Abstract
Regulator of chromosome condensation (RCC1) binding to chromatin is highly dynamic, as determined by fluorescence recovery after photobleaching analysis of GFP-RCC1 in stably transfected tsBN2 cells. Microinjection of wild-type or Q69L Ran markedly slowed the mobility of GFP-RCC1, whereas T24N Ran (defective in nucleotide loading) decreased it further still. We found significant alterations in the mobility of intranuclear GFP-RCC1 after treatment with agents that disrupt different Ran-dependent nuclear export pathways. Leptomycin B, which inhibits Crm1/RanGTP-dependent nuclear export, significantly increased the mobility of RCC1 as did high levels of actinomycin D (to inhibit RNA polymerases I, II, and III) or α-amanitin (to inhibit RNA polymerases II and III) as well as energy depletion. Inhibition of just mRNA transcription, however, had no affect on GFP-RCC1 mobility consistent with mRNA export being a Ran-independent process. In permeabilized cells, cytosol and GTP were required for the efficient release of GFP-RCC1 from chromatin. Recombinant Ran would not substitute for cytosol, and high levels of supplemental Ran inhibited the cytosol-stimulated release. Thus, RCC1 release from chromatin in vitro requires a factor(s) distinct from, or in addition to, Ran and seems linked in vivo to the availability of Ran-dependent transport cargo.
INTRODUCTION
Regulator of chromosome condensation (RCC1) stimulates guanine nucleotide exchange by the small GTPase Ran (Bischoff and Ponstingl, 1991a). Inside the cell, RCC1 is essential for the efficient conversion of RanGDP to RanGTP. RanGTP in turn is essential for several key cellular processes, including nucleocytoplasmic transport, regulation of spindle formation and nuclear envelope reassembly at mitosis, and prevention of rereplication of DNA during S phase (Dasso, 2002; Yamaguchi and Newport, 2003).
The local concentration of RanGTP is a positional cue used by nuclear import and export complexes to distinguish between the cytoplasm and nuclear interior, and this regulates the assembly and disassembly of these transport complexes in the correct compartment. RanGTP is kept high inside the nucleus by localization of RCC1 to chromatin in the nuclear interior, and low inside the cytoplasm by the RanGAP (GTPase activating protein) that is restricted to that compartment. The differential placement of these two Ran accessory factors forms a gradient of RanGTP across the nuclear pore complex essential for most nuclear transport during interphase (Izaurralde et al., 1997). All nuclear carriers of the karyopherin-β (Kap-β) (importin β) family bind RanGTP (Gorlich et al., 1997; Gorlich and Kutay, 1999). Binding of RanGTP is required for Kap-β export carriers to simultaneously bind their cargo inside the nucleus. These export complexes disassemble after encounter with the RanGAP (and a cofactor RanBP1) in the cytoplasm and the subsequent conversion of complexed RanGTP to RanGDP. Conversely, import complexes consisting of the carrier with bound cargo assemble only in the absence of RanGTP (the cytoplasm) and disassemble upon encountering RanGTP inside the nucleus. In addition, production of RanGTP at the surface of mitotic chromatin by chromatin-bound RCC1 is critical for proper placement of the mitotic spindle and reassembly of the nuclear envelope around chromatin at the end of mitosis (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Hetzer et al., 2000; Zhang and Clarke, 2000, 2001; Clarke and Zhang, 2001; Wilde et al., 2001; Dasso, 2002; Hetzer et al., 2002).
RanGTP is required for the nuclear transport of many, but not all, cargoes (Kuersten et al., 2001). Import of basic nuclear localization signal-containing nuclear proteins requires the carrier Kap-β 1 (together with an adapter Kap-α) and RanGTP. Proteins containing a leucine-rich nuclear export signal (NES) are exported from the nucleus by the export carrier Crm1, a member of the Kap-β family, and all Crm1-mediated export requires RanGTP (Fornerod et al., 1997; Fukuda et al., 1997; Askjaer et al., 1999). In addition to shuttling proteins, Crm1 export cargo includes both large and small newly formed ribosomal subunits, which assemble in nucleoli and are exported separately to the cytoplasm (Johnson et al., 2002). Leptomycin B (LMB) inhibits Crm1 binding to its cargo and thus specifically inhibits any Crm1-mediated export pathway, including the export of ribosomal subunits (Kudo et al., 1998, 1999). tRNAs are exported from the nucleus by one of two alternate Kap-β carriers, exportin-t or exportin-5, and both of these export pathways also are dependent on RanGTP (Kutay et al., 1998; Bohnsack et al., 2002; Calado et al., 2002).
In contrast to these pathways of nuclear export, mRNA export does not seem to use a carrier of the Kap-β family or RanGTP (Clouse et al., 2001). The nuclear carriers Tap and p15/NTX1, of the NTF2 family of nuclear carriers, are involved in targeting mRNPs to the nuclear pore complex; however, these carriers do not bind Ran (Santos-Rosa et al., 1998; Fribourg et al., 2001). This study examines the dynamic association of RCC1 with chromatin, how the kinetics of RCC1 binding and release from chromatin are affected by an excess of Ran and by ongoing Ran-dependent nuclear export pathways, and the requirements for RCC1 release from chromatin in vitro.
MATERIALS AND METHODS
Construction of GFP-RCC1 Fusion
Primers incorporating a 5′ HindIII site and a 3′ Bam H1 site were used to polymerase chain reaction amplify human RCC1 from a protein A-RCC1 fusion construct described previously (Talcott and Moore, 2000). RCC1 was then ligated into the green fluorescent protein (GFP) expression vector pEGFP-N1 (BD Biosciences Clontech, Palo Alto, CA), by using the HindIII and BamHI sites. Ligation resulted in RCC1 fusion to the N terminus of enhanced green fluorescent protein. A second set of primers was used to confirm the correct sequence of the GFP-RCC1 construct.
GFP-RCC1 tsBN2 Cells
tsBN2 cells were routinely cultured at 33°C as described previously (Talcott and Moore, 2000). Cells were transfected with GFP-RCC1, by using the FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN). After transfection, the cells were incubated for 24 h at 33°C and then transferred to a 40°C incubator for an additional 24 h. Fresh media containing 0.5 mg/ml G418 (Invitrogen, Carlsbad, CA) was added at this time. GFP-RCC1 tsBN2 cells have been cultured throughout all subsequent passages in the presence of G418 and at 40°C.
Biochemical Analysis of GFP-RCC1 tsBN2 Cells
GFP-RCC1 tsBN2 (40°C) and tsBN2 cells (33°C) were grown to ∼85% confluence. One flask of tsBN2 cells was shifted to 40°C for 15 h. All the cells were then trypsinized and counted. An equal number of cells from each flask were pelleted by centrifugation for 10 min at 1000 × g, washed in phosphate-buffered saline (PBS), and lysed in 0.5 M Tris, pH 6.8, 6% SDS. Lysates were precipitated with 10% trichloroacetic acid, and the protein pellets were resuspended in SDS-PAGE sample buffer, subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted with an RCC1 antibody (Schwoebel et al., 1998).
For analysis of the salt extractability of GFP-RCC1, tsBN2 and GFP-RCC1 tsBN2 cells were trypsinized, washed, counted, and pelleted. An equal number of cells was resuspended in cold 20 mM HEPES, pH 7.3, 1 mM dithiothreitol (DTT), 0.1% Triton X-100 and incubated for 15 min on ice. After centrifugation, the permeabilized cell pellets were resuspended in 20 mM HEPES, pH 7.3, 1 mM DTT containing either 0, 0.2 M, 0.4 M, or 0.6 M NaCl and incubated on ice for 15 min. After centrifugation at 3000 × g for 10 min, the supernatants and pellets were immunoblotted with an RCC1 antibody as described above.
Cell Treatments
For energy depletion studies, GFP-RCC1 tsBN2 cells were incubated in glucose-free DMEM (Invitrogen) containing penicillin (100 U/ml), streptomycin sulfate (100 μg/ml), 10 mM HEPES, pH 7.3, and 10% fetal bovine serum [hereafter referred to as gluc (-) media] containing 10 mM sodium azide and 6 mM 2-deoxy-d-glucose (Schwoebel et al., 2002). Cells were incubated 20–30 min in energy depletion buffer before fluorescence recovery after photobleaching (FRAP) analysis. Transcription inhibitors and LMB were added to the GFP-RCC1 tsBN2 cells at the concentrations indicated in the figure legends and incubation was for 5 h at 40°C before FRAP analysis.
Microinjection
Human wild-type (wt), Q69L, and T24N His-Ran recombinant proteins were expressed and purified as described previously (Izaurralde et al., 1997; Carazo-Salas et al., 1999). We assayed the recombinant wt Ran for its ability to support in vitro nuclear protein import (Schwoebel et al., 1998), and it is fully active in that assay (our unpublished data). The proteins were dialyzed against injection buffer (20 mM HEPES, pH 7.3, 120 mM KOAc, 2 mM MgOAc, 1 mM DTT) and concentrated in a Centricon-10. GFP-RCC1 tsBN2 cells were injected with a mixture of Ran at 5 mg/ml (200 μM) and Texas Red-labeled 70-kDa dextran (Molecular Probes, Eugene, OR) at 1 mg/ml in injection buffer. Before injection, protein mixtures were centrifuged at 14,000 × g for 10 min at 4°C.
Live-Cell Analysis and FRAP
Before analysis, GFP-RCC1 tsBN2 cells growing on 40-mm glass coverslips were transferred to a live-cell chamber (Bioptechs, Butler, PA) with fresh media (20 ml, containing any treatment additions) recirculated via a peristaltic pump. The live cell chamber and objective lens were continuously monitored and maintained at 37°C. For the images shown in Figure 1, cells were incubated in media containing 1 μg/ml Hoechst 33258 for 15 min to label DNA in living cells. Imaging of live cells was performed on a Deltavision restoration microscopy system (Applied Precision, Issaquah, WA). The images shown were deconvolved and the Z-series were compressed to create a single image.
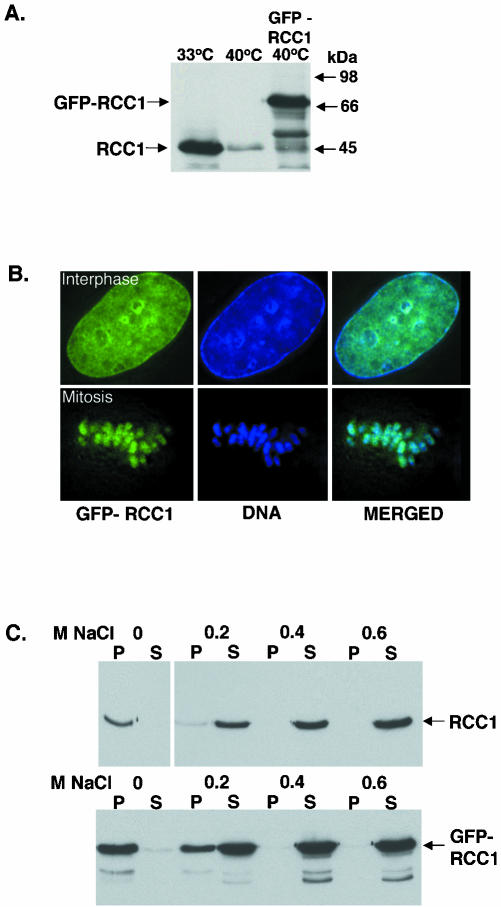
Figure 1.
Comparison of GFP-RCC1 with endogenous RCC1. (A) Anti-RCC1 immunoblot showing the expression levels of RCC1 and GFP-RCC1. Left lane, tsBN2 cells grown at permissive temperature (33°C). Middle lane, tsBN2 cells after being shifted to nonpermissive temperature (40°C) for 15 h. Right lane, GFP-RCC1 tsBN2 cells grown at 40°C. The same amount of cells is loaded on each lane. (B) GFP-RCC1 colocalizes with chromatin throughout the cells cycle. Left panel, GFP-RCC1 in a living cell. Middle panel, DNA stained with Hoescht stain in the same cell. Right panel, overlay of these two images. The top row shows an interphase cell, whereas the bottom row shows a cell in metaphase (C) RCC1 and GFP-RCC1 require the same salt concentration for elution from chromatin. Top gel, extraction of RCC1 from tsBN2 cells (grown at 33°C) with increasing concentrations of NaCl. Bottom gel, GFP-RCC1 is extracted from GFP-RCC1 cells (grown at 40°C) the same concentration of NaCl. After salt incubation, the samples were separated into supernatants (S) and pellets (P) and both were blotted with an anti-RCC1 antibody.
FRAP was performed as described previously (Stenoien et al., 2001, 2002) by using an LSM 510 confocal microscope (Carl Zeiss, Thornwood, NY). A single Z-section was imaged before and at 1-s intervals after the bleach. The photobleach was performed using the 488-nm laser line at maximum power for 75 iterations in the boxed regions. Images acquired before and after the bleach were obtained using 1% laser power and did not significantly bleach the sample. Images were exported as TIF files and final figures were generated using Adobe Photoshop, version 7 (Adobe Systems, Mountain View, CA).
Using the LSM510 software, the fluorescence recovery in a region of interest corresponding to the bleach area was determined for each bleached cell, and data were exported to Excel (Microsoft, Redmond, WA) for further analysis. Except in Figure 4B, intensity values were normalized using the following equation; It = (Xt - Y)/(Z - Y), where I is the normalized intensity at time t, X is the intensity at time t, Y is the intensity immediately after the photobleach (where t is equal to zero), and Z is the intensity at the final time point. To generate fluorescence intensity graphs, the normalized intensity values were averaged and plotted versus time. To calculate t1/2 values, the line equations for each individual recovery were created and the time at which I = 0.5 was determined. Mean t1/2 values for each condition were calculated ± SEM. Differences in mean t1/2 values were analyzed using Student's t tests. In Figure 4B, intensity values were determined using the equation Irel = It/I0, where Irel is the relative intensity at each time point normalized by dividing the actual fluorescence at each time point (It) by the initial starting fluorescence (I0).
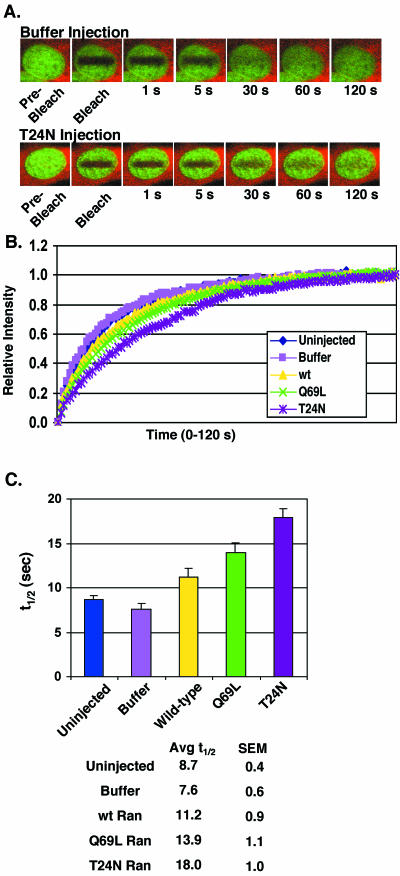
Figure 4.
Excess Ran slows GFP-RCC1 recovery after photobleaching. (A) GFP-RCC1 cells microinjected with injection buffer (top) or the mutant T24N Ran (bottom) before photobleaching and measurement of fluorescence recovery. The red fluorescence is the Texas Red 70-kDa dextran coinjected with buffer or unlabeled Ran in the cytoplasm to mark the injection site. The green fluorescence is GFP-RCC1. (B) Recovery of fluorescence after photobleaching for GFP-RCC1 interphase cells either uninjected (blue diamonds), microinjected with marker alone in injection buffer (pink squares), or marker plus wt Ran (yellow triangles), Q69L Ran (green Xs), or T24N Ran (purple asterisks). After photobleaching, the fluorescence was measured every second for 120 s. (C) The t1/2 of recovery for each condition calculated from the data points shown in B. n = 15 for each condition and the SEs are shown (SEM). The times of recovery of interphase cells injected with wt Ran, Q69L Ran, and T24N Ran were significantly different from cells injected with buffer alone (p<.05). The recovery time for T24N Ran was significantly different from that for wt Ran (p<.001), however the recovery time for Q69L Ran was not significantly different from wt Ran (p=0.076). In these experiments 5 microinjected cells from 3 separate coverslips (total of 15 cells for each condition) were blindly selected for FRAP analysis. The analyzed recovery period was extended to 2 min (compared with 1 min in the experiments shown in Figure 2) to allow complete recovery of all cells.
Immunofluorescence Microscopy
After the indicated treatments, GFP-RCC1 tsBN2 cells were rinsed in PBS, fixed in 3% paraformaldehyde in PBS for 15 min on ice, and permeabilized with 0.1% Triton X-100 for 20 min on ice. The cells were blocked in 5% serum (donkey or goat depending on the species of the second antibody) in PBS for 30 min at room temperature (RT). The first antibodies were diluted in the appropriate blocking solution, and incubated with the cells for 1 h at room temperature. After washing, the cells were incubated in labeled second antibody in blocking solution for 1 h at room temperature. After washing, the cells were fixed again in 3% paraformaldehyde for 10 min at room temperature followed by 2 × 5-min incubations in 1 mg/ml Na borohydride in PBS. After rinsing in PBS, the cells were mounted in 90% glycerol/10% PBS containing 1 mg/ml phenylenediamine. The antibodies used were: mouse anti-Ran (Transduction Laboratories, Lexington, KY), rabbit anti-Crm1 (from M. Yoshida) (Kudo et al., 1997), mouse anti-nucleophosmin/B23 (from P.K. Chan) (Yung et al., 1985), and mouse (IgM) anti-SRm160 (from J. Nickerson) (Wan et al., 1994). The labeled second antibodies were tetramethylrhodamine B isothiocyanate (TRITC)-labeled donkey anti-mouse or anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove, PA) and Texas Red-labeled goat anti-mouse IgM (Southern Biotechnologies, Birmingham, AL).
In Vitro Assay for Measuring GFP-RCC1 Release from Chromatin
GFP-RCC1 tsBN2 were cultured at 40°C on 12-mm coverslips in 24-well plates to ∼70% confluence. The cells were placed on ice, washed one time in cold buffer A (20 mM HEPES-KOH, pH 7.3, 110 mM K acetate, 2 mM Mg acetate, 1 mM EGTA, 1 mM DTT) and permeabilized for 5 min on ice in 0.1% Triton X-100 in buffer A. After rinsing in cold buffer A, the coverslips were blotted, and transferred cell side down to 20 μl of incubation mixture. The incubation mixture contained 2 mg/ml bovine serum albumin in buffer A plus the additions listed in the figure legends. Xenopus ovarian cytosol was prepared as described previously and dialyzed against buffer A (Moore and Blobel, 1992). After incubation for 20 min at room temperature, the coverslips were returned to the plate on ice, rinsed one time with cold buffer A, and fixed for 15 min on ice in 3% formaldehyde in buffer A. To amplify the rather faint GFP signal, and to minimize quenching of the signal during subsequent quantitation, the GFP-RCC1 was localized after fixation by indirect immunofluorescence microscopy with a rabbit anti-GFP first antibody (ab290) (Abcam) and a TRITC-labeled donkey anti-rabbit second antibody (Jackson ImmunoResearch Laboratories). To quantify the amount of GFP-RCC1 fluorescence bound to chromatin, the coverslips were observed on an Axiophot microscope (Carl Zeiss) equipped with a Princeton Micromax charge-couple device camera. Using MetaMorph software, a circle was drawn over a region of each nucleus and the fluorescence intensity within that circle determined. Between 30 and 90 nuclei were quantitated for each condition.
RESULTS
Most cellular RCC1 in vertebrate cells remains associated with chromatin throughout the cell cycle as indicated by biochemical fractionation (Ohtsubo et al., 1989), indirect immunofluorescent localization of RCC1 (Guarguaglini et al., 2000; Moore et al., 2002), and by the localization of GFPRCC1 in living cells (Moore et al., 2002; Trieselmann and Wilde, 2002; Li et al., 2003). To analyze the association of RCC1 with mitotic and interphase chromatin in living cells under as physiological conditions as possible, we created a stable GFP-RCC1–expressing cell line from the tsBN2 hamster cell line that contains a point mutation in RCC1 (Ohtsubo et al., 1989). When grown at the permissive temperature of 33°C, tsBN2 cells function normally. On shift to the nonpermissive temperature of 40°C; however, the mutant RCC1 is rapidly degraded rendering the cells temperature sensitive (ts) for growth (Figure 1A). We were able to rescue this ts growth defect by transfection with a GFP-RCC1 construct 24 h before the temperature shift. After continuous culture under drug selection at 40°C for approximately a month, we isolated a stably transfected tsBN2 cell line expressing GFP-RCC1.
GFP-RCC1 tsBN2 cells express GFP-RCC1 at levels only slightly above the amount of endogenous RCC1 normally contained by tsBN2 cells grown at the permissive temperature (Figure 1A). The behavior of GFP-RCC1 in this stable cell line faithfully mimics that of endogenous RCC1 in the following ways: 1) GFP-RCC1 rescued the ts growth defect of the tsBN2 cells, indicating this tagged form of RCC1 is functional; 2) the cellular localization of GFP-RCC1 at interphase and mitosis is identical to that reported for wt RCC1 (Figure 1B); and 3) the salt extractability of GFP-RCC1 from tsBN2 chromatin is identical to that of endogenous RCC1 (Figure 1C) (Ohtsubo et al., 1989). Thus, by these criteria, GFP-RCC1 tsBN2 cells provide a unique system for accurately probing the intracellular dynamics of RCC1 in living cells.
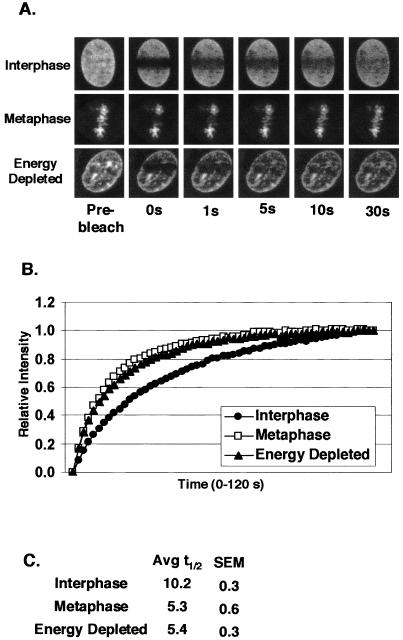
The association of GFP-RCC1 with chromatin in living tsBN2 cells was examined using FRAP. When a region of the interphase nucleus was photobleached, GFP-RCC1 fluorescence recovered within 1 min, indicating that RCC1's association with chromatin is highly dynamic (Figure 2B). When the average relative fluorescence (n = 15 cells) is plotted versus time, the recovery curve is best fitted to a single exponential suggesting that the entire GFP-RCC1 population recovers at the same rate. The recovery half-life (t1/2) of GFP-RCC1 was calculated to be 10.2 ± 0.3 s (Figure 2C). This recovery is much more rapid than proteins such as histones (t1/2 values of minutes for histone H1 to hours for the core histones) that are integrated into chromatin (Lever et al., 2000; Misteli et al., 2000; Kimura and Cook, 2001), but slower than proteins such as transcription factors that make more transient interactions with chromatin (Stenoien et al., 2001). To determine whether the mobility of GFP-RCC1 changes during the cell cycle, we photobleached GFP-RCC1 on metaphase chromatin (Figure 2). The photobleached regions in mitotic cells recovered their fluorescence approximately twice as fast as those on interphase chromatin with a t1/2 of 5.3 ± 0.6 s (Figure 2C).
Figure 2.
GFP-RCC1 is highly mobile. A rectangular region of GFP-RCC1 cells was photobleached as described in the MATERIALS AND METHODS. The fluorescence inside the photobleached rectangle was then measured every 1 s for 60 s to determine the rate of recovery. (A) A comparison of the recovery rate of GFP-RCC1 fluorescence after photobleaching in interphase nuclei (top and bottom row) and a cell in metaphase (middle row). The cell in the bottom row was first energy depleted by incubation with sodium azide and 2-deoxyglucose in gluc (-) media for 20 min before observation. (B) Quantitation of the recoveries after photobleaching of GFP-RCC1 in interphase cells, metaphase cells, and energy-depleted interphase cells. Each point represents the average of 15 cells. (C) Average t1/2 (amount of time required after photobleaching to return to 50% of the final fluorescent intensity) of interphase, metaphase, and energy-depleted cells calculated from the data shown in B. The GFP-RCC1 recovery times for metaphase and energy-depleted cells were significantly different from interphase cells, p < 0.001, n = 15).
One way that RCC1 binds chromatin is via an interaction with histones H2A and H2B, and binding of these histones stimulates a modest increase in RCC1's GEF activity toward Ran (Nemergut et al., 2001). GFP-labeled core histones, however, exchange on and off chromatin with a t1/2 of hours rather than the seconds observed here for GFP-RCC1 (Kimura and Cook, 2001). The linker histone H1 has a much faster nuclear mobility than the core histones (albeit with a mobility still markedly slower than GFP-RCC1) (Lever et al., 2000; Misteli et al., 2000), but there is no evidence that RCC1 interacts with histone H1. In addition, the nuclear mobility of GFP-histone H1 is markedly decreased upon energy depletion of the cells due to an inhibition of histone H1 phosphorylation (Dou et al., 2002).
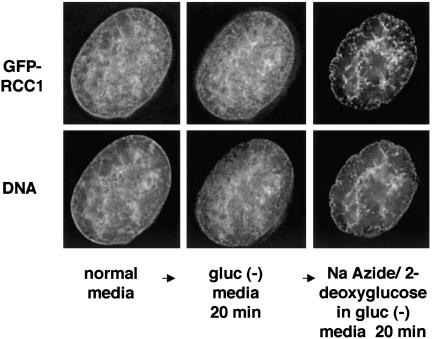
To determine whether the observed movement of GFPRCC1 on and off chromatin is affected by energy levels, cells were energy depleted by incubating them in glucose-free media containing sodium azide and 2-deoxyglucose for 20–30 min before photobleaching (Schwoebel et al., 2002). Instead of slowing down GFP-RCC1 movement, energy depletion increased the rate of fluorescence recovery after photobleaching approximately twofold compared with untreated cells (t1/2 value of 5.4 ± 0.3 s; Figure 2C). The nuclear distribution of GFP-RCC1 in interphase cells also changed significantly upon energy depletion. Figure 3 shows the distribution of GFP-RCC1 in the same living cell before and after incubation in energy depletion media for 20 min. On energy depletion, GFP-RCC1 became less finely dispersed inside the nucleus and much more clumped over nuclear regions that also stained the most intensely with Hoechst DNA stain. The apparent clumping may be due to changes in the chromatin distribution rather than a direct effect on RCC1. Note that these “clumps” of GFP-RCC1 seen after energy depletion cannot be stationary aggregates because the t1/2 for recovery after photobleaching is faster after energy depletion rather than slower as it would be if the clumped GFP-RCC1 were immobile (Figure 2).
Figure 3.
GFP-RCC1 becomes highly clumped inside the nucleus upon energy depletion. Cells expressing GFP-RCC1 and growing in normal media were assembled in a Bioptechs live-cell chamber with normal media and allowed to equilibrate in a 37°C incubator. After 20 min, the chamber was transferred to a DeltaVision microscopy work station and images were acquired. The media were changed to gluc (-) media, and the same cell was imaged again after 20 min. Finally, the media were changed to gluc (-) media containing sodium azide and deoxyglucose and the same cell was imaged after 20 min in this ATP depletion media. To show the DNA staining in live, unfixed cells, cells were incubated in 1 μg/μl Hoechst 33258 for 20 min before image acquisition. Shown in the figure are single, deconvolved z-sections.
The main cellular function of RCC1 is to stimulate nucleotide exchange by Ran to produce RanGTP from RanGDP. To investigate whether the observed movement of RCC1 on and off chromatin is affected by RCC1's interaction with Ran, we microinjected wt and mutant Rans (all in the GDP-bound form) into the cytoplasm of GFP-RCC1 tsBN2 cells. To mark the injected cells, we coinjected Texas Red-labeled 70-kDa dextran with each unlabeled Ran so that the microinjected cells could later be identified and selected for FRAP analysis. Wt Ran was compared with two mutant Rans, Q69L that is unable to hydrolyze GTP, and T24N that is defective in nucleotide uptake (Klebe et al., 1995). We estimate that the resulting intracellular concentration of microinjected Ran was ∼20 μM, which is approximately a 10-fold excess over the endogenous Ran in tissue culture cells (∼2 μM) (Bischoff and Ponstingl, 1991b). RCC1 is normally much less abundant than Ran (∼200 nM) (Bischoff and Ponstingl, 1991b), although the level of GFP-RCC1 in GFP-RCC1 tsBN2 cells seems somewhat higher than this (Figure 1). As a control, cells were also microinjected with the labeled dextran marker in injection buffer without Ran.
The injections of marker alone did not significantly alter the recovery rate of GFP-RCC1 fluorescence on photobleached interphase chromatin (Figure 4). Microinjection of wt Ran significantly slowed recovery (t1/2 of 11.2 ± 0.9 s) compared with uninjected cells (t1/2 of 8.7 ± 0.4 s) and buffer injected cells (t1/2 of 7.6 ± 0.6 s). The hydrolysis-defective mutant Q69L gave a mean recovery time slower than wt (t1/2 = 13.9 ± 1.1 s); however, this wasn't a statistically significant difference from wt Ran (p = 0.076). Strikingly, microinjection of T24N Ran dramatically decreased the recovery rate to t1/2 value of 18.0 ± 1.0 s. This reduced mobility was significantly different from that seen in cells receiving wt Ran or control cells. Thus, in contrast to energy depletion that resulted in a faster rate of recovery of GFPRCC1 fluorescence on photobleached interphase chromatin (Figure 2), increasing the intracellular concentration of wt Ran and Q69L Ran slowed the rate of recovery. This reduction in mobility was even more pronounced with T24N Ran (Figure 4, B and C). These results indicate that the transient associations of RCC1 with Ran that occur rapidly and repetitively in a living cell are capable of modulating the association of RCC1 with chromatin.
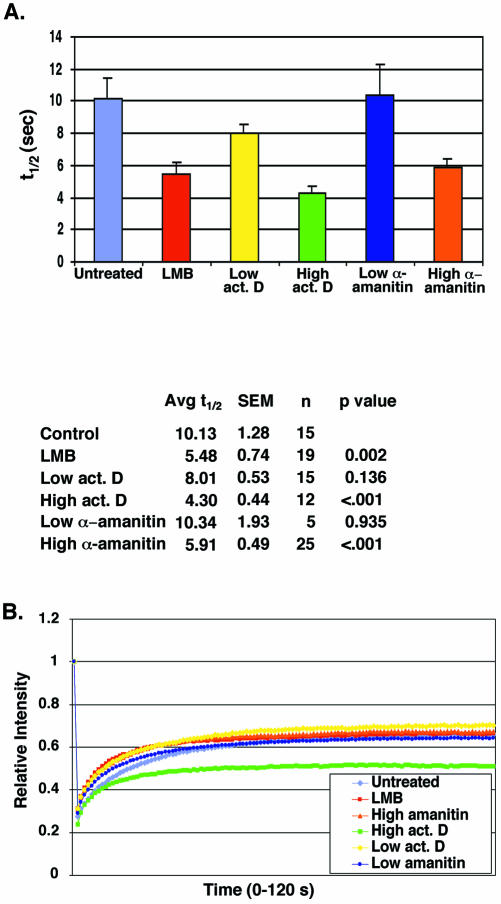
We questioned whether disrupting nuclear transport pathways that use RanGTP would alter the kinetics of RCC1 association with chromatin, possibly by altering the concentration of Ran accessible to RCC1. First, we tested the effects of LMB, which inhibits Crm1-mediated (Ran-dependent) nuclear export. We found that LMB treatment did increase the rate of fluorescence recovery after photobleaching of GFP-RCC1 approximately twofold (t1/2 of 5.48 ± 0.74 s) compared with untreated cells (t1/2 of 10.13 ± 1.28 s) (Figure 5A).
Figure 5.
Agents that disrupt various Ran-dependent export pathways also alter the mobility of GFP-RCC1. GFP-RCC1 tsBN2 cells were pretreated for 5 h (at 40°C) with the indicated agent before photobleaching. (A) Measurement of the t1/2 required for recovery of fluorescence after photobleaching after the various treatments. Cells were incubated for 5 h at 40°C before FRAP analysis with either LMB (50 ng/ml), low actinomycin D (0.02 μg/ml), high actinomycin D (3.2 μg/ml), low α-amanitin (50 μg/ml), or high α-amanitin (300 μg/ml). (B) The extent of recovery after photobleaching after no treatment (purple diamonds), LMB (red squares), high α-amanitin (orange triangles), high actinomycin D (green squares), low actinomycin D (yellow diamonds), and low α-amanitin (blue circles). The cells treated with high actinomycin D did not recover to the same level as the others, indicating the likely presence of a second, more immobile population of GFP-RCC1. In Figures 2B and 4B, 1.0 on the y-axis of the recovery graphs represents the final recovery achieved after photobleaching. In Figure 5B, 1.0 on the y-axis represents the initial fluorescence before photobleaching.
We next tested different RNA transcription inhibitors for their effects on GFP-RCC1 mobility (Bregman et al., 1995; Kamath et al., 2001). Newly synthesized ribosomal subunits and tRNAs both need Ran for export, however mRNA does not (Cullen, 2003). Three RNA polymerases carry out RNA transcription within the nucleus: pol I transcribes 5.8S, 18S, and 28S rRNA; pol II transcribes mRNA plus snoRNA and some snRNA; and pol III transcribes tRNA, 5SRNA, and some snRNA and other small RNAs. At a high concentration, actinomycin D inhibits all three polymerases but, at a lower concentration, selectively inhibits RNA pol I (Perry and Kelley, 1970). At a low concentration, α-amanitin inhibits RNA pol II and, at a higher concentration, also RNA pol III (Weinmann and Roeder, 1974).
Pretreatment with high actinomycin D or high α-amanitin (like LMB) significantly decreased the time required for recovery after photobleaching of GFP-RCC1 (Figure 5). Even though the mean recovery time was somewhat reduced after treatment with low actinomycin D (t1/2 of 8.01 ± 0.53 compared with 10.13 ± 1.28 s), the p value indicated this was not a significant difference (p = 0.136) (Figure 5A). Pretreatment of the cells with a low concentration of α-amanitin, which inhibits only mRNA transcription, had no effect on GFPRCC1 mobility. After treatment with LMB or the transcription inhibitors, a visual inspection of the cells indicated that the nuclear distribution of GFP-RCC1 did not noticeably change relative to its distribution in untreated cells (our unpublished data). Only after energy depletion (Figures 2A and 3) was there a noticeable change in GFP-RCC1 localization after any of these treatments or microinjections.
Inhibition of all three classes of RNA polymerases with high actinomycin D had an additional effect on GFP-RCC1 mobility that was not seen with any other treatment or microinjected agent. In every treatment discussed thus far, the kinetics of recovery was indicative of a single population of GFP-RCC1, and the extent of recovery was the same after all treatments even though the rate of this recovery changed. With high actinomycin D treatment, however, the final extent of recovery was reduced, indicating a second, more immobile fraction (Figure 5B). To demonstrate this graphically, intensities were normalized by dividing the actual intensity at each time point by the initial starting intensity. This normalization method clearly shows that the final fluorescence recovery levels for cells treated with high actinomycin D are lower, indicating a more immobile fraction is present. The nature of this immobile fraction of GFP-RCC1 after actinomycin D treatment is unknown.
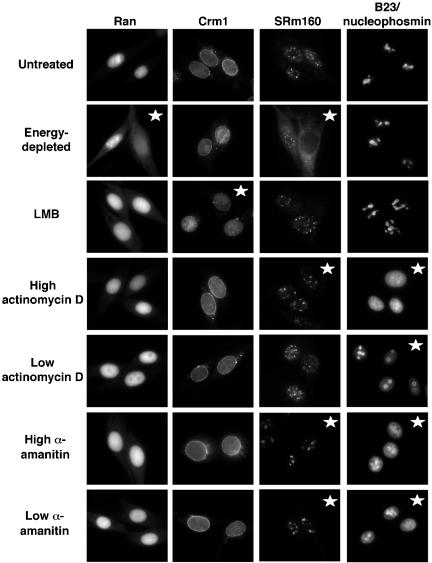
To ensure that these treatments were having their expected effects on cells and to observe the effects of these different treatments on the distribution of Ran and the nuclear export carrier Crm1, we fixed cells after treatment and performed indirect immunofluorescence microscopy with antibodies to Ran, Crm1, SRm160, or B23/nucleophosmin. SRm160 is a pre-mRNA splicing factor (Blencowe et al., 1998). Like the majority of mRNA splicing factors, SRm160 is not localized at sites of transcription but is instead enriched in nuclear domains called speckles (Wan et al., 1994; Misteli, 2000). When cells are treated with RNA pol II inhibitors, splicing activity is reduced and the speckles become fewer in number, enlarged, and rounded (Misteli et al., 1997). In GFP-RCC1 tsBN2 cells, the SRm160 containing nuclear speckles underwent these visual changes after treatment with either high actinomycin D or both concentrations of α-amanitin, indicating that these treatments were in fact inhibiting RNA transcription by pol II (Figure 6, third column). Interestingly, energy depletion resulted in a disappearance of SRm160-staining speckles from many nuclei; the reason for this is unknown.
Figure 6.
Immunofluorescence microscopy showing the effects of these various treatments on the localization of four cellular proteins. GFP-RCC1 tsBN2 cells were either energy depleted with Na azide and 2-deoxyglucose in gluc (-) media for 30 min at 40°C, or incubated at 40°C for 5 h with either LMB (50 ng/ml), low actinomycin D (0.02 μg/ml), high actinomycin D (3.2 μg/ml), low α-amanitin (50 μg/ml), or high α-amanitin (300 μg/ml). After treatment, the cells were fixed and processed for indirect immunofluorescence microscopy by using primary antibodies to Ran, the nuclear export carrier Crm1, the mRNA splicing factor SRm160, or the nucleolar protein B23/nucleophosmin followed by a TRITC- or Texas Redlabeled second antibody. Distribution patterns that differ from the pattern in untreated cells are marked with stars.
The nucleolar protein B23/nucleophosmin is well characterized with respect to its translocation in and out of nucleoli in response to changes in nucleolar and nuclear metabolism (Chan et al., 1985, 1996; Yung et al., 1985). For this reason, we used the localization of B23/nucleophosmin to monitor nucleolar changes resulting from the different treatments (Figure 6). Energy depletion and LMB had no effect on the nucleolar localization of B23/nucleophosmin; however, all of the RNA transcription inhibitors changed the distribution of this protein. After treatment with low actinomycin D to inhibit only RNA pol I, B23/nucleophosmin was found in a ring around the nucleolus, rather than throughout the nucleolus as in the untreated cells. This is a characteristic localization pattern of B23/nucleophosmin when rRNA synthesis by RNA pol I is blocked (Smetana et al., 2001). B23/nucleophosmin localization in nucleoli is also dependent on pol II transcription. When pol II is inhibited, B23/nucleophosmin is found throughout the nucleoplasm instead of being highly enriched in nucleoli (Desnoyers et al., 1996). This dispersement of B23/nucleophosmin within the nucleoplasm was observed after treatment with high actinomycin D and after treatment with both high and low concentrations of α-amanitin. Thus, these observed changes in the localization of B23/nucleophosmin and SRm160 confirmed that pol I transcription was being inhibited by low actinomycin D treatment and that pol II transcription was being inhibited by treatment with both high actinomycin D and both concentrations of α-amanitin.
The distribution of the nuclear export carrier Crm1 was unaffected by any of the transcription inhibitors or by energy depletion but was altered (not surprisingly) by LMB treatment (Figure 6). The bright nuclear rim staining of Crm1, still present after all the other treatments, was absent after LMB treatment. The cellular distribution of Ran on the other hand, was unaltered by the transcription inhibitors and LMB treatment but was changed upon energy depletion. Many energy-depleted cells showed a marked increase in the cytoplasmic pool of Ran with a corresponding decrease in the nuclear pool, and this altered Ran distribution has been observed before under conditions that block the production of RanGTP (Ren et al., 1993; Matynia et al., 1996). Notably, after energy depletion Ran did not seem to be enriched in any nuclear spots that would correspond to the “clumps” of nuclear GFP-RCC1 present in these energy-depleted cells. This may indicate GFP-RCC1 has a faster mobility in energydepleted cells because it is less likely to be associated with Ran (see DISCUSSION).
We also checked the ability of GFP-RCC1 tsBN2 cells after the various treatments to carry out Crm1-mediated nuclear export (our unpublished data). We microinjected the reporter TRITC-NES-BSA (a conjugate prepared with peptides containing the Rev leucine-rich NES) into the nuclei of untreated or treated cells (Schwoebel et al., 2002). After 30 min, untreated cells had exported most of this reporter to the cytoplasm. As expected, energy depletion and LMB treatment inhibited the export of this conjugate; however, the RNA transcription inhibitors were without effect (our unpublished data).
While this work was in progress, Zheng and coworkers reported that the association of GFP-RCC1 with chromatin in transfected 3T3 cells is highly dynamic, with kinetics of movement similar to those reported here in the GFP-RCC1 tsBN2 cells (Li et al., 2003). They also found microinjected T24N Ran inhibited GFP-RCC1 movement, and further in vitro experiments using the Xenopus egg extract system led to their conclusion that successful nucleotide exchange dissociates the RCC1–Ran complex, permitting RCC1 and Ran to release from chromatin (see DISCUSSION) (Li et al., 2003).
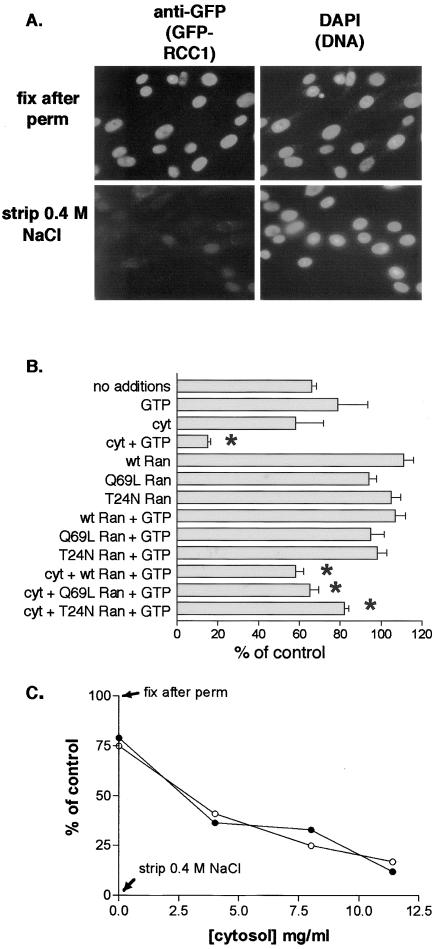
We devised an in vitro system to reconstitute the release of RCC1 from chromatin to determine the requirements for this process. GFP-RCC1 tsBN2 grown on coverslips were permeabilized with 0.1% Triton X-100, which permeabilizes both the plasma membrane and the nuclear envelope. GFPRCC1 stays bound to chromatin after permeabilization (Figure 7A, top row); however, it can be removed from the DNA with 0.4 M NaCl (Figure 7A, bottom row), consistent with the extraction properties of GFP-RCC1 and RCC1 from chromatin (Figure 1).
Figure 7.
Efficient GFP-RCC1 release from chromatin in permeabilized GFP-RCC1 tsBN2 cells requires cytosol and energy. (A) After permeabilization with 0.1% Triton X-100 and rinsing in buffer A, cells were either fixed immediately (top row) or incubated with buffer A containing 0.4 M NaCl for 20 min on ice before rinsing and fixation. GFP-RCC1 was localized with an anti-GFP first antibody and TRITC-labeled second antibody (left) and the cells were counterstained with 4,6-diamidino-2-phenylindole to localize the DNA. (B) After permeabilization, cells were incubated for 20 min at RT with the indicated additions before washing, fixation, and indirect immunofluorescence. GTP and Xenopus ovarian cytosol (cyt) were added at 1 mM and 11.5 mg/ml, respectively. wt, Q69L, or T24N Ran was added at 10 μM. Detection and quantitation of GFP-RCC1 was as described in MATERIALS AND METHODS. Shown for each condition are the mean and SEM of the nuclear fluorescence of between 30 and 90 nuclei. All values are expressed relative to the 100% (fixed after permeabilization) and the 0% (fixed after stripping with 0.4 M NaCl) controls. Note that the inclusion of 10 μM Ran decreased the loss obtained with cytosol alone (asterisks). (C) A concentration curve measuring the release of GFP-RCC1 obtained with increasing amounts of cytosol either in the presence (•) or absence (○) of 1 μM Ran. All samples contained 1 mM GTP.
To determine the requirements for GFP-RCC1 release from chromatin, coverslips after permeabilization were incubated with various additions. After rinsing and fixation, the amount of GFP-RCC1 remaining bound to the chromatin was quantitated (see MATERIALS AND METHODS). When the permeabilized cells were incubated for 20 min at RT with only buffer (34% loss), or buffer plus GTP (21% loss) there was a partial release of GFP-RCC1; however, most GFPRCC1 remained bound to chromatin throughout the incubation (Figure 7B). Notably however, when the permeabilized cells were incubated with cytosol plus GTP, the majority of GFP-RCC1 (85%) was released. This cytosolstimulated loss of GFP-RCC1 was at least partially dependent on added nucleotide, because when dialyzed cytosol was added without GTP the loss decreased to 45%.
Endogenous Ran is highly abundant in this Xenopus ovarian cytosol, representing ∼1% of the total protein in the S-100 giving a concentration of ∼4.6 μM when the cytosol is 11.5 mg/ml (Moore and Blobel, 1993). To determine whether the active component of cytosol responsible for stimulating release of GFP-RCC1 from chromatin is Ran, purified recombinant wt, Q69L, or T24N Ran were substituted for cytosol plus or minus GTP. Somewhat surprisingly, each of these Rans was unable to stimulate efficient release in the presence of GTP (Figure 7B). Furthermore, excess wt, Q69L, or T24N Ran added together with cytosol and GTP inhibited the cytosol-stimulated loss of GFP-RCC1 (asterisks). T24N Ran inhibited most strongly, decreasing the cytosol-stimulated loss from 85 to 18%, with Q69L and wt Ran decreasing the loss to 35 and 42%, respectively. This result is reminiscent of our microinjection results (Figure 4C), in which these three microinjected Rans slowed the rate of recovery after photobleaching of GFP-RCC1 with the same order of potency.
That cytosol, but not Ran alone, could stimulate release of GFP-RCC1 indicated that a necessary factor distinct from, or in addition to, Ran was present in the cytosol.
To determine whether a requirement for added Ran could be detected at a lower cytosol concentration, a concentration curve of cytosol plus or minus 1 μM Ran (plus GTP) was assayed for its ability to stimulate GFP-RCC1 release; however, the two curves were identical (Figure 7C). This result does not rule out the involvement of the cytosolic Ran in cytosol-catalyzed GFP-RCC1 release; if a requisite Ran cofactor is less abundant than Ran in the cytosol, then it is this factor that would be limiting at low protein concentrations rather than Ran.
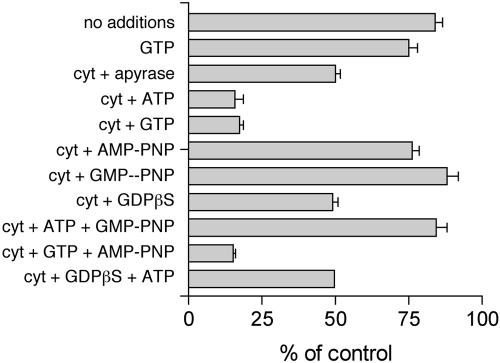
We analyzed the energy requirements for cytosol-stimulated release of GFP-RCC1 from chromatin (Figure 8). Buffer alone stimulated release of 16% of GFP-RCC1 and the inclusion of 1 mM GTP only slightly increased this loss (25% loss). As before, the inclusion of cytosol plus 1 mM GTP stimulated a much greater loss of 83%, and 1 mM ATP worked equally well (84% loss). We have previously observed that nucleoside diphosphate kinase activity remains in permeabilized cells and readily interconverts added ATP and GTP (our unpublished data). Thus, whether it is actually ATP or GTP (or both) that is required for GFP-RCC1 release can't be discerned in this experiment. Neither AMP-PNP nor GMP-PNP (nonhydrolyzable analogs of ATP and GTP, respectively) would stimulate release of GFP-RCC (unlike ATP and GTP), indicating a requirement for nucleotide hydrolysis. The addition of 1 mM GTP plus 1 mM AMP-PNP stimulated release of 85% of the GFP-RCC1; however, only 16% was released when 1 mM ATP plus 1 mM GMP-PNP were added. This result indicated that GTP hydrolysis is required for efficient GFP-RCC1 release, but ATP hydrolysis is not. Whether Ran is the GTPase hydrolyzing the GTP remains unresolved, because an excess of the hydrolysis-defective Ran mutant (Q69L) was less effective than an excess of the T24N mutant at preventing release of GFP-RCC1 either in vivo (Figure 4C) or in vitro (Figure 7B).
Figure 8.
The nucleotide requirements for GFP-RCC1 release from chromatin in permeabilized cells. Permeabilized GFP-RCC1 cells were incubated for 20 min at RT with the indicated addition before washing and fixation. Each indicated nucleotide was added at 1 mM. Cytosol (cyt) was added at 11.5 mg/ml. Detection and quantitation of GFP-RCC1 was as described in the MATERIALS AND METHODS. Between 40 and 90 nuclei were quantitated for each condition to yield the mean and SEM.
The inclusion of 1 mM GDPβ S, a GDP analog, rather than GTP in the cytosol resulted in a reduced loss of GFP-RCC1 (51% loss) relative to GTP (83% loss). This 51% loss was very similar to the loss achieved upon the inclusion of apyrase in the cytosol to deplete free nucleotides (50% loss) (Figure 8), or cytosol with no additions (45% loss) (Figure 7B). This result further implicates a GTPase in this process and indicates that the GTPase has to be in the GTP-bound form for efficient RCC1 release from chromatin. Possible explanations for these results are discussed below.
DISCUSSION
The association of RCC1 with chromatin is highly dynamic, both in interphase and mitotic cells (Figure 2, 3, 4) (Li et al., 2003). That the movement of RCC1 on and off chromatin is modulated by its association with Ran is supported by several lines of evidence. First, microinjection of wt, Q69L, or T24N Ran significantly slowed the nuclear mobility of GFP-RCC1, with T24N exhibiting the strongest effect. The T24N mutation renders Ran deficient in loading with nucleotide, an event that normally terminates the interaction between Ran and RCC1 (Klebe et al., 1995). Thus, T24N Ran remains bound to RCC1 in a nucleotide-free state. The slower mobility of GFP-RCC1 in the presence of excess Ran could be explained by the chromatin binding ability of Ran. Ran:RCC1 complexes contain two potential chromatin-interacting sites, rather than just the one of RCC1 alone, and therefore the Ran:RCC1 complex would be predicted to have a greater probability than RCC1 alone of being chromatin-bound. Notably, after energy depletion the nuclear Ran did not seem to colocalize with the clumps of RCC1 that occur after this treatment (compare Figures 3 and 6). GFP-RCC1 mobility may become faster after energy depletion because, for whatever reason, its binding to Ran is reduced.
We also observed significant effects on RCC1 mobility by treatments that disrupt various RanGTP-dependent nuclear transport pathways, including LMB and the RNA transcription inhibitors actinomycin D and α-amanitin. These treatments, however, significantly increased the mobility of GFPRCC1, unlike the microinjection of excess Ran, which decreased it. Notably, each of the treatments that increases the mobility of GFP-RCC1 would also be predicted to disrupt formation of nuclear export cargo:export carrier: RanGTP complexes.
This shortage of export cargo could be increasing GFPRCC1 mobility in the following way. Macara and colleagues have shown that RanBP3 (binding protein 3) can promote binding of Crm1 to RCC1 in the presence of Ran and that binding of RanBP3 to RCC1 increases the catalytic activity of RCC1 toward Ran ∼10-fold. The stimulatory effects of RanBP3 and histones are additive, meaning that RCC1 bound to histones and RanBP3 is a considerably more effi-cient GEF toward Ran than free RCC1. Macara and colleagues hypothesized that RanBP3 acts as a scaffold protein to promote the efficient assembly of export complexes at sites of RanGTP production (Lindsay et al., 2001; Nemergut et al., 2002). They also discussed that release of the Crm1–RanBP3–RanGTP–export cargo complex from RCC1 might possibly require additional cellular factors, because they found the addition of cargo in vitro did not promote disassembly. Even though RanBP3 is likely to increase the rate of nucleotide exchange of RCC1 to generate RanGTP such that an export complex can quickly assemble with cargo, assembly with cargo and release of that assembled complex from RCC1 may slow the release of RCC1 from chromatin. In the absence of cargo, our FRAP results indicate that the mobility of GFP-RCC1 increases, which might be explained by an absence of export complex formation that normally slows the release of GFP-RCC1 from chromatin.
How would the treatments that increased the mobility of GFP-RCC1 affect the production of export cargo? First, LMB disrupts CRM1 binding to a leucine-rich NES–containing export cargo; this cargo includes ribosomal subunits in addition to shuttling proteins (Kudo et al., 1998, 1999). LMB treatment increased GFP-RCC1 mobility (Figure 5A). Treatment with low actinomycin D (to inhibit just RNA pol I), and low α-amanitin (to inhibit just RNA pol II) did not significantly affect GFPRCC1 mobility (Figure 5A). Inhibition of RNA pol II and pol III by high α-amanitin, however, significantly increased the mobility of GFP-RCC1, as did treatment with high actinomycin D to inhibit all three RNA polymerases. Inhibition of mRNA synthesis by RNA pol II may have no effect on GFP-RCC1 mobility because mRNA export is thought not to involve Ran (Clouse et al., 2001). However, the failure of low actinomycin D (pol I inhibitor) to significantly alter the mobility of GFP-RCC1 is puzzling (Figure 5), because newly synthesized ribosomal subunits are probably a major contributor to the total RanGTP-dependent export load. Possibly, it is the total mass of Ran-dependent export cargo eliminated that is the critical variable rather than a specific type of cargo. 5S RNA as well as tRNAs are transcribed by RNA pol III, and the 5S RNA associates with the large ribosomal subunit before export of that subunit from the nucleus. To our knowledge, the effects of high α-amanitin on the export of large ribosomal subunits is unknown; however, it seems likely that it would block their export. Thus, low actinomycin D should inhibit formation of export complexes containing ribosomal subunits, whereas high actinomycin D should inhibit the formation of these, plus complexes containing tRNAs and mRNAs. Low α-amanitin should only inhibit the formation of mRNA export complexes, whereas the higher concentration should inhibit the formation of these, plus complexes containing tRNAs and large ribosomal subunits. Similarly to what has been hypothesized for Crm1 export complexes, other cofactors (similarly to RanBP3) may assemble with RCC1, Ran, other Ran-binding export carriers and cargo to facilitate their assembly at sites of RanGTP production. It is our hypothesis that the observed increases in GFP-RCC1 nuclear mobility resulting from treatment with LMB and RNA transcription inhibitors result from a decrease in export complexes assembling around RCC1, which enables GFP-RCC1 to move more rapidly off chromatin after nucleotide exchange by Ran.
Our in vitro system reconstituting GFP-RCC1 release from chromatin has revealed that this release involves more than just a simple RCC1-catalyzed exchange of guanine nucleotide by Ran (Figures 7 and 8). In a test tube, any free guanine nucleotide (e.g., GTP, GDP, GMP-PNP, GDPβS) will be taken up by Ran in the Ran:RCC1 complex and trigger dissociation of the complex (Klebe et al., 1995); however neither GDP-βS nor GMP-PNP would support efficient release in vitro (Figure 8). The inability of GMP-PNP to support release indicates a requirement for GTP hydrolysis in this process. A requirement for GTP hydrolysis by Ran would be unexpected due to the lack of RanGAP within the nucleus. Our data are actually more consistent with another GTPase distinct from Ran being involved, because the behaviors of wt Ran and the nonhydrolyzable mutant of Ran (Q69L), did not differ significantly either in vivo or in vitro (Figures 3 and 5), unlike the difference between GTP (83% loss) and GMP-PNP (12% loss) on the cytosol-mediated release of GFP-RCC1 in vitro (Figure 6). It is unclear why samples incubated with apyrase to deplete free tri- and diphosphate nucleotides from the system showed as much release as they did (50% loss) compared with samples receiving GMP-PNP (12% loss) (Figure 8). Possibly, a certain percentage of GFP-RCC1 is primed for release in such a way that additional nucleotide is unnecessary, although the exact reason for the apyrase result remains unknown. Whether the active cytosolic factors might represent components of nuclear export complexes, or factors (additional GTPases?) required for the final release of the assembled complexes from RCC1 remains to be determined. An interesting possibility is that many RCC1–RanGTP–RanBP3–Crm1–export cargo complexes are already assembled upon cell permeabilization and are awaiting the trigger (the cytosolic factor?) to release. Experiments are in progress to identify the active component(s) of cytosol required to stimulate GFP-RCC1 release from chromatin, and identification of this factor(s) will help to elucidate the mechanisms involved in this process.
Acknowledgments
This work was supported by a National Institute of Diabetes and Digestive and Kidney Diseases Molecular Endocrinology training grant National Institutes of Health DK07696 and National Institute of General Medical Sciences Initiative for Minority Student Development grant GM56929 (to I.C.), National Institutes of Health grant CA 41424 (to D.S.), and an American Heart Association grant 0355115Y (to M.S.M.). We thank Iain Mattaj for the his-Ran constructs and advice on purification of the proteins, Yoshiro Yoneda for leptomycin B, and Jeff Nickerson and Pui-Kwong Chan for antibodies.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc. E03-06-0409. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0409.
References
- Askjaer, P., et al. (1999). RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19, 6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., and Ponstingl, H. (1991a). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80-82. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., and Ponstingl, H. (1991b). Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc. Natl. Acad. Sci. USA 88, 10830-10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J., Issner, R., Nickerson, J.A., and Sharp, P.A. (1998). A coactivator of pre-mRNA splicing. Genes Dev. 12, 996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack, M.T., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E., and Gorlich, D. (2002). Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21, 6205-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman, D.B., Du, L., van der Zee, S., and Warren, S.L. (1995). Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129, 287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado, A., Treichel, N., Muller, E.C., Otto, A., and Kutay, U. (2002). Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21, 6216-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Guarguaglini, G., Gruss, O.J., Segref, A., Karsenti, E., and Mattaj, I.W. (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178-181. [DOI] [PubMed] [Google Scholar]
- Chan, P.K., Aldrich, M., and Busch, H. (1985). Alterations in immunolocalization of the phosphoprotein B23 in HeLa cells during serum starvation. Exp. Cell Res. 161, 101-110. [DOI] [PubMed] [Google Scholar]
- Chan, P.K., Qi, Y., Amley, J., and Koller, C.A. (1996). Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett. 100, 191-197. [DOI] [PubMed] [Google Scholar]
- Clarke, P.R., and Zhang, C. (2001). Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 11, 366-371. [DOI] [PubMed] [Google Scholar]
- Clouse, K.N., Luo, M.J., Zhou, Z., and Reed, R. (2001). A Ran-independent pathway for export of spliced mRNA. Nat. Cell Biol. 3, 97-99. [DOI] [PubMed] [Google Scholar]
- Cullen, B.R. (2003). Nuclear RNA export. J. Cell Sci. 116, 587-597. [DOI] [PubMed] [Google Scholar]
- Dasso, M. (2002). The Ran GTPase: theme and variations. Curr. Biol. 12, R502-R508. [DOI] [PubMed] [Google Scholar]
- Desnoyers, S., Kaufmann, S.H., and Poirier, G.G. (1996). Alteration of the nucleolar localization of poly(ADP-ribose) polymerase upon treatment with transcription inhibitors. Exp. Cell Res. 227, 146-153. [DOI] [PubMed] [Google Scholar]
- Dou, Y., Bowen, J., Liu, Y., and Gorovsky, M.A. (2002). Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158, 1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I.W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- Fribourg, S., Braun, I.C., Izaurralde, E., and Conti, E. (2001). Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 8, 645-656. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308-311. [DOI] [PubMed] [Google Scholar]
- Gorlich, D., Dabrowski, M., Bischoff, F.R., Kutay, U., Bork, P., Hartmann, E., Prehn, S., and Izaurralde, E. (1997). A novel class of RanGTP binding proteins. J. Cell Biol. 138, 65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol. 15, 607-660. [DOI] [PubMed] [Google Scholar]
- Guarguaglini, G., Renzi, L., D'Ottavio, F., Di Fiore, B., Casenghi, M., Cundari, E., and Lavia, P. (2000). Regulated Ran-binding protein 1 activity is required for organization and function of the mitotic spindle in mammalian cells in vivo. Cell Growth Differ. 11, 455-465. [PubMed] [Google Scholar]
- Hetzer, M., Bilbao-Cortes, D., Walther, T.C., Gruss, O.J., and Mattaj, I.W. (2000). GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell 5, 1013-1024. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., Gruss, O.J., and Mattaj, I.W. (2002). The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4, E177-E184. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I.W., and Gorlich, D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A.W., Lund, E., and Dahlberg, J. (2002). Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27, 580-585. [DOI] [PubMed] [Google Scholar]
- Kalab, P., Pu, R.T., and Dasso, M. (1999). The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9, 481-484. [DOI] [PubMed] [Google Scholar]
- Kamath, R.V., Leary, D.J., and Huang, S. (2001). Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol. Biol. Cell 12, 3808-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H., and Cook, P.R. (2001). Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153, 1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe, C., Bischoff, F.R., Ponstingl, H., and Wittinghofer, A. (1995). Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34, 639-647. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Khochbin, S., Nishi, K., Kitano, K., Yanagida, M., Yoshida, M., and Horinouchi, S. (1997). Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J. Biol. Chem. 272, 29742-29751. [DOI] [PubMed] [Google Scholar]
- Kudo, N., Matsumori, N., Taoka, H., Fujiwara, D., Schreiner, E.P., Wolff, B., Yoshida, M., and Horinouchi, S. (1999). Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96, 9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E.P., Yoneda, Y., Yanagida, M., Horinouchi, S., and Yoshida, M. (1998). Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242, 540-547. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Ohno, M., and Mattaj, I.W. (2001). Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11, 497-503. [DOI] [PubMed] [Google Scholar]
- Kutay, U., Lipowsky, G., Izaurralde, E., Bischoff, F.R., Schwarzmaier, P., Hartmann, E., and Gorlich, D. (1998). Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1, 359-369. [DOI] [PubMed] [Google Scholar]
- Lever, M.A., Th'ng, J.P., Sun, X., and Hendzel, M.J. (2000). Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408, 873-876. [DOI] [PubMed] [Google Scholar]
- Li, H.Y., Wirtz, D., and Zheng, Y. (2003). A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160, 635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, M.E., Holaska, J.M., Welch, K., Paschal, B.M., and Macara, I.G. (2001). Ran-binding protein 3 is a cofactor for crm1-mediated nuclear protein export. J. Cell Biol. 153, 1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matynia, A., Dimitrov, K., Mueller, U., He, X., and Sazer, S. (1996). Perturbations in the spi1p GTPase cycle of Schizosaccharomyces pombe through its GTPase-activating protein and guanine nucleotide exchange factor components result in similar phenotypic consequences. Mol. Cell. Biol. 16, 6352-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. (2000). Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J. Cell Sci. 113, 1841-1849. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Caceres, J.F., and Spector, D.L. (1997). The dynamics of a premRNA splicing factor in living cells. Nature 387, 523-527. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Gunjan, A., Hock, R., Bustin, M., and Brown, D.T. (2000). Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877-881. [DOI] [PubMed] [Google Scholar]
- Moore, M.S., and Blobel, G. (1992). The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell 69, 939-950. [DOI] [PubMed] [Google Scholar]
- Moore, M.S., and Blobel, G. (1993). The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365, 661-663. [DOI] [PubMed] [Google Scholar]
- Moore, W., Zhang, C., and Clarke, P.R. (2002). Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 12, 1442-1447. [DOI] [PubMed] [Google Scholar]
- Nemergut, M.E., Lindsay, M.E., Brownawell, A.M., and Macara, I.G. (2002). Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J. Biol. Chem. 277, 17385-17388. [DOI] [PubMed] [Google Scholar]
- Nemergut, M.E., Mizzen, C.A., Stukenberg, T., Allis, C.D., and Macara, I.G. (2001). Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540-1543. [DOI] [PubMed] [Google Scholar]
- Ohba, T., Nakamura, M., Nishitani, H., and Nishimoto, T. (1999). Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356-1358. [DOI] [PubMed] [Google Scholar]
- Ohtsubo, M., Okazaki, H., and Nishimoto, T. (1989). The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109, 1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R.P., and Kelley, D.E. (1970). Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell Physiol. 76, 127-139. [DOI] [PubMed] [Google Scholar]
- Ren, M., Drivas, G., D'Eustachio, P., and Rush, M.G. (1993). Ran/TC 4, a small nuclear GTP-binding protein that regulates DNA synthesis. J. Cell Biol. 120, 313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Pante, N., and Hurt, E. (1998). Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18, 6826-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwoebel, E.D., Ho, T.H., and Moore, M.S. (2002). The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 157, 963-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwoebel, E.D., Talcott, B., Cushman, I., and Moore, M.S. (1998). Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J. Biol. Chem. 273, 35170-35175. [DOI] [PubMed] [Google Scholar]
- Smetana, K., Busch, R., Chan, P.K., Smetana, K., Jr., and Busch, H. (2001). Immunocytochemical localization of nucleophosmin and RH-II/Gu protein in nucleoli of HeLa cells after treatment with actinomycin D. Acta Histochem. 103, 325-333. [DOI] [PubMed] [Google Scholar]
- Stenoien, D.L., Mielke, M., and Mancini, M.A. (2002). Intranuclear ataxin1 inclusions contain both fast- and slow-exchanging components. Nat. Cell Biol. 4, 806-810. [DOI] [PubMed] [Google Scholar]
- Stenoien, D.L., Patel, K., Mancini, M.G., Dutertre, M., Smith, C.L., O'Malley, B.W., and Mancini, M.A. (2001). FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol. 3, 15-23. [DOI] [PubMed] [Google Scholar]
- Talcott, B., and Moore, M.S. (2000). The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin alpha3/Qip. J. Biol. Chem. 275, 10099-10104. [DOI] [PubMed] [Google Scholar]
- Trieselmann, N., and Wilde, A. (2002). Ran localizes around the microtubule spindle in vivo during mitosis in Drosophila embryos. Curr. Biol. 12, 1124-1129. [DOI] [PubMed] [Google Scholar]
- Wan, K.M., Nickerson, J.A., Krockmalnic, G., and Penman, S. (1994). The B1C8 protein is in the dense assemblies of the nuclear matrix and relocates to the spindle and pericentriolar filaments at mitosis. Proc. Natl. Acad. Sci. USA 91, 594-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann, R., and Roeder, R.G. (1974). Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc. Natl. Acad. Sci. USA 71, 1790-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, A., Lizarraga, S.B., Zhang, L., Wiese, C., Gliksman, N.R., Walczak, C.E., and Zheng, Y. (2001). Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3, 221-227. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., and Newport, J. (2003). A Role for Ran-GTP and Crm1 in blocking re-replication. Cell 113, 115-125. [DOI] [PubMed] [Google Scholar]
- Yung, B.Y., Busch, R.K., Busch, H., Mauger, A.B., and Chan, P.K. (1985). Effects of actinomycin D analogs on nucleolar phosphoprotein B23 (37, 000 daltons/pI 5.1). Biochem. Pharmacol. 34, 4059-4063. [DOI] [PubMed] [Google Scholar]
- Zhang, C., and Clarke, P.R. (2000). Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288, 1429-1432. [DOI] [PubMed] [Google Scholar]
- Zhang, C., and Clarke, P.R. (2001). Roles of Ran-GTP and Ran-GDP in precursor vesicle recruitment and fusion during nuclear envelope assembly in a human cell-free system. Curr. Biol. 11, 208-212. [DOI] [PubMed] [Google Scholar]