Abstract
Catalysed by members of the adenosine deaminase acting on RNA (ADAR) family of enzymes, adenosine-to-inosine (A-to-I) editing converts adenosines in RNA molecules to inosines, which are functionally equivalent to guanosines. Recently, global approaches to studying this widely conserved phenomenon have emerged. The use of bioinformatics, high-throughput sequencing and other approaches has increased the number of known editing sites by several orders of magnitude, and we now have a greater understanding of the control and the biological significance of editing. This Progress article reviews some of these recent global studies and their results.
RNA editing is a post-transcriptional process that covalently alters the sequence of an RNA molecule. A well-studied form of such editing is adenosine-to-inosine (A-to-I) RNA editing, which involves the conversion of adenosine residues to inosine residues, specifically in dsRNA structures1–3. A-to-I RNA editing is conserved from humans to sea anemones4 and is catalysed by enzymes of the adenosine deaminase acting on RNA (ADAR) family. Inosine preferentially base pairs with cytidine and is therefore functionally equivalent to guanosine. Thus, A-to-I editing can change the fate of an RNA molecule in numerous ways, including recoding its codons, creating or destroying splice sites and altering its role in the RNAi phenomenon.
It is therefore not surprising that malfunction of the editing machinery has been implicated in various human ailments5. Depression and schizophrenia, amyotrophic lateral sclerosis (ALS) and neuronal apoptosis following disruption of blood flow to the brain are all associated with abnormal editing patterns5. Furthermore, ADAR2 (also known as ADARB1) is needed to prevent hyperuricaemia6 in certain tissues, and heterozygosity for functional null mutations in the ADAR1 (also known as DRADA) gene results in dyschromatosis symmetrica hereditaria5, a human pigmentary genodermatosis. These examples provide many incentives for the study of A-to-I editing.
In recent years the application of global approaches to the study of A-to-I editing — including bioinformatics and high-throughput sequencing — has led to substantial discoveries that would have been impossible by previous means. These new discoveries range from the identification of a vastly greater number of editing sites to elucidation of the patterns in which editing takes place in a transcript, the regulation of the editing activity and the functional significance of the editing. This article reviews some of these recent global approaches to A-to-I RNA editing and their results.
Identification of editing sites
The existence of widespread A-to-I editing is suggested by the high inosine content of RNA extracts7. However, by 2003 only a handful of editing sites had been discovered, fortuitously, by comparing cDNA sequences derived from expressed RNAs to genomic DNA sequences. As inosine is reverse transcribed as guanosine, the identification of guanosine in cDNA and adenosine in genomic DNA was considered to be a hallmark of A-to-I editing1–3. To close the gap between the putative and the known number of editing sites, new and innovative approaches were needed.
Bioinformatics approaches
Before 2003, the lack of a means to predict editing sites from their sequence context hampered their identification. That year, Hoopengardner et al.8 discovered that many editing sites are situated in especially conserved regions. Formation of a dsRNA structure that is suitable as an ADAR substrate requires an editing-site complementary sequence (ECS) lying elsewhere in the transcript to base pair with the sequence surrounding the editing site. Because even a synonymous mutation would disrupt this base pairing, many editing sites that convey evolutionary advantages are in conserved sequences that are recognizable by comparative genomics. By this approach, Hoopengardner et al. identified and verified 16 novel editing sites in Drosophila melanogaster and one in humans; more recent applications of their method9 have extended these results. However, the total number of known editing sites remained limited, and not all editing sites are located within such conserved regions.
Because an A-to-I RNA editing event is detected as an A-to-G change in the cDNA sequence, one can also identify candidate editing sites by aligning and comparing publicly available cDNA sequences from expressed RNAs with their corresponding genomic sequences (FIG. 1a). The problem is to separate true editing events from the background noise of A-to-G changes that are caused by SNPs, somatic mutations, pseudogenes and sequencing errors. This background noise can be minimized by ignoring A-to-G changes that correspond to validated SNPs and discarding cDNA sequences that are indicated to be of low quality by the presence of many mismatches in the cDNA–genome alignments. However, the false-discovery rate, as estimated by comparing the number of G-to-A and A-to-G mismatches detected in the same alignment, is still intolerable.
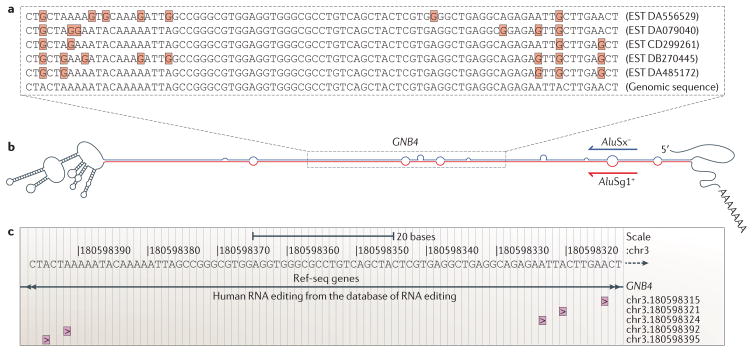
Figure 1. Adenosine-to-inosine editing and bioinformatics.
Recently, bioinformatics has been important in the field of adenosine-to-inosine (A-to-I) editing both as integral parts of many studies and in disseminating the results. a | When comparing publicly available ESTs to genomic sequences, multiple editing leads to clusters of adenosine-to-guanosine differences (highlighted by the Gs in red boxes) that are unlikely to originate from sources other than RNA editing10–12. b | Reversely oriented Alu sequences within the same transcript can form highly double-stranded RNA structures that are edited at multiple positions1–3, as exemplified by part of the 3′ untranslated region of the transcript encoding guanine nucleotide binding protein (G protein), beta polypeptide 4 (GNB4), the structure of which is shown here as predicted by mfold38. c | Editing sites identified in this and other ways have now been gathered in the new database, the Database of RNA Editing (DARNED)37, which can be accessed through the UCSC genome browser. As shown here, five out of 14 editing-site candidates inferable from the EST data shown are currently in DARNED.
A strategy developed in 2004 overcame this problem by considering only clustered editing-site candidates10–12. Within such clusters, which are caused by especially long and stable dsRNA structures (FIG. 1b), editing sites could be predicted with reasonable confidence, and this raised the number of known sites by orders of magnitude. Variations on this strategy have also been the basis for later studies: a recent paper identified many editing sites in humans, mice and Xenopus tropicalis that had previously been overlooked, by estimating the quality of each cDNA sequence from its sequence chromatogram, as available from the NCBI Trace Archive, rather than from the cDNA–genome alignment13.
The disadvantage of focusing on editing-site clusters is that they localize almost exclusively to the promiscuously edited double-stranded regions that are formed between two reversely oriented Alu sequences in the same transcript. These primate-specific, 300 nucleotide sequences, of which the human genome contains ~1.4 million, localize mostly to non-coding regions, and the biological significance of editing within such regions largely remains to be established.
By comparison, the significance of editing sites found within coding sequences can be tested more easily, and therefore much recent effort has focused on identifying such sites. One study did this by examining the Alu-less mouse genome14. A search within the coding sequences of more than 10,000 well-annotated mouse genes identified ~2,500 conserved regions with the potential to form dsRNA structures. Deep sequencing of 45 regions that were considered particularly promising found ten new editing sites, eight of which recode codons. However, each of these sites is only edited in 0.6% to 2.4% of transcripts. With no evidence of a gain of function, which could make any minute level of editing significant, the editing sites of known biological significance remain few.
So, although bioinformatics approaches have raised the number of known editing sites by orders of magnitude, they also have short-comings. They are blind to editing events that are not represented in the raw data, the confidence with which they report editing sites is often low, and their screening procedures can bias results. For example, screening for clustering causes over-representation of Alu editing. Bioinformatics methods are, however, well complemented by high-throughput sequencing technology.
High-throughput sequencing approaches
As explained above, a bioinformatics search for editing sites that is not based on clustering will report many candidates without a bias towards Alu sequences, albeit each with intolerably low confidence. For these putative editing sites, Li et al.15 developed a large-scale verification procedure using high-throughput sequencing.
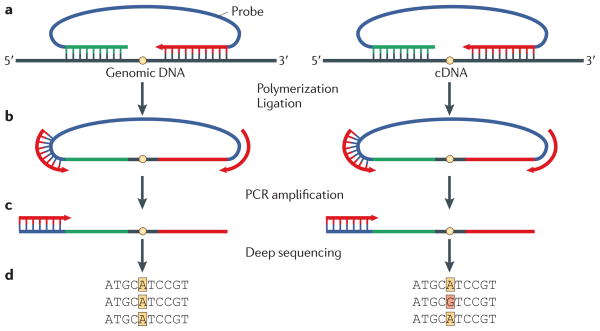
Using samples from a single human individual, 36,000 editing-site candidates lying outside repetitive sequences were captured by padlock probes, amplified and sequenced (FIG. 2). This was done in parallel for cDNA from seven tissues and genomic DNA. In this way, 239 sites — 55 in coding sequences, of which 38 caused recoding events — were identified for which only adenosine was seen in genomic DNA, whereas at least 5% of cDNA reads were guanosine in at least two of the seven tissues. A more detailed analysis of 18 of these sites verified 14, indicating a false-discovery rate of about one in five.
Figure 2. Padlock capture, amplification and sequencing of editing-site candidates.
To validate or reject an editing-site candidate, shown as a yellow dot on the genomic DNA or transcript-derived cDNA, the padlock approach can be used15. a | A padlock probe is made to anneal to both sides of a candidate site on both genomic DNA and cDNA, and it is then extended by a DNA polymerase and ligated by a DNA ligase. b | This forms a circular piece of DNA that contains the complement of the candidate site, which is amplified by a PCR reaction. c | Individual strands of the PCR product are then sequenced by deep sequencing. d | If only adenosine is seen in the genomic DNA-derived site, but a fraction of the cDNA-derived site contains guanosine, this indicates A-to-I editing. Figure modified, with permission, from REF. 15 © (2009) American Association for the Advancement of Science.
Although this error rate is an improvement on previous methods, it is still significant. In particular, caution is needed when looking for sites that are edited to a small extent or in few tissues. As our ability to carry out large-scale sequencing improves, experiments of this kind will probably have an increasing role in A-to-I editing research. Even so, a place remains for research using less standardized methods to identify large sets of edited transcripts.
Other approaches
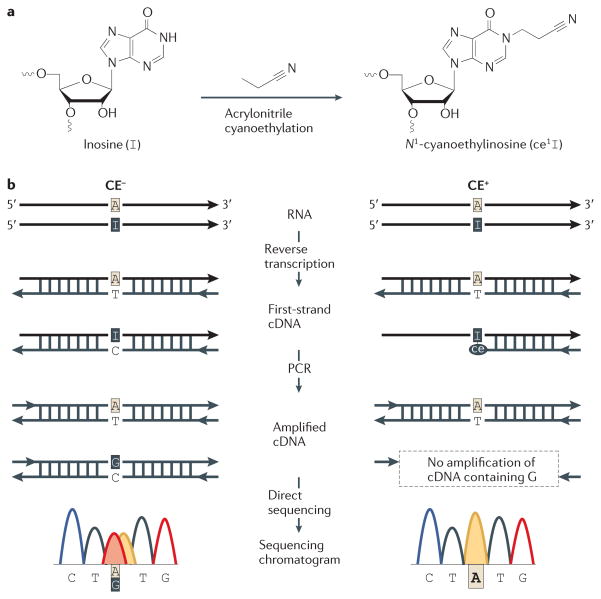
A difficulty with any of the research discussed above is separating true editing sites from false discoveries. Recently, Sakurai et al.16 reported a biochemical method to overcome this problem. The method, named inosine chemical erasing (ICE), uses inosine-specific cyanoethylation (FIG. 3a), reverse transcription, PCR and direct sequencing to detect true editing sites (FIG. 3b). In sequencing chromatograms, it enables guanosines that originate from inosine to be distinguished from those that originate from SNPs, somatic mutations, pseudogenes or sequencing errors — without requiring genomic DNA for reference or the isolation of total RNA from cells in which ADAR is inactivated by genetic depletion or RNAi. Application of this method to 1,277 predicted editing sites in human transcripts demonstrated its scalability. On the basis of sequencing chromatograms, 716 sites had guanosines in more than 10% of transcripts. The ICE method validated 677 of these guanosine peaks as originating from editing, whereas the remaining 39 seem to stem from other sources. Further searches for editing sites led to the identification and ICE validation of 4,395 novel human editing sites.
Figure 3. Verification of editing sites by the inosine chemical erasing (ICE) method.
a | Inosine can be cyanoethylated by acrylonitrile, an ability that is not shared by the Watson–Crick bases. This can be used to validate editing sites. b | Sequencing of cDNA that has been reverse transcribed from an edited transcript will give a mixed adenosine–guanosine peak at the editing site (CE−). However, this mixed peak can also have other origins. Cyanoethylation before reverse transcription prevents cDNA from being made from the edited transcripts. A true editing site can therefore be validated by the disappearance of the guanosine peak in the sequencing chromatogram following cyanoethylation (CE+)16. Figure modified, with permission, from REF. 16. © (2010) Macmillan Publishers Ltd. All rights reserved.
Innovative, non-standard ways to examine RNA editing such as ICE could both improve and complement the tools that are currently used. Much of current research relies on known editing sites, which makes the ways we identify more of these sites worthy of optimization. That said, the positions of editing sites is only one of the interesting facets of ADAR editing.
Editing patterns
One other aspect of A-to-I editing that we would like to understand is whether several editing sites in the same transcript are edited independently or in a coordinated manner. To answer this question, Ensterö et al.17 applied deep sequencing to analyse the sequences of ~570 individual transcripts from each of three selectively edited mouse genes. Interestingly, positive coupling was found between editing sites separated by 11–14 nucleotides; it seemed that editing first takes place at a highly edited site, and only afterwards on the coupled sites. This suggests that, initially, one ADAR binds an especially favourable editing site, and it then attracts other ADARs to the nearby coupled sites by protein–protein interactions. Short chains of ADARs could thus bind an RNA molecule in register. Notably, this pattern of coupled editing does not seem to extend to promiscuously edited Alu sequences18,19.
Regulation of editing
The spatiotemporal regulation of A-to-I editing is another enigma, because the extent of editing is not well correlated with ADAR expression levels1,2. By detailing changes in the extent of editing, two complementary large-scale studies have recently been applied to this problem.
First, Wahlstedt et al.20 tracked the extent of editing at 29 editing sites across 11 selectively edited transcripts during mouse brain development. Editing at most of these sites gradually increased from embryogenesis to adulthood. At a few sites the change was less gradual, and at some point in development it makes a jump, but editing increases over developmental time overall.
Second, Osenberg et al.21 looked at editing levels during differentiation of human embryonic stem cells. Using both direct sequencing and Sequenom MassARRAY technology to assess the levels of editing within eight Alu sequences and the coding regions of five genes, they found that both promiscuous and selective editing decreased during differentiation to different extents in different transcripts.
By detailing changes in the extent of editing, these studies could illuminate how yet unknown regulating factors are involved in the editing mechanism and what biological functions editing serves.
Biological significance
Most recently, global approaches have also helped us to improve our understanding of the biological significance of A-to-I editing. This editing has been implicated in many phenomena, including tuning of RNAi activity, modulation of neuronal signalling, control of microRNA (miRNA) function, and establishment of higher brain function in humans1–3. Global approaches have been applied particularly to study the last two processes.
Control of miRNA expression and functions
miRNAs are endogenous non-coding RNAs that suppress translation of transcripts. Primary transcripts of miRNA genes (pri-miRNAs) are processed sequentially by nuclear Drosha and cytoplasmic Dicer to form the ~21 nucleotide mature miRNAs that are loaded onto the RNA-induced silencing complex (RISC)22,23.
The involvement of A-to-I RNA editing in the control of the miRNA biogenesis pathway and the expression of edited mature miRNAs has been reported1,24–26. Both ADAR1 and ADAR2 edit specific adenosine residues of certain pri-miRNAs, such as human pri-miR-142 (REF. 27), pri-miR-151 (REF. 28), pri-miR-376 cluster RNAs6 and Epstein–Barr virus pri-ebv-miR-BART6 (REF 29). Editing of these pri-miRNAs and consequent alterations in their dsRNA structures suppresses their processing by Drosha or Dicer27,28, prevents their loading onto RISC29, or results in expression of miRNAs with altered sequences. This results in silencing of genes different from those targeted by the unedited miRNAs6.
Global and systematic screening for miRNA editing sites has been conducted. Linsen et al.30 examined small rat RNAs and found six miRNAs edited in at least 10% of reads, one being miR-377 and the remaining five being mature products of pri-miR-376. By sequencing small RNAs, Landgraf et al.31 found that 2.2% of human miRNAs showed A-to-G transitions. Interestingly, this editing rate is fourfold higher than the 0.5% rate that they observed for mice. This finding is supported by results from Chiang et al.32, who found no significant over-representation of A-to-G changes compared with G-to-A changes in mature mouse miRNAs, and from de Hoon et al.33, who showed that the vast majority of over-represented RNA–genome mismatches at specific positions in the mouse genome were caused by incorrect mapping of RNA to the genome rather than by RNA editing.
Taken together, these results indicate rates of miRNA editing that are surprisingly low when compared to the number of known functionally significant miRNA editing events. This could be because editing often prevents the maturation of the miRNA26 and because editing may lead to the degradation of the miRNA27, which would prevent detection of the editing in these screens.
Modulation of miRNA target sites
Transcripts become targets of a miRNA by containing ‘target sites’ to which the miRNA is complementary. Because, as described above, this complementarity can be affected by editing of the miRNA, one may reasonably predict that it could also be modulated by editing of the target site. Owing to the huge number of potential target sites, investigation of this prediction has been done using global-scale methods.
Two studies in particular have bioinformatically assessed how many known editing sites fall within target sites that could be destroyed or perfected by an A-to-G change. Liang and Landweber34 found ~500 such sites in humans, although they point out that these results are reported with low confidence; this is because editing sites tend to be within primate-specific Alu sequences and, without taking cross-species conservation into account, target sites cannot currently be predicted accurately. Furthermore, they noted that the tissues in which the editing took place often differed from those in which the relevant miRNA was expressed. Discounting target site conservation and tissue specificity, Borchert et al.35 identified more than 3,000 human target sites that were potentially created by editing. Among these, they noticed one particularly common 13 nucleotide motif that they verified as being targetable by miRNA-513 and miRNA-769-3p only when edited.
Although this information itself is valuable, it is clear that a further understanding of the scope and the consequences of miRNA target-site editing will follow further progress in target-site and editing- site identification. It will therefore be interesting to see how the knowledge revealed by these two early studies will be extended in the future.
Higher brain function
As we share most of our genomes with other primates, the biological differences underlying our higher brain functions are a great mystery. Particularly because many A-to-I editing sites are identified in transcripts of the nervous system1–3, editing could potentially have a role in supporting human capabilities for reasoning. To investigate this, a recent study compared our currently available data on editing in Alu sequences in humans with those in chimpanzees and rhesus monkeys19. Although editing levels in all three species are higher than in non-primate mammals, editing levels were found to be higher by twofold in humans than in chimpanzees or rhesus monkeys, even when the sequence surrounding the editing site was conserved. Furthermore, among human and chimpanzee genes with newly inserted Alu sequences, genes encoding proteins central to the nervous system are over-represented. This raises the exciting possibility that human A-to-I RNA editing is important to our higher brain functions.
Towards an inosinome
The emergence of biological databases and high-throughput sequencing technology has enabled global experiments that were previously impossible. This has led to the identification of unprecedented numbers of editing sites and the elucidation of some of their functions. It is clear that global experiments will continue to influence A-to-I editing research. Although we now know relatively many editing sites, we know little about how their editing varies by tissue, developmental stage and other parameters. For instance, the procedure developed by Li et al.15 to identify editing sites could be used to compare editing patterns in patients with dyschromatosis symmetrica hereditaria, who are heterozygous for a functional null mutation in the ADAR1 gene5, to those in non-affected controls. This could reveal the specific mechanism underlying the disease. In an early version of such a study, Paz et al.36 looked at ESTs and found that the extent of editing is nearly halved in brain tumours compared with normal brain tissue, implicating editing by ADARs in the control of proliferation rate.
These emerging global methods do not replace traditional approaches, but rather complement them. Global approaches showing that ADARs have propensities to edit sites in particular patterns call for structural and functional studies of how this propensity comes about. Global approaches demonstrating that editing by ADARs is developmentally regulated would benefit from searches for the regulatory factors.
With the surge of global-scale experiments that generate vast amounts of information, the need arises for a unified way to access this data. So far, scientists using such data have had to access files in various formats from individual publications. The recent creation of the common editing-site database, the Database of RNA Editing (DARNED)37 is therefore exciting. DARNED aims to map all editing sites to their corresponding positions on the human genome (FIG. 1c) and to provide relevant information such as their environment (for example, coding, untranslated region or intron), the tissues in which editing has been observed and the supporting evidence.
We believe that such a database, if it continues to grow with and adjust to new experimental results, will become a valuable tool for scientists whether they study RNA editing or other fields. Furthermore, as abnormalities in A-to-I editing have been implicated in various human ailments, a database such as DARNED could become valuable as a diagnostic tool in the future. The assembly of our experimental data into an integrated database is reasonable, given that we are dealing with a single phenomenon — A-to-I editing — that has implications for all processes in the cells of humans and many animals.
Acknowledgments
B.-E.W. is a recipient of a University of Pennsylvania Vagelos scholarship. M.S. is supported in part by a fellowship from the Japan Society for the Promotion of Science. K.N. is supported by grants from the US National Institutes of Health, the Ellison Medical Foundation and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jepson JE, Reenan RA. RNA editing in regulating gene expression in the brain. Biochim Biophys Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, Zhang W, Li Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life. 2009;61:572–578. doi: 10.1002/iub.207. [DOI] [PubMed] [Google Scholar]
- 5.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 9.Levanon EY, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 11.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaranek AW, Levanon EY, Zecharia T, Clegg T, Church GM. A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing. PLoS Genet. 2010;6:e1000954. doi: 10.1371/journal.pgen.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enstero M, et al. A computational screen for site selective A-to-I editing detects novel sites in neuron specific Hu proteins. BMC Bioinformatics. 2010;11:6. doi: 10.1186/1471-2105-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 16.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nature Chem Biol. 2010;6:733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- 17.Enstero M, Daniel C, Wahlstedt H, Major F, Ohman M. Recognition and coupling of A-to-I edited sites are determined by the tertiary structure of the RNA. Nucleic Acids Res. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barak M, et al. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–6915. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paz-Yaacov N, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahlstedt H, Daniel C, Enstero M, Ohman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osenberg S, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS ONE. 2010;5:e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 24.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blow MJ, et al. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawahara Y, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iizasa H, et al. Editing of Epstein-Barr Virus-encoded BART6 MicroRNAs Controls Their Dicer Targeting and Consequently Affects Viral Latency. J Biol Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linsen SE, de Wit E, de Bruijn E, Cuppen E. Small RNA expression and strain specificity in the rat. BMC Genomics. 2010;11:249. doi: 10.1186/1471-2164-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Hoon MJ, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang H, Landweber LF. Hypothesis: RNA editing of microRNA target sites in humans? RNA. 2007;13:463–467. doi: 10.1261/rna.296407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 2009;18:4801–4807. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paz N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 38.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]