Abstract
MTM1, MTMR2, and SBF2 belong to a family of proteins called the myotubularins. X-linked myotubular myopathy, a severe congenital disorder characterized by hypotonia and generalized muscle weakness in newborn males, is caused by mutations in MTM1 (Laporte et al., 1996). Charcot-Marie-Tooth types 4B1 and 4B2 are severe demyelinating neuropathies caused by mutations in MTMR2 (Bolino et al., 2000) and SBF2/MTMR13 (Senderek et al., 2003), respectively. Although several myotubularins are known to regulate phosphoinositide-phosphate levels in cells, little is known about the actual cellular process that is defective in patients with these diseases. Mutations in worm MTM-6 and MTM-9, myotubularins belonging to two subgroups, disorganize phosphoinositide 3-phosphate localization and block endocytosis in the coelomocytes of Caenorhabditis elegans. We demonstrate that MTM-6 and MTM-9 function as part of a complex to regulate an endocytic pathway that involves the Arf6 GTPase, and we define protein domains required for MTM-6 activity.
INTRODUCTION
Myotubularins are a family of phosphatases that can be divided into at least four subgroups based on sequence and phylogenetic analysis (Laporte et al., 2001; Wishart et al., 2001). They have conserved domains that include an N-terminal GRAM domain, which could mediate protein or lipid binding (Doerks et al., 2000), a Rac1-induced localization domain (Laporte et al., 2002a), a phosphatase domain containing the conserved CSDGWDR required for phosphatase activity (Laporte et al., 2001; Wishart et al., 2001), and a SET-interacting domain of unknown function (Laporte et al., 2001). Key residues required for phosphatase activity are absent in the “antiphosphatase” subgroup (which includes SBF2). Furthermore, “substrate-trapping” mutants of catalytically active myotubularins have been made by substituting the conserved nucleophilic cysteine with alanine or serine; these mutants bind more tightly, but not irreversibly, to their substrate (Flint et al., 1997; Blondeau et al., 2000; Laporte et al., 2002a). Finally, some myotubularins contain additional motifs (Laporte et al., 2001), including FYVE domains and plekstrin homology domains, which mediate interactions with specific phosphoinositides (PIs).
Several myotubularins specifically dephosphorylate the 3′-phosphate of phosphoinositide 3-phosphate [PI(3)P] (Blondeau et al., 2000; Taylor et al., 2000; Walker et al., 2001; Zhao et al., 2001; Berger et al., 2002; Schaletzky et al., 2003), and some have been shown to also act on Phosphatidylinositol 3,5-bisphosphate (Walker et al., 2001; Schaletzky et al., 2003). Although, the overexpression of human MTMR3 results in enlarged vacuoles in Saccharomyces cerevisiae (Walker et al., 2001), and the overexpression of human MTM1, MTMR2, or MTMR3 results in enlarged vacuoles in Schizosaccharomyces pombe (Blondeau et al., 2000; Laporte et al., 2002b), no actual defect in uptake has been reported in these cells. Furthermore, overexpression of MTM1 in mammalian cells does not noticeably affect vesicular trafficking (Laporte et al., 2002a). Despite this indirect evidence in support of myotubularin function in membrane traffic, no defect in a specific cellular process has so far been identified in cells lacking an endogenous myotubularin.
By screening for mutants unable to internalize green fluorescent protein (GFP) secreted into the body cavity, we previously identified mutations in 14 cup (coelomocyte up-take defective) genes that disrupt endocytosis by Caenorhabditis elegans coelomocytes, scavenger cells with extremely high endocytic activity (Fares and Greenwald, 2001a). We describe here the molecular identification of CUP-6 and CUP-10 as two of the six myotubularins found in worms (Laporte et al., 2001; Wishart et al., 2001). We also demonstrate a function for these two proteins in an ARF-6- and RME-1-mediated endocytic pathway.
MATERIALS AND METHODS
C. elegans Strains and Methods
Standard methods were used for genetic analysis (Brenner, 1974). Extrachromosomal arrays were generated by coinjecting restriction digested plasmids and marker DNA, each at 1–10 μg/ml along with EcoRV digested genomic DNA at 100 μg/ml into the germline (Mello and Fire, 1995). Several transgenic lines were generated and analyzed from each injection. RNA-mediated interference (RNAi) was carried out as described previously (Xue et al., 2003) by microinjection of double-stranded RNA synthesized in vitro or by feeding bacteria that produce it. Markers used mtm-6(ar513) III (Fares and Greenwald, 2001a), mtm-6(ar515) III (Fares and Greenwald, 2001a), mtm-6(ok330) III (Gene Knockout Consortium), mtm-9(ar479) V (Fares and Greenwald, 2001a), and arIs37[pmyo-3::ssGFP] I (Fares and Greenwald, 2001b). cdIs5 I is similar to arIs37 but uses the red fluorescent protein DsRed2 (BD Biosciences Clonetech, Palo Alto, CA) instead of GFP.
Molecular Methods
Standard methods were used for the manipulation of recombinant DNA (Sambrook et al., 1989). All enzymes were from New England Biolabs (Beverly, MA), unless otherwise indicated. Site-directed mutagenesis of MTM-6 was done using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA).
cDNA Sequencing
yk clones (Kohara, personal communication) corresponding to MTM-6 and MTM-9 cDNAs were sequenced in their entirety. The 5′ ends of the MTM-6 and MTM-9 mRNAs were determined using 5′ rapid amplification of cDNA ends (Roche Diagnostics, Indianapolis, IN).
Plasmids
All DNA constructs were cloned in front of a coelomocyte-specific promoter (Fares and Greenwald, 2001b). GFP(S65T) (Chalfie et al., 1994) or mRFP1 (Campbell et al., 2002), a monomeric form of DsRed, was used for making all fusion proteins; these markers were introduced at the N terminus of RME-1, or at the C terminus of MTM-6, MTM-9, ARF-6, and 2 × FYVE(Hrs). Y116A8C.12 is the C. elegans ARF-6 (Fares, unpublished data). MTM-6, MTM-9, and RME-1 fusion proteins rescued their corresponding mutants. Endocytosis was normal in coelomocytes expressing ARF-6 (for which no mutation exists) and 2 × FYVE(Hrs) fusion proteins. Details of plasmid constructions are available upon request.
Immunoprecipitation
GFP-MTM-6A and Flg-MTM-9 were transfected into human 293 tissue culture cells either alone or together. Total cell lysate (250 mg) was subjected to immunoprecipitation with antibodies against GFP or Flg. The immunoprecipitates were washed three times with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.2, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 1% Trasylol), separated by SDS gel electrophoresis (10%), and after transfer to nitrocellulose, Western blotted with antibodies to either GFP or Flg. Total cell lysates (50 μg) were run as a control to show equal levels of protein expression in the transfected cells.
Bovine Serum Albumin-Rhodamine (BSA-Rhod) Endocytosis As described previously (Fares and Greenwald, 2001b; Zhang et al., 2001), BSA-Rhod (Sigma-Aldrich, St. Louis, MO) dissolved at 1 mg/ml in water was injected into the pseudocoelom of young adult hermaphrodites at 20°C. At the indicated times, the worms were mounted in a drop of ice-cold 1% formaldehyde, and slides were kept on ice until viewing.
Microscopy
Worms were fixed in 1% formaldehyde. Deconvolution images were recorded as digital images (with Z-series containing 0.15-μm sections) by using a Series 300 charge-coupled device camera (Photometrics, Tucson, AZ) and deconvolved using DeltaVision software (Applied Precision, Issaquah, WA). Confocal images were taken with a Nikon PCM 2000, by using HeNe 543 excitation for the red dye and argon 488 for the green dye.
Accession Numbers
Accession numbers were MTM-6A (AY313177), MTM-6B (AY313179), and MTM-9 (AY313178).
Supplemental Material
Figures showing the gene structures of mtm-6 and mtm-9 are available on the Molecular Biology of the Cell Web site.
RESULTS
Identification of CUP-6 and CUP-10 as Myotubularins
Mutations in cup-6 and in cup-10 result in decreased coelomocyte endocytosis and hence increased accumulation of GFP in the body cavity of GFP-secreting pmyo-3::ssGFP worms (Fares and Greenwald, 2001a; Figure 1). Starting with the map positions of cup-6 and of cup-10 (Fares and Greenwald, 2001a), we determined that CUP-6 and CUP-10 correspond to C. elegans myotubularins (worm open reading frames F53A2.8 and Y39H10A.3, respectively) based on phenocopy by RNAi (Fire et al., 1998; Xue et al., 2003), transgenic rescue of the mutant phenotypes and sequence analysis of the various alleles (see below). It should be noted that coelomocytes are partially resistant to RNAi, such that the endocytosis defect in coelomocytes after cup-6 or cup-10 RNAi is not as severe or as penetrant compared with mutations in the endogenous genes (Xue et al., 2003; Fares, unpublished data).
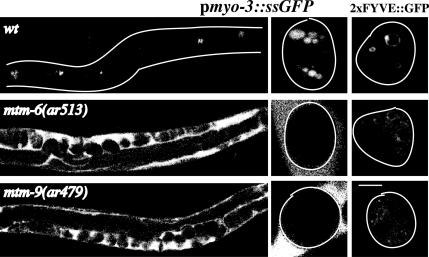
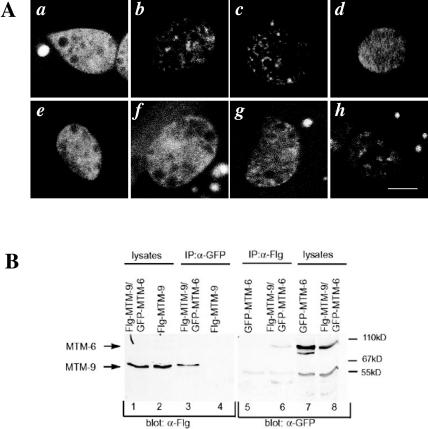
Figure 1.
Endocytosis defects of mtm mutant strains. Shown are confocal images of wild-type, mtm-6(ar513), and mtm-9(ar479) worms either expressing pmyo-3::ssGFP (left two columns) or 2 × FYVE::GFP in their coelomocytes (right column). Worms carrying pmyo-3::ssGFP secrete GFP from their muscles into the body cavity. The coelomocytes (bright spots in the wild-type worms) endocytose the GFP from the body cavity. Defects in endocytosis result in GFP accumulating in the body cavity. The left column is a low-magnification image; the middle and last column are higher magnification images showing individual coelomocytes. Outlines of worms and coelomocytes are drawn when needed. Bar, 5 μm for the high-magnification images.
Specific effects of cup-6 and cup-10 mutations on phosphoinositides indicate functional conservation of C. elegans and mammalian myotubularins. Direct visualization of intracellular PI(3)P by using a GFP-tagged, PI(3)P reporter 2 × FYVE(Hrs)::GFP (Gillooly et al., 2000) showed that both genes are required for PI(3)P localization in vivo (Figure 1). Thus, mutations in cup-6 and in cup-10, but not in other members of a large collection of cup mutants (Fares and Greenwald, 2001a), caused severe disruption of the PI(3)P staining pattern in vivo (Figure 1; our unpublished data). Given the strong conservation of sequence (Laporte et al., 2001) and function, we renamed cup-6 as mtm-6 and cup-10 as mtm-9 to conform to both the mammalian and the worm gene nomenclature.
Defective Uptake of Fluid by the Coelomocytes of mtm-6 and mtm-9 Mutants
We determined that mtm-6(ar513) and mtm-9(ar479) coelomocytes were unable to endocytose BSA-Rhod introduced into the body cavity (Figure 2). There are two main advantages of using BSA-Rhod instead of GFP expressed from the pmyo-3::ssGFP transgene for a detailed examination of the endocytic pathway in coelomocytes: 1) we can inject the BSA-Rhod at a higher concentration, thus allowing us to monitor early events in endocytosis; and 2) whereas the GFP is degraded in late endosomes and lysosomes, the rhodamine moiety in BSA-Rhod is not affected, thus allowing us to visualize these compartments.
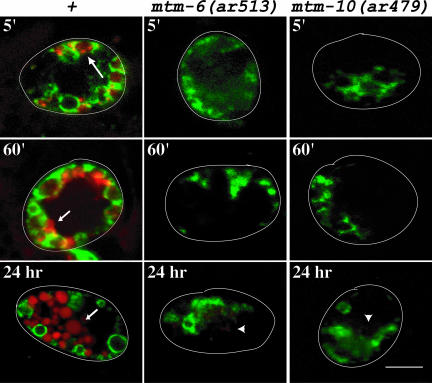
Figure 2.
Time course of uptake of BSA-Rhod by coelomocytes expressing RME-8::GFP in wild-type, mtm-6(ar513), or mtm-9(ar479) worms. The images are overlays of GFP (green) and rhodamine (red) fluorescence images taken with a confocal microscope. The time the pictures were taken after injection are indicated. The large arrow indicates concentrations of the BSA-Rhod in endosomes labeled with RME-8::GFP. Small arrows indicate lysosomes that are not labeled with RME-8::GFP. The arrowheads indicate extremely faint rhodamine staining in the coelomocytes of the mutant worms after 24 h of uptake. Outlines of worms and are drawn. Bar, 5 μm in all images.
We injected the BSA-Rhod into the body cavity of worms and monitored its uptake by coelomocytes expressing the endosomal marker RME-8::GFP (Zhang et al., 2001). RME-8::GFP labels endosomes, but not lysosomes in coelomocytes. In a wild-type cell, the BSA-Rhod occurs in RME-8::GFP–labeled compartments by 5 min after its injection into the body cavity. At this time, some of the BSA-Rhod occurs as concentrations, presumed to be nascent lysosomes, budding from these RME-8::GFP–labeled compartments (Fares and Greenwald, 2001b; Zhang et al., 2001; Figure 2, large arrow). By 60 min, the BSA-Rhod is concentrated in lysosomes that are not labeled by RME-8::GFP, and remains there for the duration of the experiment (small arrows in Figure 2). In contrast, no BSA-Rhod is seen in mtm-6(ar513) and mtm-9(ar479) coelomocytes by the 5-min and 60-min time point. Indeed, even after 24 h of incubation in BSA-Rhod, relatively small amounts of the BSA-Rhod are taken up by these cells (Figure 2, arrowhead).
This indicates that the rate of endocytosis in mtm-6(ar513) and mtm-9(ar479) coelomocytes is sharply reduced, very likely at an early step in the pathway. Also note that the RME-8::GFP staining pattern in mtm-6 and mtm-9 worms seems more disorganized than in wild-type cells (Figure 2).
Functional Domains of MTM-6
Molecular analysis revealed that mtm-6 encodes two isoforms, MTM-6A that is sufficient to rescue the endocytic defect of mtm-6(ar513) worms (Figure 3), and MTM-6B, which contains additional amino acids at the amino terminus with no obvious motifs (see Supplemental Material). In contrast to MTM-6A::GFP (see below), we could not detect MTM-6B::GFP when overexpressed in coelomocytes, suggesting that it is unstable, nor did it rescue the endocytic defect of mtm-6(ar513) worms or disrupt endocytosis in a wild-type cell (our unpublished data). Furthermore, the absence of MTM-6B does not have an obvious effect on coelomocyte endocytosis because simply expressing MTM-6A rescues the coelomocyte endocytosis defect of mtm-6 mutations that cannot make either form of the protein. Further analysis was therefore done using MTM-6A. mtm-9 encodes a predicted protein that lacks key residues required for phosphatase activity (Figure 3; Supplemental Material).
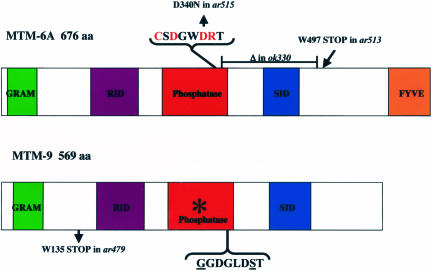
Figure 3.
MTM-6– and MTM-9–predicted proteins and mutant alleles. Schematic representation of the proteins, and their various domains, predicted from the cDNA sequences. The catalytic amino acids of the phosphatase domain of MTM-6 are indicated in red and the corresponding substituted amino acids of MTM-9 are underlined. The asterisk denotes that the phosphatase domain of MTM-9 lacks residues thought to be required for activity. The predicted changes in the various mutant alleles are shown. mtm-6(ok330) and mtm-9(ar479) represent the molecular null alleles. All the alleles of mtm-6 gave identical phenotypes.
The phosphatase domain of MTM-6 is essential for its in vivo function. This is supported by two lines of evidence: first, mtm-6(ar515), a mutation with a phenotype as severe as an mtm-6 null, contains an amino acid substitution of a conserved aspartic acid residue required by the myotubularin to bind to PIs (Wishart et al., 2001; Figure 3). Second, transgenic expression of a substrate-trapping mutant (Flint et al., 1997; Blondeau et al., 2000; Laporte et al., 2002a), MTM-6(C335S), causes dominant inhibition of endocytosis (Figure 4).
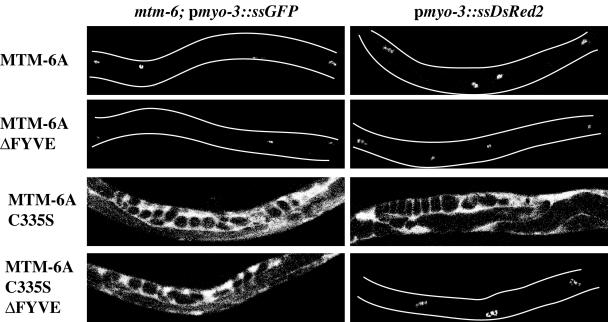
Figure 4.
Effect of MTM-6 alleles on endocytosis by coelomocytes. Shown are low-magnification confocal images of mtm-6(ar513); pmyo-3::ssGFP or of pmyo-3::ssDsRed2 worms carrying transgenes expressing GFP fusions of various forms of MTM-6A. Bright spots in the worms are coelomocytes normally endocytosing the fluid-phase marker. Bright fluorescence in the body cavity indicates absence of endocytosis. Outlines of worms are drawn when needed.
Analysis of specific deletion mutants suggests that the FYVE domain of MTM-6A is required for MTM-6 to maintain its membrane localization. Overexpression of either MTM-6A::GFP or MTM-6AΔFYVE::GFP (fusion protein completely lacking the FYVE domain) rescues the endocytosis defect of mtm-6, indicating that, at least under these conditions, the FYVE domain is not required by MTM-6 for its targeting to membranes or for its activity (Figure 4). However, in contrast to MTM-6A(C335S), MTM-6A(C335S)ΔFYVE did not interfere with coelomocyte endocytosis (Figure 4), indicating that the FYVE domain increases the avidity rather than the affinity of MTM-6A to specific membranes.
Subcellular Localization of MTM-6A and MTM-9
Both MTM-6A::GFP and MTM-6AΔFYVE::GFP are largely cytosolic, with some membrane localization (Figure 5A, a and e). In agreement with other myotubularin substrate-trapping mutants (Flint et al., 1997; Blondeau et al., 2000; Laporte et al., 2002a), MTM-6A(C335S)::GFP shows a punctate staining pattern and localizes to the plasma membrane and to compartments close to the plasma membrane in coelomocytes (Figure 5A, b). This membrane association does not require the presence of MTM-9 because it remains unchanged in mtm-9(ar479) worms (Figure 5A, c). In contrast, and in support of its inability to give a dominant negative phenotype, MTM-6A(C335S)ΔFYVE::GFP is mostly cytoplasmic (Figure 5A, f).
Figure 5.
Subcellular localization and interaction of MTM-6A and MTM-9. (A) High-magnification confocal images of the GFP fusion protein expressed in the coelomocytes of the following strains (Ex signifies the transgenic array from which the indicated protein is expressed): a, +; Ex[MTM-6A::GFP]; b, +; Ex[MTM-6A(C335S):: GFP]; c, mtm-9(ar479); Ex[MTM-6A(C335S)::GFP]; d, rme-1(b1045); Ex[MTM-6A(C335S)::GFP]; e, +; Ex[MTM-6AΔFYVE::GFP]; f, +; Ex[MTM-6A (C335S)ΔFYVE::GFP]; g, +; Ex[MTM9::GFP]; and h, Ex[MTM-6A(C335S)]; Ex[MTM9::GFP]. Bar, 5 μm in all images. (B) Coimmunoprecipitation of GFP-MTM-6A and Flg-MTM-9 expressed in human 293 cells. Antibodies against GFP or Flg were used to immunoprecipitate the corresponding fusion protein indicated above the gel. Blots were probed for the presence of the other fusion protein by using the antibodies indicated below the gel.
Like, MTM-6, the phosphatase-inactive MTM-9 protein is also predominantly localized to the coelomocyte cytoplasm, although as for MTM-6, a functional MTM-9::GFP fusion protein reveals some membrane association (Figure 5A, g).
Physical Association of MTM-6A and MTM-9
Given previous reports of physical interactions between other catalytically active and inactive forms of human and mouse MTM proteins (Kim et al., 2003; Mochizuki and Majerus, 2003; Nandurkar et al., 2003), and the requirement for both MTM-6 and MTM-9 for coelomocyte endocytosis, we thought it possible that MTM-9 could interact with MTM-6A to regulate its activity. Expression of the membrane-associated MTM-6A(C335S) mutant protein increased membrane association of MTM-9, providing in vivo evidence that MTM-6A localization or activity regulates MTM-9 localization (Figure 5A, h).
This regulation of MTM-9 by MTM-6 may be direct, because we determined that MTM-6A and MTM-9 physically associate and/or are part of the same complex. GFP-MTM-6A and Flg-MTM-9 were expressed in human 293 cells either alone, or together, and the ability of the two proteins to coimmunoprecipitate was determined. We found that MTM-6A and MTM-9 coimmunoprecipitated using antibodies to either GFP or Flg (Figure 5B, lanes 3 and 6). The coimmunoprecipitation was specific because it only occurred when both proteins were expressed in the cells (Figure 5B, lanes 4 and 5).
The above-mentioned observations, taken together, provide strong evidence for functional interactions between the catalytically active and inactive forms of MTM proteins in regulating fluid phase endocytosis by coelomocytes, probably through endocytic organelles visualized using the substrate-trapping MTM-6A(C335S)::GFP fusion protein.
Association of MTM-6 with an ARF-6/RME-1 Endocytic Pathway
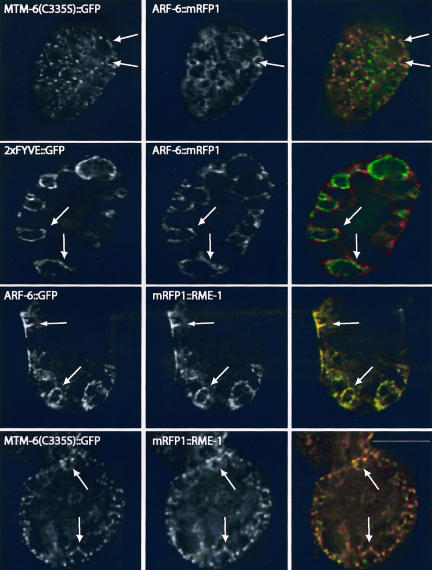
To better characterize the MTM-6 and MTM-9 endocytic pathway, we further examined the identity of the MTM-6A(C335S)::GFP labeled membranes. Adenosine diphosphate-ribosylation factor 6 (Arf6) is required in some mammalian tissues for nonclathrin-dependent forms of membrane traffic (Naslavsky et al., 2003). C. elegans ARF-6 is strongly expressed in coelomocytes (our unpublished data) and gives a pattern of localization similar to MTM-6A(C335S)::GFP (Figure 6). Indeed, MTM-6A(C335S)::GFP and ARF-6::mRFP1 show significant colocalization in coelomocytes (Figure 6). Consistent with a function of MTM-6 in PI(3)P metabolism, 2 × FYVE::GFP and ARF-6::mRFP1 also partially colocalize (Figure 6). Furthermore, using deconvolution microscopy to better visualize membrane-associated protein, we find that MTM-9::GFP and ARF-6::mRFP1 significantly colocalize in coelomocytes (our unpublished data).
Figure 6.
Deconvolution images of wild-type worms expressing the indicated proteins from transgenes. Panels in the right column are overlays of the corresponding GFP (green) and mRFP1 (red) images. Arrows indicate some of the membranes to which both proteins colocalize. Bar, 5 μm in all images.
These studies indicate that MTM-6/9-proteins function in a specialized endocytic pathway that involves Arf6. We therefore analyzed other components of the Arf6 pathway in C. elegans. Rme1/Ehd1/Pincher is an epsin homology domain-containing protein that is required for endocytosis or endocytic trafficking in some mammalian cells (Lin et al., 2001), including the Arf6 pathway in some cell types (Caplan et al., 2002). RME-1 is the C. elegans homolog of Ehd1 (Grant et al., 2001) and is required for endocytosis by coelomocytes (Fares and Greenwald, 2001a; Grant et al., 2001). Indeed, like their mammalian counterparts (Caplan et al., 2002), ARF-6::GFP and mRFP1::RME-1 show extensive colocalization in coelomocytes (Figure 6). Consistent with the above results, MTM-6A(C335S)::GFP and mRFP1::RME-1 largely colocalize in coelomocytes (Figure 6). This colocalization is functionally significant because the membrane association of MTM-6A(C335S)::GFP is severely disrupted in rme-1(b1045) mutant worms (Figure 5A, d).
DISCUSSION
We provide in this study our analysis of two biochemically distinct myotubularins and we show, for the first time, essential roles for the two proteins in a newly identified PI(3)P and Arf6/Rme1-associated endocytic pathway.
The identification of mutations in two myotubularins in an unbiased screen for endocytosis mutants strongly argues that a primary role of myotubularins is to regulate endocytosis. Indeed, RNAi analysis suggests that in addition to MTM-6 and MTM-9, worm MTM-1 and MTM-3 are also required for endocytosis by coelomocytes (Xue et al., 2003). This indicates that the myotubularins have largely nonoverlapping functions in regulating this process, perhaps by regulating PI levels at distinct membranes. This is in agreement with previous studies on human MTM1 and MTMR2 (Laporte et al., 2002b).
At which step(s) of the coelomocyte endocytic pathway are MTM-6 and MTM-9 required? When we monitored endocytosis in coelomocytes after injecting BSA-Rhod into the body cavity of worms, we saw little/no uptake of the dye in mtm-6 and mtm-9 coelomocytes. The simplest interpretation of these data is that MTM-6 and MTM-9 are required very early in the endocytic pathway in coelomocytes. We favor this model because it is consistent with the colocalization of the proteins with ARF-6 and RME-1 close to and on the plasma membrane. Indeed, in mammalian cells ARF-6 is required at the plasma membrane and in specialized early endosomes (Naslavsky et al., 2003), whereas a mammalian homolog of RME-1 localizes to cell surface ruffles (Shao et al., 2002). MTM-6 and MTM-9 could also be required in later endocytic compartments by regulating the levels and/or localization of other essential PI-phosphates directly, or indirectly because PI(3)P is a substrate for making other PI-phosphates. This is consistent with our observation that the RME-8–labeled compartments seem more disorganized in mtm-6 and mtm-9 coelomocytes than in wild-type cells. Note that we could detect very little colocalization between RME-8, and ARF-6, RME-1, or CUP-6(C335S) (Fares, unpublished data), indicating that RME-8 localizes to distinct endocytic compartments.
We had previously shown that reducing mtm-6 levels in the intestine results in the appearance of PI(3)P at the apical membrane rather than on internal vesicles (Xue et al., 2003). This indicates that in this tissue, MTM-6 is required to maintain low levels of PI(3)P at the plasma membrane. Although we do observe a severe disorganization of PI(3)P in coelomocytes, we do not observe PI(3)P at the plasma membrane of coelomocytes in mtm-6 and mtm-9 mutants. This is probably due to some functional redundancy among the myotubularins in coelomocytes and/or because of differences in the endocytic pathways used by these two tissues. Previous studies on the role of myotubularins in regulating membrane-associated PI(3)P levels relied on the overexpression of MTM1 and MTMR2 in mammalian COS cells and yielded conflicting results (Kim et al., 2002; Laporte et al., 2002a).
In coelomocytes, PI(3)P levels do not simply increase in the absence of MTM-6 or MTM-9; rather, PI(3)P becomes disorganized. We believe this is due to the involvement of other myotubularins in coelomocytes, such that in the absence of MTM-6 or MTM-9, there is misregulation rather than simple reduction of phosphatase activity. Furthermore, FYVE domains are known to bind to PI(3)P. We could not genetically analyze the requirement for the MTM-6 FYVE domain in the wild-type protein because overexpression of the MTM-6ΔFYVE displayed similar cytoplasmic localization and rescue of mtm-6 mutants as MTM-6. However, although overexpressed MTM-6(C335S) localized to membranes and inhibited endocytosis by coelomocytes, these effects were absent if the FYVE domain was reduced. This suggests that the FYVE domain is required for continued localization to membranes and that its function is probably critical for the function of the endogenous protein, that is, under conditions when the protein is not overexpressed.
MTM-6(C335S) and MTM-9 colocalize with ARF-6 and with RME-1. In the absence of a mutation in C. elegans ARF-6, we tried to lower ARF-6 levels in coelomocytes through RNAi. We were unsuccessful because we could not detect any changes in the levels of ARF-6::GFP expressed in these cells after RNAi (Fares, unpublished data). However, RME-1 is required for endocytosis by coelomocytes, and we showed that the colocalization of MTM-6(C335S) with RME-1 is significant because the elimination of RME-1 function in these cells disrupted the membrane association of MTM-6(C335S).
Our conclusion that MTM-6, and probably other myotubularins, functions in ARF-6/RME-1–mediated endocytosis helps explain two previous observations. 1) Human MTM1, MTMR2, MTMR3, and MTMR9 localize, in part, to Rac1-induced membrane ruffles (Laporte et al., 2002a,b; Nandurkar et al., 2003), sites of clathrin-independent endocytosis. Interestingly, Arf6 colocalizes with Rac1 and can influence its ability to effect plasma membrane remodeling (Radhakrishna et al., 1999; Boshans et al., 2000). Furthermore, Rme1/Ehd1/Pincher associates with membrane ruffles in PC12 cells to regulate a clathrin-independent endocytic pathway (Shao et al., 2002). 2) Overexpression of Gas3/PMP22, a component of the compact peripheral myelin, results in a peripheral neuropathy, Charcot-Marie-Tooth type 1A, which like type 4B, is characterized by myelination abnormalities, although other pathological details differ among the two diseases. It has recently been shown that overexpressed Gas3/PMP22 specifically affects Arf6 membrane trafficking and that some of the endocytosed Gas3/PMP22 localizes to Arf6 compartments (Chies et al., 2003).
Links between Arf6 and phosphoinositides are also known. Arf6 regulates phosphatidylinositol 4,5-bisphosphate levels at the plasma membrane and in endosomes (Brown et al., 2001), and PI(3)P is required to transport endocytosed material from Arf6-endosomes to classic Rab5-labeled early endosomes (Naslavsky et al., 2003). Myotubularins most likely regulate PI(3)P (and probably other forms of PI) in endosomal compartments, including Arf6-endosomes. Furthermore, given the association of myotubularins with the plasma membrane in mammalian cells and in worms, they could also act to reduce PI(3)P levels at the plasma membrane that may interfere with the initial steps of endocytosis.
We propose that similar disruptions in an Arf6 pathway is at least a partial cause for the development of diseases stemming from mutations in human myotubularins. This and future studies will allow us to better probe the molecular basis for myotubular myopathy and Charcot-Marie-Tooth type 4B and will give us a better understanding of the process of endocytosis in general.
Supplementary Material
Acknowledgments
We thank R. Tsien for the gift of the mRFP1 plasmid, Y. Kohara for yk clones, and the C. elegans Gene Knockout Consortium for the deletion allele of mtm-6. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by a National Institute of General Medical Sciences grant to H.F.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–08–0605. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0605.
Abbreviations used: Arf6, adenosine diphosphate-ribosylation factor 6; GFRP, green fluorescent protein; mRFP1, monomeric red fluorescent protein; PI, phosphoinositide; PI(3)P, phosphoinositide 3-phosphate; RNAi, RNA-mediated interference; BSA-Rhod, bovine serum albumin conjugated to rhodamine.
Online version of this article contains supplementary material for some figures. Online version is available at www.molbiolcell.org.
References
- Berger, P., Bonneick, S., Willi, S., Wymann, M., and Suter, U. (2002). Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum. Mol. Genet. 11, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Blondeau, F., Laporte, J., Bodin, S., Superti-Furga, G., Payrastre, B., and Mandel, J.L. (2000). Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet. 9, 2223–2229. [DOI] [PubMed] [Google Scholar]
- Bolino, A., et al. (2000). Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat. Genet. 25, 17–19. [DOI] [PubMed] [Google Scholar]
- Boshans, R.L., Szanto, S., van Aelst, L., and D'Souza-Schorey, C. (2000). ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, F.D., Rozelle, A.L., Yin, H.L., Balla, T., and Donaldson, J.G. (2001). Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154, 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, S., Naslavsky, N., Hartnell, L.M., Lodge, R., Polishchuk, R.S., Donaldson, J.G., and Bonifacino, J.S. (2002). A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W., and Prasher, D.C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802–805. [DOI] [PubMed] [Google Scholar]
- Chies, R., Nobbio, L., Edomi, P., Schenone, A., Schneider, C., and Brancolini, C. (2003). Alterations in the Arf6-regulated plasma membrane endosomal recycling pathway in cells overexpressing the tetraspan protein Gas3/PMP22. J. Cell Sci. 116, 987–999. [DOI] [PubMed] [Google Scholar]
- Doerks, T., Strauss, M., Brendel, M., and Bork, P. (2000). GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem. Sci. 25, 483–485. [DOI] [PubMed] [Google Scholar]
- Fares, H., and Greenwald, I. (2001a). Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares, H., and Greenwald, I. (2001b). Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 28, 64–68. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Flint, A.J., Tiganis, T., Barford, D., and Tonks, N.K. (1997). Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 94, 1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly, D.J., Morrow, I.C., Lindsay, M., Gould, R., Bryant, N.J., Gaullier, J.M., Parton, R.G., and Stenmark, H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, B., Zhang, Y., Paupard, M.C., Lin, S.X., Hall, D.H., and Hirsh, D. (2001). Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573–579. [DOI] [PubMed] [Google Scholar]
- Kim, S.A., Taylor, G.S., Torgersen, K.M., and Dixon, J.E. (2002). Myotubularin and MTMR2, phosphatidylinositol 3-phosphatases mutated in myotubular myopathy and type 4B Charcot-Marie-Tooth disease. J. Biol. Chem. 277, 4526–4531. [DOI] [PubMed] [Google Scholar]
- Kim, S.A., Vacratsis, P.O., Firestein, R., Cleary, M.L., and Dixon, J.E. (2003). Regulation of myotubularin-related (MTMR)2 phosphatidylinositol phosphatase by MTMR5, a catalytically inactive phosphatase. Proc. Natl. Acad. Sci. USA 100, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte, J., Blondeau, F., Buj-Bello, A., and Mandel, J.L. (2001). The myotubularin family: from genetic disease to phosphoinositide metabolism. Trends Genet. 17, 221–228. [DOI] [PubMed] [Google Scholar]
- Laporte, J., Blondeau, F., Gansmuller, A., Lutz, Y., Vonesch, J.L., and Mandel, J.L. (2002a). The PtdIns3P phosphatase myotubularin is a cytoplasmic protein that also localizes to Rac1-inducible plasma membrane ruffles. J. Cell Sci. 115, 3105–3117. [DOI] [PubMed] [Google Scholar]
- Laporte, J., Hu, L.J., Kretz, C., Mandel, J.L., Kioschis, P., Coy, J.F., Klauck, S.M., Poustka, A., and Dahl, N. (1996). A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 13, 175–182. [DOI] [PubMed] [Google Scholar]
- Laporte, J., Liaubet, L., Blondeau, F., Tronchere, H., Mandel, J.L., and Payrastre, B. (2002b). Functional redundancy in the myotubularin family. Biochem. Biophys. Res. Commun. 291, 305–312. [DOI] [PubMed] [Google Scholar]
- Lin, S.X., Grant, B., Hirsh, D., and Maxfield, F.R. (2001). Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat. Cell Biol. 3, 567–572. [DOI] [PubMed] [Google Scholar]
- Mello, C., and Fire, A. (1995). DNA transformation. Methods Cell Biol. 48, 451–482. [PubMed] [Google Scholar]
- Mochizuki, Y., and Majerus, P.W. (2003). Characterization of myotubularin-related protein 7 and its binding partner, myotubularin-related protein 9. Proc. Natl. Acad. Sci. USA 100, 9768–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandurkar, H.H., Layton, M., Laporte, J., Selan, C., Corcoran, L., Caldwell, K.K., Mochizuki, Y., Majerus, P.W., and Mitchell, C.A. (2003). Identification of myotubularin as the lipid phosphatase catalytic subunit associated with the 3-phosphatase adapter protein, 3-PAP. Proc. Natl. Acad. Sci. USA 100, 8660–8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky, N., Weigert, R., and Donaldson, J.G. (2003). Convergence of nonclathrin- and clathrin-derived endosomes involves Arf6 Inactivation and changes in phosphoinositides. Mol. Biol. Cell 14, 417–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna, H., Al-Awar, O., Khachikian, Z., and Donaldson, J.G. (1999). ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112, 855–866. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schaletzky, J., Dove, S.K., Short, B., Lorenzo, O., Clague, M.J., and Barr, F.A. (2003). Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr. Biol. 13, 504–509. [DOI] [PubMed] [Google Scholar]
- Senderek, J., Bergmann, C., Weber, S., Ketelsen, U.P., Schorle, H., Rudnik-Schoneborn, S., Buttner, R., Buchheim, E., and Zerres, K. (2003). Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum. Mol. Genet. 12, 349–356. [DOI] [PubMed] [Google Scholar]
- Shao, Y., Akmentin, W., Toledo-Aral, J.J., Rosenbaum, J., Valdez, G., Cabot, J.B., Hilbush, B.S., and Halegoua, S. (2002). Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J. Cell Biol. 157, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.S., Maehama, T., and Dixon, J.E. (2000). myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D.M., Urbe, S., Dove, S.K., Tenza, D., Raposo, G., and Clague, M.J. (2001). Characterization of MTMR3. an inositol lipid 3-phosphatase with novel substrate specificity. Curr. Biol. 11, 1600–1605. [DOI] [PubMed] [Google Scholar]
- Wishart, M.J., Taylor, G.S., Slama, J.T., and Dixon, J.E. (2001). PTEN and myotubularin phosphoinositide phosphatases: bringing bioinformatics to the lab bench. Curr. Opin. Cell Biol. 13, 172–181. [DOI] [PubMed] [Google Scholar]
- Xue, Y., Fares, H., Grant, B., Li, Z., Clark, S.G., and Skolnik, E.Y. (2003). Genetic analysis of the myotubularin family of phosphatases in Caenorhabditis elegans. J. Biol. Chem. 278, 34380–34386. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Grant, B., and Hirsh, D. (2001). RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell 12, 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., Qi, Y., Chen, J., and Zhao, Z.J. (2001). FYVE-DSP2, a FYVE domain-containing dual specificity protein phosphatase that dephosphorylates phosphotidylinositol 3-phosphate. Exp. Cell Res. 265, 329–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.