Abstract
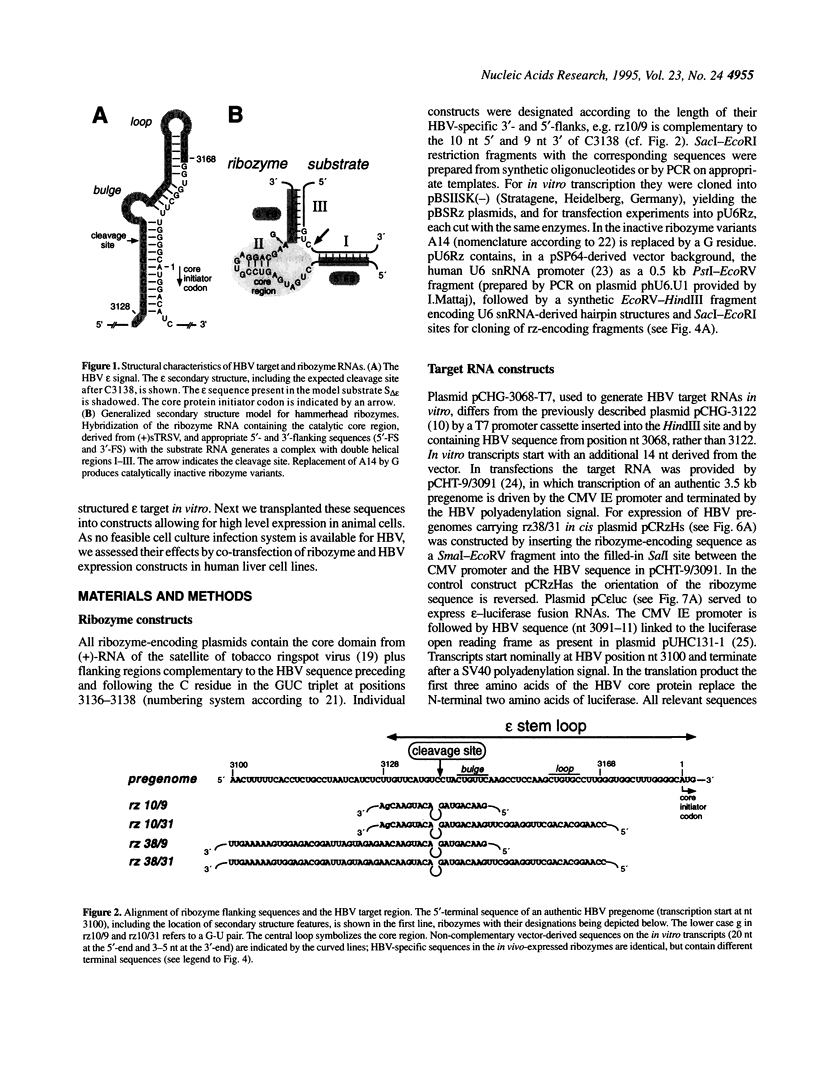
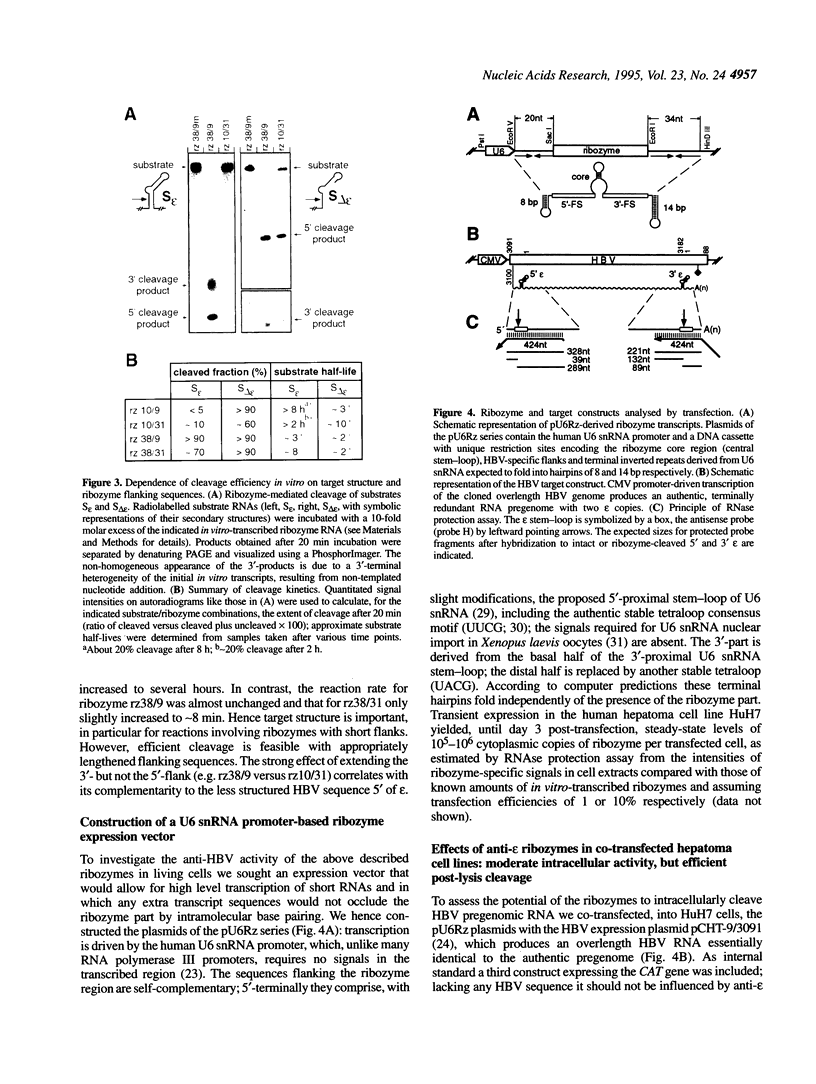
Hepatitis B virus (HBV), the causative agent of B-type hepatitis in man, is a small enveloped DNA virus that replicates through reverse transcription of an RNA intermediate, the terminally redundant RNA pregenome. An essential highly conserved cis-element present twice on this RNA is the encapsidation signal epsilon, a stem-loop structure that is critical for pregenome packaging and reverse transcription. Epsilon is hence an attractive target for antiviral therapy. Its structure, however, is a potential obstacle to antivirals whose action depends on hybridization, e.g. ribozymes. Here we demonstrate effective in vitro cleavage inside epsilon by hammerhead ribozymes containing flanking sequences complementary to an adjacent less structured region. Upon co-transfection with a HBV expression construct corresponding ribozymes embedded in a U6 snRNA context led to a significant, though modest, reduction in the steady-state level of HBV pregenomes. Inactive ribozyme mutants revealed that antisense effects contributed substantially to this reduction, however, efficient epsilon cleavage by the intracellularly expressed ribozymes was observed in Mg(2+)-supplemented cell lysates. Artificial HBV pregenomes carrying the ribozymes in cis and model RNAs lacking all HBV sequences except epsilon exhibited essentially the same behaviour. Hence, neither the absence of co-localization of ribozyme and target nor a viral component, but rather a cellular factor(s), is responsible for the strikingly different ribozyme activities inside cells and in cellular extracts.
Full text
PDF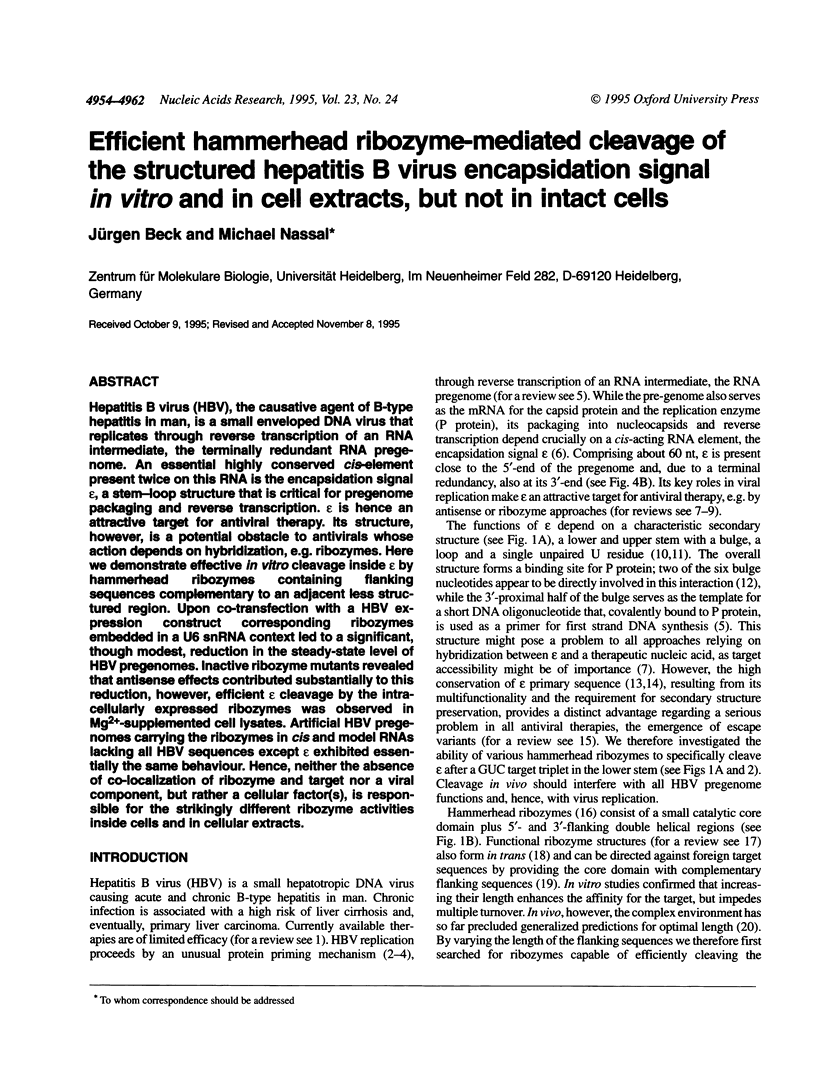






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertrand E., Pictet R., Grange T. Can hammerhead ribozymes be efficient tools to inactivate gene function? Nucleic Acids Res. 1994 Feb 11;22(3):293–300. doi: 10.1093/nar/22.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A. L., Gossen M., Bujard H. Photinus pyralis luciferase: vectors that contain a modified luc coding sequence allowing convenient transfer into other systems. Gene. 1994 Apr 8;141(1):75–77. doi: 10.1016/0378-1119(94)90130-9. [DOI] [PubMed] [Google Scholar]
- Cantor G. H., McElwain T. F., Birkebak T. A., Palmer G. H. Ribozyme cleaves rex/tax mRNA and inhibits bovine leukemia virus expression. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):10932–10936. doi: 10.1073/pnas.90.23.10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J. M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995 Jan 27;267(5197):483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Cotten M., Birnstiel M. L. Ribozyme mediated destruction of RNA in vivo. EMBO J. 1989 Dec 1;8(12):3861–3866. doi: 10.1002/j.1460-2075.1989.tb08564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisell P., Thompson S., James W. Inhibition of HIV-1 replication by ribozymes that show poor activity in vitro. Nucleic Acids Res. 1993 Nov 11;21(22):5251–5255. doi: 10.1093/nar/21.22.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A. Hammerhead ribozyme structure: U-turn for RNA structural biology. Structure. 1995 Aug 15;3(8):747–750. doi: 10.1016/s0969-2126(01)00208-8. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G., Bratty J., Chen H., Cedergren R. A hammerhead ribozyme inhibits ADE1 gene expression in yeast. Gene. 1995 Mar 21;155(1):45–50. doi: 10.1016/0378-1119(94)00891-u. [DOI] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J. 1989 Dec 20;8(13):4179–4187. doi: 10.1002/j.1460-2075.1989.tb08603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990 Oct 5;63(1):109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hertel K. J., Pardi A., Uhlenbeck O. C., Koizumi M., Ohtsuka E., Uesugi S., Cedergren R., Eckstein F., Gerlach W. L., Hodgson R. Numbering system for the hammerhead. Nucleic Acids Res. 1992 Jun 25;20(12):3252–3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann M., Tzortzakaki S., Rittner K., Sczakiel G., Tabler M. Incorporation of the catalytic domain of a hammerhead ribozyme into antisense RNA enhances its inhibitory effect on the replication of human immunodeficiency virus type 1. Nucleic Acids Res. 1993 Jun 25;21(12):2809–2814. doi: 10.1093/nar/21.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker-Niepmann M., Bartenschlager R., Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990 Oct;9(10):3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehntopf M., Esquivel E. L., Brach M. A., Herrmann F. Clinical applications of ribozymes. Lancet. 1995 Apr 22;345(8956):1027–1031. doi: 10.1016/s0140-6736(95)90762-9. [DOI] [PubMed] [Google Scholar]
- Knaus T., Nassal M. The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 1993 Aug 25;21(17):3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskus T., Rakela J., Persing D. H. The stem-loop structure of the cis-encapsidation signal is highly conserved in naturally occurring hepatitis B virus variants. Virology. 1994 May 1;200(2):809–812. doi: 10.1006/viro.1994.1247. [DOI] [PubMed] [Google Scholar]
- Lok A. S., Akarca U., Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):4077–4081. doi: 10.1073/pnas.91.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok A. S. Treatment of chronic hepatitis B. J Viral Hepat. 1994;1(2):105–124. doi: 10.1111/j.1365-2893.1994.tb00110.x. [DOI] [PubMed] [Google Scholar]
- London R. E. Methods for measurement of intracellular magnesium: NMR and fluorescence. Annu Rev Physiol. 1991;53:241–258. doi: 10.1146/annurev.ph.53.030191.001325. [DOI] [PubMed] [Google Scholar]
- Nassal M., Junker-Niepmann M., Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990 Dec 21;63(6):1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- Nassal M., Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993 Sep;1(6):221–228. doi: 10.1016/0966-842x(93)90136-f. [DOI] [PubMed] [Google Scholar]
- Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992 Jul;66(7):4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonberg S. B., Scott G. K., Garovoy M. R., Benz C. C., Hunt C. A. In vivo generation of highly abundant sequence-specific oligonucleotides for antisense and triplex gene regulation. Nucleic Acids Res. 1994 Jul 25;22(14):2830–2836. doi: 10.1093/nar/22.14.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offensperger W. B., Blum H. E., Gerok W. Molecular therapeutic strategies in hepatitis B virus infection. Clin Investig. 1994 Oct;72(10):737–741. doi: 10.1007/BF00180539. [DOI] [PubMed] [Google Scholar]
- Pasek M., Goto T., Gilbert W., Zink B., Schaller H., MacKay P., Leadbetter G., Murray K. Hepatitis B virus genes and their expression in E. coli. Nature. 1979 Dec 6;282(5739):575–579. doi: 10.1038/282575a0. [DOI] [PubMed] [Google Scholar]
- Pollack J. R., Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993 Jun;67(6):3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger A., Nassal M. Distinct requirements for primary sequence in the 5'- and 3'-part of a bulge in the hepatitis B virus RNA encapsidation signal revealed by a combined in vivo selection/in vitro amplification system. Nucleic Acids Res. 1995 Oct 11;23(19):3909–3915. doi: 10.1093/nar/23.19.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J., Appel B., Digweed M., Lührmann R. Localization of a base-paired interaction between small nuclear RNAs U4 and U6 in intact U4/U6 ribonucleoprotein particles by psoralen cross-linking. J Mol Biol. 1985 Oct 20;185(4):721–731. doi: 10.1016/0022-2836(85)90057-9. [DOI] [PubMed] [Google Scholar]
- Rossi J. J. Controlled, targeted, intracellular expression of ribozymes: progress and problems. Trends Biotechnol. 1995 Aug;13(8):301–306. doi: 10.1016/S0167-7799(00)88969-6. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Sczakiel G., Nedbal W. The potential of ribozymes as antiviral agents. Trends Microbiol. 1995 Jun;3(6):213–217. doi: 10.1016/s0966-842x(00)88927-1. [DOI] [PubMed] [Google Scholar]
- Sczakiel G., Oppenländer M., Rittner K., Pawlita M. Tat- and Rev-directed antisense RNA expression inhibits and abolishes replication of human immunodeficiency virus type 1: a temporal analysis. J Virol. 1992 Sep;66(9):5576–5581. doi: 10.1128/jvi.66.9.5576-5581.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M. Interaction between tumour necrosis factor alpha ribozyme and cellular proteins. Involvement in ribozyme stability and activity. J Mol Biol. 1994 Oct 7;242(5):619–629. doi: 10.1006/jmbi.1994.1612. [DOI] [PubMed] [Google Scholar]
- Steinecke P., Herget T., Schreier P. H. Expression of a chimeric ribozyme gene results in endonucleolytic cleavage of target mRNA and a concomitant reduction of gene expression in vivo. EMBO J. 1992 Apr;11(4):1525–1530. doi: 10.1002/j.1460-2075.1992.tb05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. Small catalytic RNAs. Annu Rev Biochem. 1992;61:641–671. doi: 10.1146/annurev.bi.61.070192.003233. [DOI] [PubMed] [Google Scholar]
- Tang X. B., Hobom G., Luo D. Ribozyme mediated destruction of influenza A virus in vitro and in vivo. J Med Virol. 1994 Apr;42(4):385–395. doi: 10.1002/jmv.1890420411. [DOI] [PubMed] [Google Scholar]
- Tavis J. E., Ganem D. Expression of functional hepatitis B virus polymerase in yeast reveals it to be the sole viral protein required for correct initiation of reverse transcription. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4107–4111. doi: 10.1073/pnas.90.9.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Wang G. H., Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993 Nov;67(11):6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L. Antisense treatment of viral infection. Adv Virus Res. 1994;44:267–303. doi: 10.1016/s0065-3527(08)60331-0. [DOI] [PubMed] [Google Scholar]
- Xing Z., Whitton J. L. Ribozymes which cleave arenavirus RNAs: identification of susceptible target sites and inhibition by target site secondary structure. J Virol. 1992 Mar;66(3):1361–1369. doi: 10.1128/jvi.66.3.1361-1369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Poeschla E., Wong-Staal F. Progress towards gene therapy for HIV infection. Gene Ther. 1994 Jan;1(1):13–26. [PubMed] [Google Scholar]
- von Weizsäcker F., Blum H. E., Wands J. R. Cleavage of hepatitis B virus RNA by three ribozymes transcribed from a single DNA template. Biochem Biophys Res Commun. 1992 Dec 15;189(2):743–748. doi: 10.1016/0006-291x(92)92264-x. [DOI] [PubMed] [Google Scholar]






