutive transcriptional activity when the LBD is deleted from an SR bound to its response element DNA site (25,56,58). Also relevant are the extensive data showing the differing strength and gene-selective activities of the shorter and longer forms of the NTD, naturally found in each of the SRs (25–27). The kinetic and dynamic behavior of an SR observed in cells reveals rapid interactions with various coregulatory proteins in the nucleus, and with chromatin and DNA (88). There are reports showing that the chromatin/DNA interactions involving SRs do not always produce productive transcription complexes (25,88,89). In recent years, a number of protein binding partners for SRs AF1 have been identified (25,29). Some of these protein binding partners can bind both AF1 and AF2 (25,90). Differing sets of binding partners come together in response to agonist- or antagonist- bound LBD, and an agonist in one cell type can act as an antagonist in another cell type (22), suggesting that either differing binding partner proteins exist in a particular cell type or that there is differing use of the same partners under the influence of other cellular conditions (Figure 3).
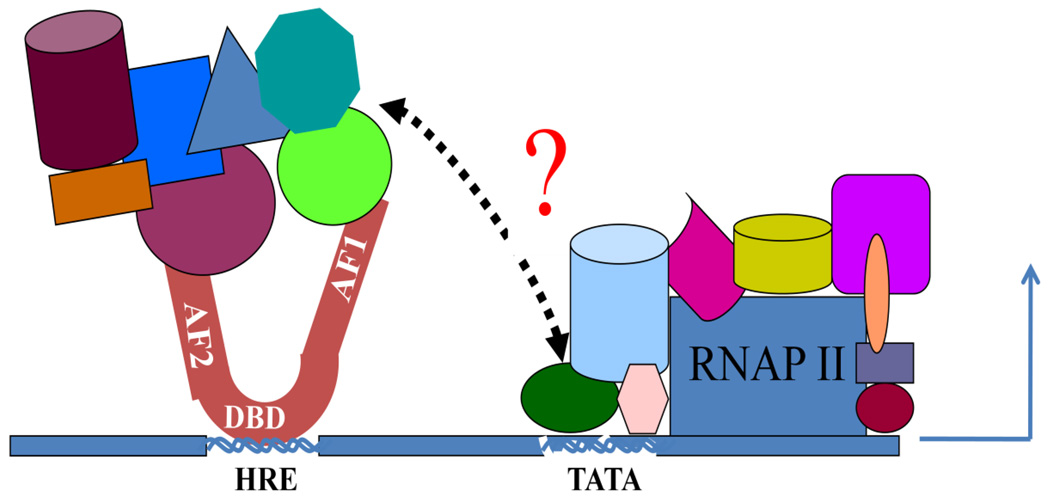
Figure 3. A model for the regulation of transcription by SR:coregulator assemblies.
The SR bound to its hormone response element (HRE) recruits certain specific coregulatory proteins (shown by different colors and shapes) mainly through AF1 and/or AF2 regions. Some of these coregulators may have been bound to both AF1 and AF2 through different binding motifs that may lead to formation of a bridge between AF1 and AF2 through these and/or other cofactor(s). By doing so, the signal is passed between AF1 and AF2, facilitating receptor activity. Precise relationships and locations of these cofactor proteins interactions are not accurate and may differ in different SRs. In certain cases, AF1 and AF2 also can interact directly through intra-molecular cross talk. The precise assembly of these coregulatory proteins may be determined by their levels of expression and concentrations in specific cell types and by the conformational state of the SR, particularly its ID AF1 domain. The complex may alter local chromatin structure, and affect the stabilization of the basal transcription pre-initiation complex through TBP bound to its TATA box and/or with other sub-units of the complex. This may well vary within different members of the SR family. The SR:coregulatory protein complex somehow bound to DNA enhancer sites, recruits and regulates Pol II via accumulations of specific proteins, which act as a functional bridge between the receptor and Pol II.
It is also possible that the affinity and kinetics of binding to partner proteins may differ depending upon cell type. This could be due to the influence of a specific cellular environment that generates receptor surfaces that either include or exclude specific binding partners, and this could be of great importance in case of the ID AF1 domain. The overall picture is not yet clear, and points to the existence of a very complex and dynamic cascade controlled by the SR for its interactions with a variety of other binding partners, on and off various DNA sites in a promoter dependent manner. To a certain extent, the structural changes observed in the AF2 due to preferential binding of coactivator or corepressor binding partners have helped in the understanding of the mechanisms involved therein (15,37). However, the data, so far, fall short of explaining the full action of the SR. The main reason for this is the lack of our understanding of the structural dynamics of the AF1, and its subsequent functional activity under physiological conditions. This information may provide an opportunity to understand critical questions related to cell- and promoter-specific actions of the SRs. We and others are actively pursuing research to address this complex and important issues related to SR action. We believe that the AF1’s ID nature plays an important role in this process. The precision of the limits of the primary amino acid sequences that define the AF1 varies between SRs, yet interspecies comparisons show that for a particular receptor, there is high constancy of AF1 sequences (25).
Since proteins with differing primary sequences can fold to give very similar condensed structures, the available data on SR AF1s raises several questions that must be addressed: i) does each AF1 fold to a single, unique conformer or to multiple unique conformers?; and ii) among SRs, do the AF1s come to a single common conformer, to a set of shared conformers, or to a subset of shared conformers, with additional conformers unique to each SR type? The answer to these and several other fundamental questions will be available only when the three-dimensional structure of each folded AF1 conformation is determined. The wide variability in AF1 suggests that there should be some differences in folded functionally active structures, but does not rule out a shared central conformer or conformers, common to all under physiological conditions. Since ID AF1s can rapidly adopt a number of conformations, and there are various factors, such as binding of receptor to a specific promoter that might bring differential folding of AF1, it is possible that AF1conformation plays an important role in cell- and promoter-specific effects of SRs in the regulation of a target gene. The ultimate understanding of the mechanisms of action of AF1 in activating transcription will require multiple levels of knowledge, eventually reaching the level that will explain how they act in complex with many other proteins in chromatin. A comprehensive knowledge of the structure and binding functions of AF1 will contribute to a new level of understanding of this complicated mechanism. In addition to the various types of binding partners in different cells contributing to different responses, differences in specific cell signaling may also play an important role. Both the receptors and most of their coactivators are phospho-proteins, and differences in function and difficulty in determining the structure of ID AF1 may in part be due to phosphorylation.
Potential means by which ID AF1 domain of the steroid receptors adopts a functionally active three dimensional conformation
It is generally presumed that AF1 must bind, directly or via co-activators/co-repressors, to some part of the primary transcription machinery. In the absence of data supporting its 3-D structure, several theories have been advanced to explain how AF1 may function. One suggests that AF1 acts without any definite structure; that it merely forms a cloud of negatively charged amino acids. Based on observations that negatively charged areas are important for transactivation function in the case of some acidic activators (5,58), it was proposed that this would be sufficient to cause transcription initiation. At least in case of the glucocorticoid receptor, mutational analysis suggests that this is not the case (58), because mutations that eliminated or replaced acidic amino acids with neutral ones had little effect on the ability of the AF1 to activate transcription (58,59). On the other hand, mutations of hydrophobic amino acids of AF1 strongly reduced the AF1 activity (59). It is therefore hypothesized that an induced conformation or set of conformations occurs in AF1 in order for it to carry out its transcription function (25,52). What causes these conformational changes, and how are they physiologically important? The answer to this may lie with induced-fit mechanism of folding (69,70). Induced fit might occur only when the AF1 domain encounters its proper binding partner. If it happens through critical coregulatory proteins for AF1, the possibility of a physiologically functional conformation(s) is great. It is known that the AF1 makes physical and functional interactions with other coregulatory proteins in order to transactivate gene(s) and that the induced AF1 conformation is important for several of these interactions (25,52,59,60). Thermodynamic considerations predict that if one or more conformers of AF1 lacking any significant structure bind specific protein partners with high affinity, interaction of the AFI with that partner at appropriate concentrations should cause AF1s to fold (25). This could occur either by simple mass action or by direct induced fit following initial, relatively non-specific binding partner interactions with unfolded conformer(s) of AF1 (25).
We have shown, in case of the glucocorticoid receptor, that a direct interaction between AF1 and the TATA Box-binding protein (TBP) leads to formation of tertiary structure in AF1 (73). In vivo, we have documented that AF1:TBP interactions measured by fluorescence resonance energy transfer (FRET), correlates with enhanced transcription of both co-transfected and endogenous genes (87). Deletions of AF1 or its core region caused loss of TBP interaction and of tanscriptional activity (87). These observations are consistent with the fact that TBP can be limiting and drawn to sites to be transcribed by various transcription factors. Similar results have been shown for the induced folding of AF1 of the estrogen receptor (ER) through TBP (54). While both ERα and ERβ possess ID AF1/NTD, TBP binding induces structure formation only in ERα, suggesting that TBP-induced folding in ID AF1 may be specific and dependent upon its ability to work as a coregulator for that particular receptor. We also have shown that when ID AF1 of the AR binds to a protein from the basal transcription machinery complex, RAP74 (a subunit of TFIIF complex), structure is formed in AF1 (75). Another type of induced binding/folding event in the PR AF1 occurs through the binding interaction between jun dimerization protein 2 (JDP-2) and DBD (55). It has been shown that JDP-2 binds specifically to the DBDs of all SHRs and in so doing, stimulates transcription (55). The effects of JDP-2 on transcription are gene-specific, suggesting specific actions through the NTD and therefore possibly specific AF1/NTD conformations. Structural consequences of the DBD:JDP-2 interactions have been observed in the PR NTD (55). Binding of JDP-2 with the two-domain NTD-DBD polypeptide (consisting of the entire NTD plus DBD) resulted in a substantial gain in structure formation in the AF1/NTD without any significant secondary/tertiary structural changes within the DBD (55). Thus, JDP-2 binding to the PR DBD affects the structure and transcriptional function of its AF1/NTD. These results indicate that induced binding/folding of AF1 can occur even when the binding partner is not directly interacting with AF1 but rather through inter-domain communication. Such inter-domain communications have been reported for several transcription factors, including SRs (51,56,25). In fact, we also have shown that secondary and tertiary structure are induced in the AF1 region when a two-domain GR fragment (GR1–500) comprising its entire N-terminal- (NTD) through the DNA-binding domain (DBD) is bound to a DNA oligomer containing a palindromic glucocorticoid response element (51).
Taken together, it is logical to build a working model on how the ID AF1 domain of the SHRs regulates the transcriptional activity by providing an ideally suited flexible and dynamic protein surfaces for critical coregulatory proteins, essential for gene regulation by SRs (89–91). We propose, under physiological conditions, that AF1 in the holo-SHR exists as a partially folded structure due to inter-domain interactions including but not limited to DNA and ligand bindings through respective domains, and due to the presence of specific intracellular milieu (including chemical chaperones such as certain polyols, sugars, amino acids, and methylamines). The resulting structurally modified forms of AF1 facilitate it for varied interactions with other critical coregulatory proteins, and possibly additional modulations in receptor structure essential for gene regulation by the receptor. These interactions give final folded structures to AF1 and form the basis for the multi-protein assemblies involved in SR-mediated regulation of transcription (Figure 4). How AF1 folds and what kind of functional folded conformation it adopts are open questions, and we and others have been pursuing answers to these long-standing problems.
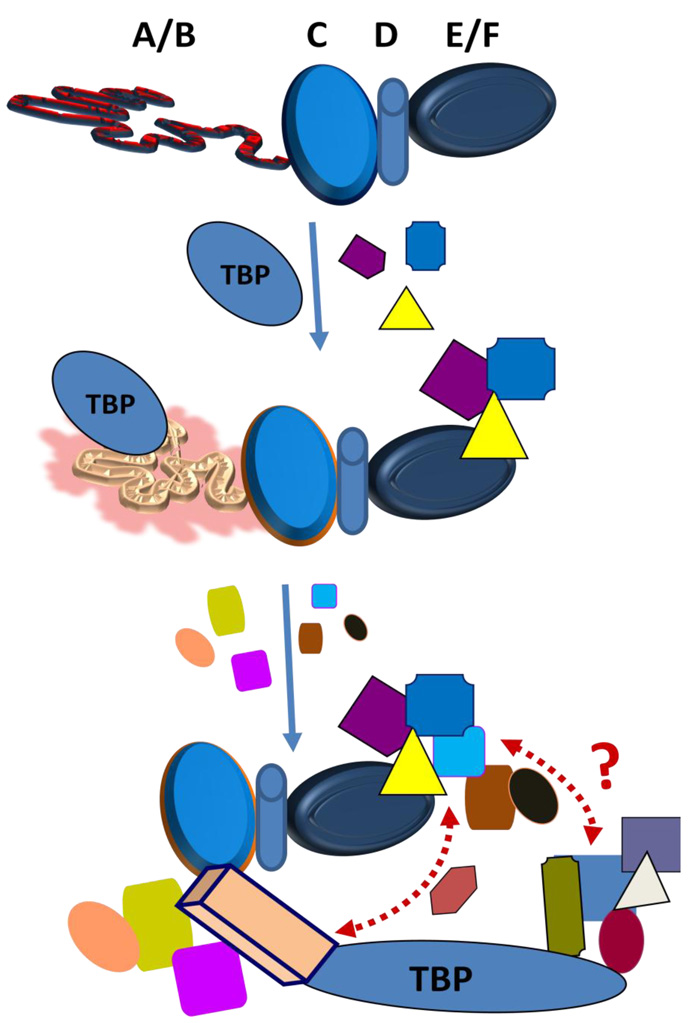
Figure 4. A hypothetical model of folding of the SRs’ AF1 domain in the context of full length receptor under physiological conditions.
The ID AF1 domain interacts with one or more proteins from the basal transcription initiation machinery complex, e.g. TBP and in doing so acquires a set of conformations that allows AF1 surfaces to create a platform for interaction with other coregulatory proteins to carry out its function. The resulting structurally modified forms of AF1 may activate it for interactions with various other critical coregulatory proteins essential for gene regulation by the receptor. These interactions give the final folded structure to AF1 and form the basis for the multi-protein assemblies involved in SR-mediated regulation of transcription. This induced fit model of AF1 folding infers that AF1 is not fully structured in vivo until it binds one or another key partner molecule (shown by different colors and shapes). This induced-conformation or limited set of conformations in AF1 is needed in order for it to carry out its transcription function. Formally, induced fit could occur by initial non-specific interactions between the unfolded AF1 domain and the BP. In this version of the model, when such interactions occur, the proximity of the two proteins leads to rapid acquisition of the proper structure in AF1. Alternatively, there initially could be more specific interactions of coregulatory proteins with a partially folded AF1, or even with a tiny pool of fully folded AF1 molecules, creating a kinetic “sink” into which the general population falls.
Summary and Perspectives
In last several years we have learned a great deal about how SRs act, however our efforts have failed to explain several outstanding questions related to the structural and functional relationships of SRs. To fully understand how signals from a ligand are passed through SRs to a specific target gene, we must understand how ID AF1s regulate interactions between the vast arrays of expressed proteins. In recent years, from the work carried out in various laboratories on protein:protein and protein:DNA interactions involving SRs and their relationships with target gene regulations, it has been shown that dynamic macromolecular ensembles involving SRs AF1 and AF2 domains are key elements in the regulation of many biological systems (18,25,29,56,59,87). Identification of the ID nature of AF1 suggests that direct protein:protein interaction may be an essential step in producing properly folded and functionally active structure in the ID region leading to transactivation activity (85,91). However, the actual three-dimensional structure of the functionally folded AF1 domain, ideally in the full length receptor remains to be determined. A recently published crystal structure of the PPAR has, for the first time, indicated the possibility of solving SR’s AF1 structure. Site-specific phosphorylation represents an important regulatory mechanism in the activities of several SRs. Many known functionally important phosphorylation sites are located in the ID AF1/NTD of the SRs. It has been predicted that generally ID proteins have a higher frequency of known phosphorylation sites than ordered regions. It is logical to hypothesize that site-specific phosphorylation of the ID AF1 may lead to changes in its conformations that are important for AF1’s interaction with other critical coregulatory proteins, and subsequent transcriptional activity. The characterization of phosphorylation–induced structure formation in AF1s and its role in facilitating protein:protein interactions should be of particular importance in understanding the mechanism by which kinase(s) regulate the actions of SRs. We and others are actively pursuing the question of the kind of functional ordered conformation(s) AF1 domains adopt, and whether multiple folded conformations are generated depending upon the nature of binding partner(s) involved. Biophysical and molecular biological approaches, so far, have eluded the mechanism by which SRs’ AF1 domains adopt ordered conformation(s) under physiological conditions. However, more studies are needed to determine whether there is a unified mechanism through which the ID AF1 domain in each receptor acquires a well defined structure. Protein:protein interactions involving the SRs and other coregulatory proteins have been extremely useful in providing targets for the development of suitable steroids and other small molecules that can modulate such interactions. Proper knowledge of the importance of coupled binding and folding of the ID AF1 domains in the regulation of the SRs actions has the potential to significantly advance our understanding of the mechanisms involved in passing signals from a steroid to specific target gene. Finally, structural analyses of the AF1 domain combined with its functional behavior will allow design and development of multifunctional approaches to produce more active steroids, and more efficient and better targeted therapeutic tools.
Acknowledgements
This work was supported by a grant from the NIH (RO1 DK058829 to RK)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beato M, Klug J. Steroid hormone receptors: an update. Human reproduction update. 2000;6:225–236. doi: 10.1093/humupd/6.3.225. [DOI] [PubMed] [Google Scholar]
- 2.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 3.Li X, O'Malley BW. Unfolding the action of progesterone receptors. J Biol Chem. 2003;278:39261–39264. doi: 10.1074/jbc.R300024200. [DOI] [PubMed] [Google Scholar]
- 4.Lonard DM, O'Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64:310–319. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 6.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- 8.Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- 9.Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci USA. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 11.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 2003;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene network. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 14.Simons SS., Jr . Vitamins and Hormones. Academic Press, Inc; 1999. Function/activity of specific amino acids in glucocorticoid receptors; pp. 49–130. [DOI] [PubMed] [Google Scholar]
- 15.Danielian PS, White R, Lees JA, Parker MG. The structural basis of estrogen receptor/coactivator recognition and the antagonism of the interaction by tamoxifen. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simental JA, Sar M, Lane MV, French FS, Wilson EM. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 17.Danielian PS, White R, Lees JA, Parker MG. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 19.Metivier R, Penot G, Flouriot G, Pakdel F. Synergism between ERalpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 alpha-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol. 2001;15:1953–1970. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- 20.Simental JA, Sar M, Lane MV, French FS, Wilson EM. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 21.Montano MM, Ekena K, Krueger KD, Keller AL, Katzenellenbogen BS. Human estrogen receptor ligand activity inversion mutants: receptors that interpret antiestrogens as estrogens and estrogens as antiestrogens and discriminate among different antiestrogens. Mol Endocrinol. 1996;10:230–242. doi: 10.1210/mend.10.3.8833652. [DOI] [PubMed] [Google Scholar]
- 22.Kraus WL, McInerney EM, Katzenellenbogen BS. Ligand-dependent, transcriptionally productive association of the amino- and carboxyl-terminal regions of a steroid hormone nuclear receptor. Proc Natl Acad Sci USA. 1995;92:12314–12318. doi: 10.1073/pnas.92.26.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 24.Tetel MJ, Giangrande PH, Leonhardt SA, McDonnell DP, Edwards DP. Hormone-dependent interaction between the amino- and carboxyl-terminal domains of progesterone receptor in vitro and in vivo. Mol Endocrinol. 1999;13:910–924. doi: 10.1210/mend.13.6.0300. [DOI] [PubMed] [Google Scholar]
- 25.Kumar R, Thompson EB. Transactivation functions of the N-terminal domains of nuclear hormone receptors: protein folding and coactivator interactions. Mol Endocrinol. 2003;17:1–10. doi: 10.1210/me.2002-0258. [DOI] [PubMed] [Google Scholar]
- 26.Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol Cell Biol. 1999;19:6085–6097. doi: 10.1128/mcb.19.9.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He B, Lee LW, Minges JT, Wilson EM. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J Biol Chem. 2002;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- 28.Bommer M, Benecke A, Gronemeyer H, Rochette-Egly C. TIF2 mediates the synergy between RARalpha 1 activation functions AF-1 and AF-2. J Biol Chem. 2002;277:37961–37966. doi: 10.1074/jbc.M206001200. [DOI] [PubMed] [Google Scholar]
- 29.Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- 30.Glass CK, Rose DW, Rosenfeld MG. Nuclear receptor coactivators. Current Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 31.Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 32.Loven MA, Muster N, Yates JR, Nardulli AM. A novel estrogen receptor alpha-associated protein, template-activating factor I beta, inhibits acetylation and transactivation. Mol Endocrinol. 2003;17:67–78. doi: 10.1210/me.2002-0280. [DOI] [PubMed] [Google Scholar]
- 33.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto KR, Darimont BD, Wagner RL, Iniguez-Lluhi JA. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harbor Symposia Quantitative Biology. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- 35.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 38.Fagart J, Wurtz JM, Souque A, Hellal-Levy C, Moras D, Rafestin-Oblin ME. Antagonism in the human mineralocorticoid receptor. EMBO J. 1998;17:3317–3325. doi: 10.1093/emboj/17.12.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanenbaum DM, Wang Y, Williams SP, Sigler PB. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc Natl Acad Sci USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 41.Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem. 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- 42.Kauppi B, Jakob C, Farnegardh M, Yang J, Ahola H, Alarcon M, Calles K, Engstrom O, Harlan J, Muchmore S. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem. 2003;278:22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 43.Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hard T, Kellenbach E, Boelens R, Maler BA, Dahlman K, Freedman LP, Carlstedt-Duke J, Yamamoto KR, Gustafsson JA, Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- 45.Schwabe JW, Neuhaus D, Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- 46.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 47.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci USA. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Claessens F, Gewirth DT. DNA recognition by nuclear receptors. Essays Biochem. 2004;40:59–72. doi: 10.1042/bse0400059. [DOI] [PubMed] [Google Scholar]
- 49.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baskakov IV, Kumar R, Srinivasan G, Ji YS, Bolen DW, Thompson EB. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J Biol Chem. 1999;274:10693–10696. doi: 10.1074/jbc.274.16.10693. [DOI] [PubMed] [Google Scholar]
- 51.Kumar R, Baskakov IV, Srinivasan G, Bolen DW, Lee JC, Thompson EB. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J Biol Chem. 1999;274:24737–24741. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- 52.Kumar R, Lee JC, Bolen DW, Thompson EB. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J Biol Chem. 2001;276:18146–18152. doi: 10.1074/jbc.M100825200. [DOI] [PubMed] [Google Scholar]
- 53.Kumar R, Serrette JM, Khan SH, Miller AL, Thompson EB. Effects of different osmolytes on the induced folding of the N-terminal activation domain (AF1) of the glucocorticoid receptor. Arch Biochem Biophys. 2007;465:452–460. doi: 10.1016/j.abb.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warnmark A, Wikstrom A, Wright AP, Gustafsson JA, Hard T. The N-terminal regions of estrogen receptor alpha and beta are unstructured in vitro and show different TBP binding properties. J Biol Chem. 2001;276:45939–45944. doi: 10.1074/jbc.M107875200. [DOI] [PubMed] [Google Scholar]
- 55.Wardell SE, Kwok SC, Sherman L, Hodges RS, Edwards DP. Regulation of the amino-terminal transcription activation domain of progesterone receptor by a cofactor-induced protein folding mechanism. Mol Cell Biol. 2005;25:8792–8808. doi: 10.1128/MCB.25.20.8792-8808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of the human progesterone A-receptor. Structural analysis and the influence of the DNA binding domain. J Biol Chem. 2000;275:7313–7320. doi: 10.1074/jbc.275.10.7313. [DOI] [PubMed] [Google Scholar]
- 57.Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391:449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dahlman-Wright K, Baumann H, McEwan IJ, Almlof T, Wright APH, Gustafsson JA, Hard T. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci USA. 1995;92:1699–1703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almlof T, Wallberg AE, Gustafsson JA, Wright APH. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor tau1-core activation domain and target factors. Biochem. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 60.Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. J Biol Chem. 2002;277:20079–20086. doi: 10.1074/jbc.M201003200. [DOI] [PubMed] [Google Scholar]
- 61.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 62.Fink AL. Natively unfolded proteins. Current Opinion Str Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 63.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 64.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochem. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hegyi H, Schad E, Tompa P. Structural disorder promotes assembly of protein complexes. BMC Str Biol. 2007;7:65. doi: 10.1186/1472-6807-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 68.Garza AS, Ahmad N, Kumar R. Role of intrinsically disordered protein regions/domains in transcriptional regulation. Life Sci. 2009;84:189–193. doi: 10.1016/j.lfs.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 69.Dunker A, Uversky V. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 70.Dyson H, Wright P. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 71.Hoffman R, Sykes B. Isoform-specific variation in the intrinsic disorder of troponin I. Proteins. 2008 doi: 10.1002/prot.22063. [DOI] [PubMed] [Google Scholar]
- 72.Minezaki Y, Homma K, Kinjo A, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 73.Kumar R, Volk D, Li J, Lee J, Gorenstein D, Thompson E. TATA box binding protein induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc Natl Acad Sci U S A. 2004;101:16425–16430. doi: 10.1073/pnas.0407160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mark W, Liao J, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A, Arrowsmith C. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J Mol Biol. 2005;345:275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 75.Kumar R, Betney R, Li J, Thompson E, McEwan I. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochem. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 76.Shen F, Triezenberg S, Hensley P, Porter D, Knutson J. Transcriptional activation domain of the herpesvirus protein VP16 becomes conformationally constrained upon interaction with basal transcription factors. J Biol Chem. 1996;271:4827–4837. doi: 10.1074/jbc.271.9.4827. [DOI] [PubMed] [Google Scholar]
- 77.Shoemaker B, Portman J, Wolynes P. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh G, Dash D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins. 2007;68:602–605. doi: 10.1002/prot.21497. [DOI] [PubMed] [Google Scholar]
- 79.Benjamin AS, Portman J, Wolynes J. P.G. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97 doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McEwan I, Dahlman-Wright K, Ford J, Wright A. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: evidence for an induced fit model of transactivation domain folding. Biochem. 1996;35:9584–9593. doi: 10.1021/bi960793v. [DOI] [PubMed] [Google Scholar]
- 81.Bienkiewicz E, Adkins J, Lumb K. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1) Biochem. 2002;41:752–759. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]
- 82.Dunker A, Brown C, Lawson J, Iakoucheva L, Obradović Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 83.Uversky V. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fink A. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 85.Dyson H, Wright P. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 86.Sartorius CA, Melville MY, Hovland AR, Tung L, Takimoto GS, Horwitz KB. A third transactivation function (AF3) of human progesterone receptors located in the unique N-terminal segment of the B-isoform. Mol Endocrinol. 1994;8:1347–1360. doi: 10.1210/mend.8.10.7854352. [DOI] [PubMed] [Google Scholar]
- 87.Copik AJ, Webb MS, Miller AL, Wang Y, Kumar R, Thompson EB. Activation function 1 of glucocorticoid receptor binds TATA-binding protein in vitro and in vivo. Mol Endocrinol. 2006;20:1218–1230. doi: 10.1210/me.2005-0257. [DOI] [PubMed] [Google Scholar]
- 88.Keeton EK, Fletcher TM, Baumann CT, Hager GL, Smith CL. Glucocorticoid receptor domain requirements for chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter in different nucleoprotein contexts. J Biol Chem. 2002;277:28247–28257. doi: 10.1074/jbc.M203898200. [DOI] [PubMed] [Google Scholar]
- 89.Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O'Malley BW, Mancini MA. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nature Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- 90.Lazennec G, Ediger TR, Petz LN, Nardulli AM, Katzenellenbogen BS. Mechanistic aspects of estrogen receptor activation probed with constitutively active estrogen receptors: correlations with DNA and coregulator interactions and receptor conformational changes. Mol Endocrinol. 1997;11:1375–1386. doi: 10.1210/mend.11.9.9983. [DOI] [PubMed] [Google Scholar]
- 91.McEwan IJ, Lavery D, Fischer K, Watt K. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal. 2007;5 doi: 10.1621/nrs.05001. e001. [DOI] [PMC free article] [PubMed] [Google Scholar]