Abstract
The plasticity of aging suggests that longevity may be controlled epigenetically by specific alterations in chromatin state. The link between chromatin and aging has mostly focused on histone deacetylation by the Sir2 family1,2, but less is known about the role of other histone modifications in longevity. Histone methylation plays a crucial role during development and in maintaining stem cell pluripotency in mammals3. Regulators of histone methylation have been associated with aging in worms4,5,6,7 and flies8, but characterization of their role and mechanism of action has been limited. Here we identify the ASH-2 trithorax complex9, which trimethylates histone H3 at lysine 4 (H3K4), as a regulator of lifespan in C. elegans in a directed RNAi screen in fertile worms. Deficiencies in members of the ASH-2 complex–ASH-2 itself, WDR-5, and the H3K4 methyltransferase SET-2 extend worm lifespan. Conversely, the H3K4 demethylase RBR-2 is required for normal lifespan, consistent with the idea that an excess of H3K4 trimethylation–a mark associated with active chromatin–is detrimental for longevity. Lifespan extension induced by ASH-2 complex deficiency requires the presence of an intact adult germline and the continuous production of mature eggs. ASH-2 and RBR-2 act in the germline, at least in part, to regulate lifespan and to control a set of genes involved in lifespan determination. These results suggest that the longevity of the soma is regulated by an H3K4 methyltransferase/demethylase complex acting in the C. elegans germline.
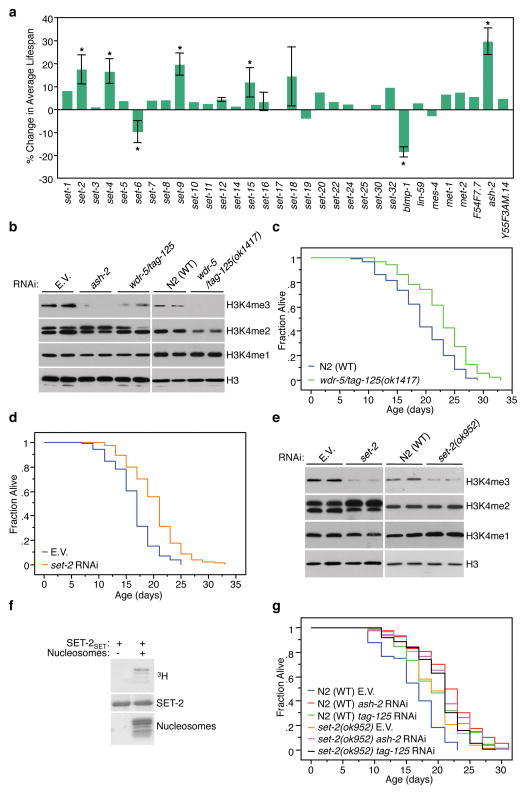
Genome-wide RNAi screens for genes that regulate lifespan in C. elegans have been previously performed in worms in which progeny production was inhibited4,10,11, which could mask the effect of some genes on lifespan. We performed a targeted RNAi screen in fertile worms by selecting genes that encode known worm methyltransferases, proteins containing the enzymatic domain of methyltransferases (SET domain), or orthologs of regulators of histone methylation. As previously reported4, set-9 and set-15 knock-down extended lifespan (Fig. 1a). We also found that ash-2, set-2, and set-4 knock-down extended fertile worm lifespan, with ash-2 knock-down having the most significant effect (23.1–30.9%, p<0.0001) (Fig. 1a, Supplementary Fig. 1a). ASH-2 is a member of an H3K4 trimethylation (H3K4me3) complex in yeast, flies, and mammals9,12,13 and is important for the conversion of H3K4 dimethylation (H3K4me2) to H3K4me3 in mammals14. ash-2 knock-down decreased global H3K4me3 levels at larval stage L3 (Fig. 1b), indicating that ASH-2 promotes histone H3 trimethylation at lysine 4 in C. elegans. WDR-5/TAG-125 is a WD40-repeat protein that interacts with ASH-2 in mammals, and is important for the mono-, di-, and tri- methylation of H3K4 in mammals and worms15,16. wdr-5/tag-125 knock-down or wdr-5/tag-125 deletion also decreased H3K4me3 levels (Fig. 1b) and significantly extended C. elegans lifespan (~30%, p<0.0001 and 16–28%, p<0.0001, respectively) (Fig. 1c, Supplementary Fig. 1b). Thus, ASH-2 and WDR-5 promote H3K4 trimethylation and normally limit lifespan in C. elegans.
Fig. 1. The ASH-2, WDR-5, and SET-2 function together to regulate H3K4me3 and lifespan in C. elegans.
a, Percent change in average lifespan in worms treated with RNAi to the indicated genes compared with empty vector. *: p<0.05. b, Western blots on L3 worm extracts (representative of 4 independent experiments). c, wdr-5/tag-125(ok1417) mutant worms have an extended lifespan. d, set-2 knock-down extends lifespan. e, Western blot of L3 worm extracts after SET-2 knock-down or deletion (representative of 3 independent experiments). f, The methyltransferase domain of worm SET-2 (SET-2SET) methylates histone H3 in vitro. e, ash-2 and wdr-5/tag-125 knock-down slightly extend the lifespan of set-2(ok952) mutant worms, but significantly less than in the WT (N2) background. ash-2 mRNA was efficiently knocked-down by RNAi in set-2(ok952) mutant worms (Supplementary Fig. 3a). E.V.: empty vector. Statistics are presented in Supplementary Tables 1 and 2.
In mammals, ASH-2 and WDR-5 form a complex with several H3K4me3 methyltransferases of the SET1/mixed lineage leukemia (MLL) family17. There are four SET1/MLL orthologues in C. elegans: SET-1, SET-2, SET-12, and SET-16 (Supplementary Fig. 1c). Of these, only SET-2, which is similar to mammalian SET1A/SET1B, regulated worm lifespan (set-2 knock-down: ~20%, p<0.0001; set-2 deletion set-2(ok952): 17-26%, p<0.0001) (Fig. 1a, Fig. 1d, Supplementary Fig. 1d, e). As previously reported16, set-2(ok952) mutant worms or worms treated with set-2 RNAi had reduced H3K4me3 levels (Fig. 1e). SET-2 was relatively specific in regulating lifespan and H3K4me3 in worms, as neither SET-9 nor SET-15 affected global H3K4me3 levels (Supplementary Fig. 1f), even though they both regulate lifespan4. The worm SET-2 methyltransferase domain expressed in bacteria directly methylated histone H3 at lysine 4 in vitro (Fig. 1f). SET2 generated H3K4me2 but not H3K4me3 in vitro (Supplementary Fig. 1g), consistent with the fact that ASH-2 is required for the conversion of H3K4me2 to H3K4me318. To test if ASH-2, WDR-5, and SET-2 act together to regulate lifespan, we performed epistasis experiments. Lifespan extension by ash-2 or wdr-5/tag-125 knock-down was significantly less pronounced in set-2(ok952) mutants than in wildtype (WT) worms (Fig. 1g, combined two-way ANOVA p<0.0001). Similarly, ash-2 knock-down did not further extend the lifespan of wdr-5/tag-125(ok1417) mutant worms (Supplementary Fig. 1h, p=0.1534). Thus, ASH-2, WDR-5, and SET-2 likely act in the same pathway, perhaps in a complex, to limit lifespan. As ash-2 and wdr-5/tag-125 knock-down slightly extended set-2(ok952) mutant worm lifespan (Fig. 1g, Supplementary Table 2), other methyltransferases may also complex with ASH-2 and WDR-5 to regulate lifespan.
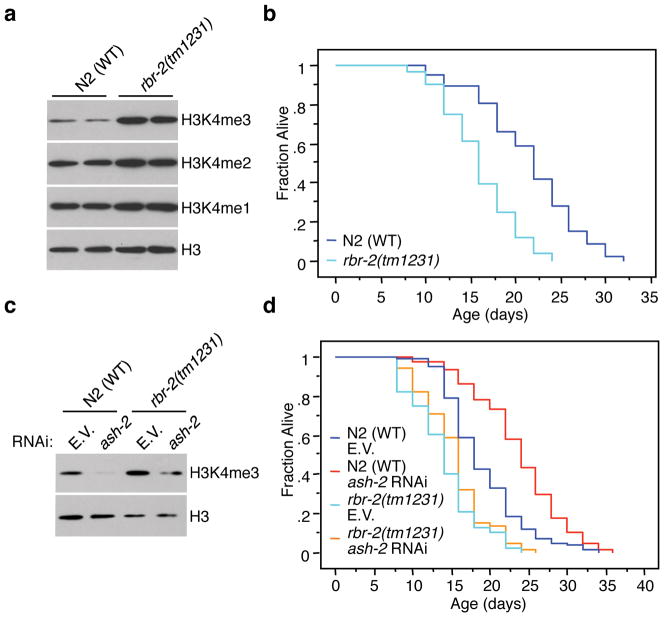
RBR-2 is an H3K4me3 demethylase involved in vulva formation in worms19 and is homologous to human RBP2 and PLU1, two H3K4me3 demethylases of the JARID family19. Consistent with a published report19, rbr-2(tm1231) mutant worms showed increased H3K4me3 levels (Fig. 2a). Both rbr-2(tm1231) mutation and rbr-2 knockdown significantly decreased lifespan (15–25%, p<0.0001 and 10–24.2%, p<0.0001, respectively) (Fig. 2b, Supplementary Fig. 2a), indicating that RBR-2 is necessary for normal longevity. The decrease of H3K4me3 induced by ash-2 knock-down was less pronounced in rbr-2(tm1231) mutant worms than in WT worms (Fig. 2c). In addition, ash-2 knock-down no longer extended rbr-2(tm1231) mutant worms lifespan (Fig. 2d, p=0.4673). Similarly, the lifespan of wdr-5/tag-125(ok1415); rbr-2(tm1231) and set-2(ok952); rbr-2(tm1231) double mutants was similar to that of rbr-2(tm1231) single mutants (Supplementary Fig. 2b, c, p=0.2121 and p=0.6943, respectively). Together, these results indicate that RBR-2 counteracts the effects of ASH-2/WDR-5/SET-2 on H3K4me3 and lifespan.
Fig. 2. RBR-2 is an H3K4me3 demethylase that counteracts the effect of the ASH-2 methyltransferase complex.
a, Western blot of rbr-2(tm1231) mutant worms (representative of 3 independent experiments). b, rbr-2(tm1231) mutant worms have a decreased lifespan. c, Western blots of rbr-2(tm1231) mutant worms treated with ash-2 RNAi (representative of 2 independent experiments). d, ash-2 knock-down does not extend the lifespan of rbr-2(tm1231) mutant worms. ash-2 mRNA was efficiently knocked-down by RNAi in rbr- 2(tm1231) mutant worms (Supplementary Fig. 3a). E.V.: empty vector. Statistics are presented in Supplementary Table 3.
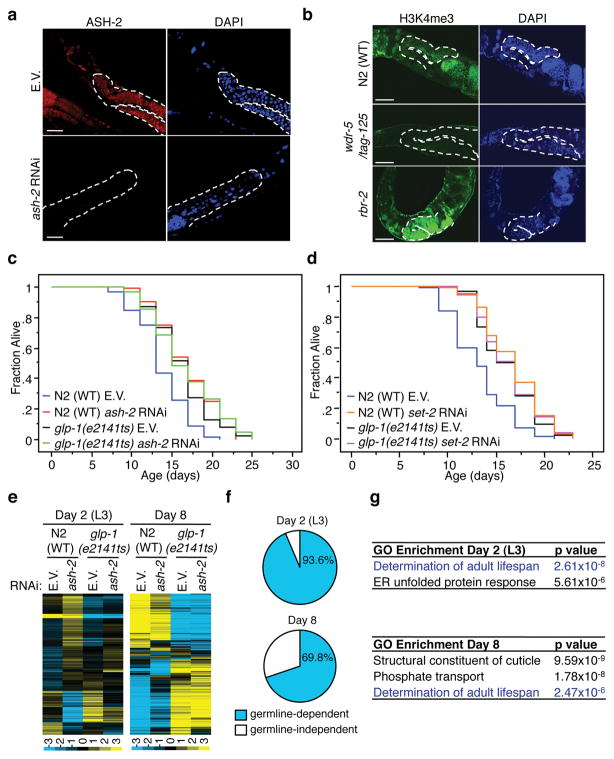
Whole-mount immunocytochemistry with a monoclonal ASH-2 antibody (Supplementary Fig. 4a) revealed that ASH-2 is expressed in the nuclei of cells from many tissues, and is highly expressed in the germline (Fig. 3a). Visualization of an ASH-2::GFP fusion driven by the endogenous ash-2 promoter in low-copy or high-copy transgenic lines confirmed that ASH-2 is highly expressed in the germline and in newly formed eggs (Supplementary Fig. 4b, c). Visualization of an RBR-2::GFP fusion driven by the endogenous rbr-2 promoter in low-copy or high-copy transgenic lines showed expression of RBR-2 in the nuclei of cells from many tissues, although RBR-2 was not particularly enriched in the germline or eggs (Supplementary Fig. 4d, data not shown). The H3K4me3 mark was present in the nuclei of cells from all tissues, with high abundance in the germline in L3 and young adult worms (Fig. 3b, Supplementary Fig. 4e), consistent with the high levels of ASH-2 and SET-220 in the germline. As expected, wdr-5/tag-125(ok1417) and set-2(ok952) mutant worms had markedly reduced H3K4me3 levels (Fig 3b, Supplementary Fig. 4e). Conversely, rbr-2(tm1231) mutant worms showed increased H3K4me3 levels, particularly in the germline (Fig 3b, Supplementary Fig. 4e). These results confirm that members of the ASH-2 complex and RBR-2 regulate H3K4me3, and that this mark is abundant in the germline.
Fig. 3. An intact germline is necessary for longevity and gene expression control by the H3K4me3 regulatory complex.
a, b, Whole-mount immunofluorescence of young adult worms stained with an ASH-2 antibody (a) or an H3K4me3 antibody (b). Dashed lines marks the germline. Scale bars: 50 μm. c, ash-2 knock-down does not further extend the long lifespan of glp-1(e2141ts) mutant worms that were shifted to the restrictive temperature at the L1 stage. d, set-2 knock-down does not further extend the long lifespan of germline-deficient glp-1(e2141ts) mutant worms that were shifted to the restrictive temperature at the L1 stage. Statistics are presented in Supplementary Table 4. e, Microarray clusters of genes that change significantly upon ash-2 knock-down in wildtype worms, but not in glp-1(e2141ts) mutants. Experimental values are presented in Supplementary Tables 8 and 9. f, Percentage of genes regulated by ASH-2 only in WT worms but not in glp-1(e2141ts) mutants. g, Top gene ontology (GO) terms for genes regulated by ASH-2 in wildtype worms, but not in glp-1(e2141ts) mutants. E.V.: empty vector.
To determine if the presence of an intact germline is necessary for lifespan regulation by ASH-2, we examined the effects of ash-2 knock-down in glp-1(e2141ts) mutant worms, which develop only 5-15 meiotic germ cells instead of ~1,500 when shifted to the restrictive temperature at the L1 stage21,22. ash-2 knock-down did not further extend the long lifespan of glp-1(e2141ts) mutant worms (Fig. 3c). We verified that at the permissive temperature, ash-2 knock-down extended glp-1(e2141ts) mutant worm lifespan (Supplementary Fig. 5a, b). Consistent with these observations, ash-2 knockdown did not extend the lifespan of sterile glp-4(bn2ts) or pgl-1(bn101ts) mutant worms (Supplementary Fig. 5c, d). The presence of an intact germline was also necessary for lifespan regulation by WDR-5, SET-2, and RBR-2 (Fig. 3d, Supplementary Fig. 5e, f, g). In contrast, lifespan regulation by SET-9 and SET-15 did not depend on an intact germline (Supplementary Fig. 6a, b), suggesting that not all methyltransferases require the germline to control lifespan.
We performed a genome-wide analysis to identify ASH-2 regulated genes that are dependent on the presence of an intact germline. ash-2 knock-down led to changes in the expression of 220 genes at the L3 stage and 847 genes at day 8 of life (day 5 of adulthood) in WT worms (Fig. 3e, Supplementary Tables 6–9). The majority of ASH-2 controlled genes were regulated in a germline-dependent manner, as their expression was affected in WT worms, but not in glp-1(e2141ts) mutant worms (93.6% at L3 and 69.8% at day 8 of life) (Fig. 3f). Interestingly, one of the top gene ontology (GO) enrichment categories for genes regulated by ASH-2 at L3 and day 8 was ‘determination of adult lifespan’ (Fig. 3g, p=2.61×10−8 and p=2.47×10−6, respectively). Furthermore, at both L3 and day 8 of life, ASH-2 regulated genes were also significantly enriched (2 fold, p=5×10−6 and 1.5 fold, p=6×10−6, respectively) for genes whose expression changed during aging23.
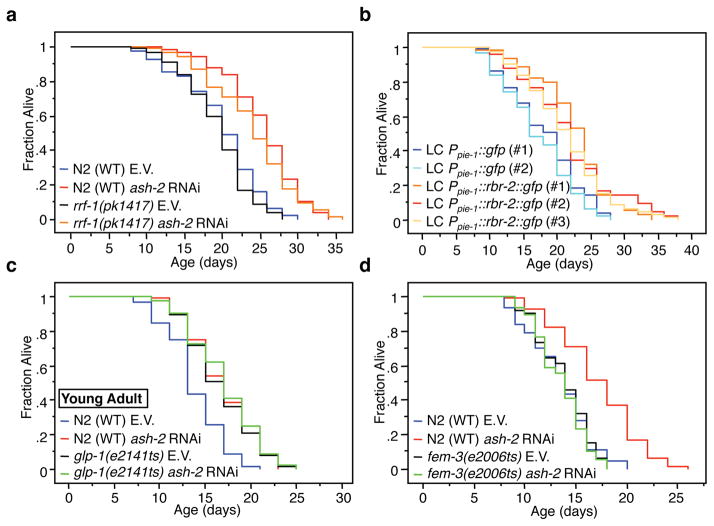
To determine if members of the ASH-2 complex function in the germline itself to regulate lifespan, we used rrf-1(pk1417) mutant worms in which RNAi is efficient in the germline, but not in somatic cells24. ash-2 or set-2 knock-down still extended lifespan in rrf-1(pk1417) mutants (23.6%, p<0.0001 and 22.5%, p<0.0001, respectively) (Fig. 4a, Supplementary Fig. 7a), indicating that ASH-2 and SET-2 function in the germline to regulate lifespan. Conversely, to test if RBR-2 expression in the germline was sufficient to extend lifespan, we generated low-copy integrants of RBR-2::GFP driven by the germline specific pie-1 promoter (Ppie-1::rbr-2::gfp). Three independent Ppie-1::rbr-2::gfp lines had an extended lifespan compared to two independent Ppie-1::gfp lines (17.9%-30.7%, p<0.005) (Fig. 4b), indicating that expression of RBR-2 in the germline is sufficient to prolong worm lifespan. High-copy Prbr-2::rbr-2::gfp transgenic worms also had a modest increase in lifespan (10-14%, p<0.005) (Supplementary Fig. 7b). Because high-copy transgenic worms tend to have suppression of transgene expression in the germline25, these results suggest that expression of RBR-2 in somatic cells may also contribute to lifespan extension, or that very low amounts of RBR-2 expressed in the germline in these high-copy transgenics may be sufficient to extend lifespan. Thus, the ASH-2 methyltransferase complex and the RBR-2 demethylase act in the germline, at least in part, to regulate lifespan.
Fig. 4. ASH-2/RBR-2 function primarily in the germline to regulate lifespan and require the continuous production of mature eggs for lifespan extension.
a, ash-2 RNAi extends the lifespan of rrf-1(pk1417) mutant worms, which are deficient for RNAi in the soma, to a similar extent as in WT (N2) worms. b, Three independent lines of low-copy (LC) integrant Ppie-1::rbr-2::gfp transgenic worms have increased lifespan compared to two independent lines of Ppie-1::gfp transgenic worms. c, ash-2 knock-down does not further extend the long lifespan of glp-1(e2141ts) mutant worms that were shifted to the restrictive temperature at the young adult stage. d, ash-2 knockdown does not extend the lifespan of fem-3(e2006ts) mutant worms, which do not produce mature eggs. ash-2 mRNA was efficiently knocked-down by RNAi in fem-3(e2006ts) mutants (Supplementary Fig. 3b). E.V.: empty vector. Statistics are presented in Supplementary Tables 4 and 5.
The ASH-2 complex and RBR-2 do not appear to regulate lifespan by simply affecting progeny production. ash-2 knock-down worms were fertile and did not have significant differences in number of eggs laid (Supplementary Fig. 8a), number of live progeny (Supplementary Fig. 8b), germline cell number (Supplementary Fig. 8c), and germline morphology (data not shown). Similarly, set-2(ok952), wdr-5/tag-125(ok1417), and rbr-2(tm1231) mutant worms did not have significant differences in egg number and were mostly fertile (Supplementary Fig. 8d).
We next asked if ASH-2 and RBR-2 require an intact adult or developing germline to regulate lifespan. Knocking-down ash-2 and rbr-2 in young adults, after the germline has developed, still affected lifespan (ash-2: +26%, p<0.0001; rbr-2: −13.5%, p<0.0001) (Supplementary Fig. 9a, b). Furthermore, ash-2 knock-down no longer extended lifespan in glp-1(e2141ts) mutant worms switched to the restrictive temperature at the young adult stage(Fig. 4c), whereas ash-2 knock-down extended lifespan in glp-1(e2141ts) mutant worms that were switched to the restrictive temperature after the egg laying period (Supplementary Fig. 9c). Similar observations were made with glp-4(bn2ts) mutant worms (Supplementary Table 4). Consistently, ash-2 knock-down no longer extended lifespan in another glp-1 mutant, glp-1(e2142ts), which has a fully developed germline, but no adult germline stem cells or eggs (Supplementary Fig. 9d). Finally, ash-2 knockdown did not further extend the long lifespan of worms treated with 5-fluorodeoxyuridine (FUdR), a drug that inhibits the proliferation of germline stem cells and the production of intact eggs in adults26 (Supplementary Fig. 9e). Similarly, rbr-2 knock-down did not decrease–and even slightly extended–worm lifespan in the presence of FUdR (Supplementary Fig. 9f), consistent with a previous study27. Thus, ASH-2 regulates lifespan in adult worms by a mechanism that depends on the presence of adult germline stem cells and/or the continuous production of eggs.
To distinguish if ASH-2 regulation of lifespan was dependent on intact germline stem cells or the continuous production of intact eggs, we used fem-3(e2006ts) mutant worms, which have functional germline stem cells, but do not produce fertilized eggs28. ash-2 knock-down no longer extended the lifespan of fem-3(e2006ts) mutant worms (Fig. 4d), suggesting that the production of mature eggs, rather than germline stem cells, is necessary for ash-2 knock-down to extend lifespan. Furthermore, ash-2 knock-down still significantly extended the lifespan of daf-16(mu86) and daf-12(m20) mutant worms, two genes involved in signaling pathways that are critical for germline stem cells to regulate lifespan21,22,29 (Supplementary Fig. 10a, b). Consistently, set-2(ok952) mutant worms displayed lifespan extension even in the presence of daf-16 RNAi (Supplementary Table 2). These results suggest that the ability of members of the ASH-2 complex to regulate lifespan depends on the continuous production of mature eggs, but not on the germline stem cell signaling pathway.
Here we show that members of an H3K4me3 methyltransferase complex play a pivotal role in the regulation of longevity in C. elegans. Our results suggest that the ASH-2 complex and RBR-2 regulate aging at least in part by controlling trimethylation of H3K4, although they might also act by controlling the methylation of non-histone proteins. ASH-2 regulates the expression of a specific subset of genes enriched for lifespan determination genes, but more global changes in chromatin state and gene expression–and therefore quantity of protein produced–may also be responsible for the modulation of lifespan by ASH-2. The observation that the continuous production of mature eggs throughout adulthood is necessary for ASH-2 to regulate lifespan in C. elegans may explain why the ASH-2 trimethylase complex was not identified in earlier screens for aging genes in C. elegans, in which the production of mature eggs was inhibited4,10,11. ASH-2/RBR-2 may function in the germline–perhaps in the maturing–eggs to control endocrine hormones that would, in turn, regulate longevity of the soma. Alternatively, ASH-2/RBR-2 may control the expression of genes involved in lifespan determination, and additional signals from the germline/mature eggs may be required to allow lifespan extension. The regulation of H3K4me3 by specific methyltransferase complexes may regulate aging in a germline-dependent manner in other species. Male flies heterozygous for the H3K4me3 methyltransferase trx had a normal lifespan8, but trx may not be the specific H3K4me3 methyltransferase that regulates lifespan, or the H3K4me3 regulatory complex may be more important in females/hermaphrodites than in males for controlling longevity. The finding that aging can be regulated epigenetically raises the intriguing possibility that aspects of the aging process could be reversed.
Methods Summary
Lifespan assays
Worm lifespan assays were performed at 20°C, without FUdR, as described previously30 unless noted otherwise. For each lifespan assay, 90 worms per condition were used in three plates (30 worms per plate), except for the initial RNAi screen, which was performed with 30-60 worms per condition.
Statistical analysis
Statistical analyses of lifespan were performed on Kaplan-Meier survival curves in StatView 5.0.01 by Logrank (Mantel-Cox) tests. For statistical comparison of independent replicates, Fisher’s combined probability tests were performed. To compare the interaction between genotype and RNAi, two-way ANOVA tests were performed.
Microarray analysis
N2 (WT) and glp-1(e2141ts) mutant worms were hatched at 16°C and switched to bacteria with control empty vector (E.V.) or ash-2 RNAi and moved to the restrictive temperature of 25.5°C at the L1 stage. RNA was isolated at larval stage L3 or day 8 of life. Each condition was done in independent triplicate with ~1000 worms per triplicate for L3 worms, and ~200 worms picked every day after adulthood to fresh plates per triplicate for day 8 worms. Total RNA was isolated using RNAqueous kit (Ambion). Microarray hybridization was performed at the Stanford Protein and Nucleic Acid facility with oligonucleotide arrays (Affymetrix, GeneChip C. elegans Genome Arrays). Background adjustment and normalization was performed with RMA (Robust Multiarray Analysis).
Full methods accompany this paper.
Supplementary Material
Acknowledgments
We are grateful to A. Fire, K. Helin, S. Kim, K. Shen, T. Stiernagle and the Caenorhabditis Genetics Center, M.W. Tan, and A. Villeneuve for gifts of strains, reagents, and antibodies. We thank S. Kim, G. Seydoux, C. Slightman, and K. Shen and for advice on worm transgenesis, and S. Kim, A. Morgan, and Y. Kobayashi for help with microarray analysis. We thank A. Fire, S. Kim, G. Seydoux, M.W. Tan, and J. Wysocka for helpful discussions. We thank members of the Brunet lab, M. Kaeberlein, J. Lieberman, J. Sage, and J. Wysocka for critical reading of the manuscript. This work was supported by NIH grant R01-AG31198 to A.B.. E.L.G was supported by NIH training grant T32-CA009302, an NSF graduate fellowship, and by NIH ARRA-AG31198. T.J.M. was supported by NIH grant T32-HG000044. D.S.L. and E.M.G. were supported by NIH training grant T32-CA009302. S.H. was supported by a Stanford graduate fellowship. G.S.M. was supported by a Human Frontier Science Program post-doctoral fellowship. M.R.B was supported by NIH fellowship F31-AG032837. O.G. was supported by a Searle Scholar award.
Footnotes
Author Contributions
E.L.G. and A.B. conceived and planned the study. E.L.G. performed the experiments and wrote the paper with the help of A.B.. T.J.M. completed Fig. 3a, b, Supplementary Fig. 4c, e, and Supplementary Fig. 8c, and generated all low-copy integrant transgenic worm lines. A.G.H. helped with Fig. 1b, e, and Supplementary Fig. 1f. E.M.G. completed Fig. 1f and Supplementary Fig. 1g. D.S.L. helped with Fig. 2a. G.S.M. generated all high-copy transgenic worm lines. S.H. helped with Supplementary Fig. 8c. M.R.B. generated the Prbr-2::rbr-2::gfp construct. All authors discussed the results and commented on the manuscript.
References
- 1.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 2.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton B, et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, et al. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McColl G, et al. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, et al. The conserved NAD(H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc Natl Acad Sci USA. 2009;106:1496–1501. doi: 10.1073/pnas.0802674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siebold AP, et al. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci USA. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature genetics. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 11.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS genetics. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller T, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papoulas O, et al. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 14.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 15.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Simonet T, Dulermo R, Schott S, Palladino F. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol. 2007;312:367–383. doi: 10.1016/j.ydbio.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 17.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Schneider J, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Strome S. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics. 2001;159:1019–1029. doi: 10.1093/genetics/159.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 22.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 23.Budovskaya YV, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 25.Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell DH, Stiles JW, Santelli J, Sanadi DR. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34:28–36. doi: 10.1093/geronj/34.1.28. [DOI] [PubMed] [Google Scholar]
- 27.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 28.Haag ES, Wang S, Kimble J. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr Biol. 2002;12:2035–2041. doi: 10.1016/s0960-9822(02)01333-7. [DOI] [PubMed] [Google Scholar]
- 29.Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- 30.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.