Abstract
Receptor-mediated trafficking of cholesterol between lipoproteins and cells is a fundamental biological process at the organismal and cellular levels. In contrast to the well-studied pathway of LDL receptor-mediated endocytosis, little is known about the trafficking of high-density lipoprotein (HDL) cholesterol by the HDL receptor, scavenger receptor BI (SR-BI). SR-BI mediates HDL cholesteryl ester uptake in a process in which HDL lipids are selectively transferred to the cell membrane without the uptake and degradation of the HDL particle. We report here the cell surface locale where the trafficking of HDL cholesterol occurs. Fluorescence confocal microscopy showed SR-BI in patches and small extensions of the cell surface that were distinct from sites of caveolin-1 expression. Electron microscopy showed SR-BI in patches or clusters primarily on microvillar extensions of the plasma membrane. The organization of SR-BI in this manner suggests that this microvillar domain is a way station for cholesterol trafficking between HDL and cells. The types of phospholipids in this domain are unknown, but SR-BI is not strongly associated with classical membrane rafts rich in detergent-resistant saturated phospholipids. We speculate that SR-BI is in a more fluid membrane domain that will favor rapid cholesterol flux between the membrane and HDL.
INTRODUCTION
Receptor-mediated trafficking of cholesterol between lipoproteins and cells is a fundamental biological process at both the organismal and cellular levels. At the organismal level, these processes determine plasma cholesterol levels and are major factors in the development of atherosclerotic cardiovascular disease. At the cellular level, receptor-mediated lipoprotein trafficking provides cholesterol for membrane biogenesis, for maintenance of membrane fluidity and function, and for the synthesis of steroid hormones and bile acids. The best studied cholesterol trafficking pathway is that defined by the low-density lipoprotein (LDL) receptor and its related family members (Brown and Goldstein, 1986; Krieger and Herz, 1994). LDL receptors mediate the hepatic uptake and processing of LDL and other atherogenic lipoproteins via classical receptor-mediated endocytosis in which the bound lipoprotein is concentrated in clathrin-coated pits, internalized, and degraded in the endosomallysosomal pathway to release cholesterol for metabolism in the cell (Brown and Goldstein, 1986).
High-density lipoprotein (HDL) is the other major lipoprotein involved in the delivery of plasma cholesterol to the liver and the trafficking of cholesterol between HDL and many peripheral cells. Scavenger receptor class B, type I (SR-BI) is a cell surface receptor for HDL (Acton et al., 1996; for reviews see Krieger, 1999; Williams et al., 1999; Silver and Tall, 2001). SR-BI is expressed at high levels in the liver and in steroidogenic cells where it mediates the selective uptake of cholesteryl ester (CE) from HDL. Studies with gene-targeted mice and mice in which SR-BI was over expressed in the liver show that SR-BI determines the level of plasma HDL and mediates the uptake of both HDL CE and free cholesterol (FC) into the liver for transport into bile (Kozarsky et al., 1997; Rigotti et al., 1997; Varban et al., 1998; Wang et al., 1998; Ueda et al., 1999). SR-BI protects against the development of atherosclerosis in mice (Arai et al., 1999; Trigatti et al., 1999; Kozarsky et al., 2000; Ueda et al., 2000; Braun et al., 2002). In steroidogenic cells, SR-BI is regulated by tropic hormones (Landschulz et al., 1996; Rigotti et al., 1996) and is the major route for delivery of HDL-cholesterol to the steroidogenic pathway (Temel et al., 1997). Thus, SR-BI plays a key role in cellular and systemic cholesterol metabolism and is important in the prevention of atherosclerotic vascular disease.
The mechanisms by which SR-BI mediates cholesterol movement between HDL and cells are not well understood. HDL CE selective uptake is defined as the movement of CE from HDL into target cells without significant internalization and degradation of the HDL particle (Gwynne and Hess, 1980; Glass et al., 1983; Stein et al., 1983; Reaven et al., 1984; Glass et al., 1985; Pittman et al., 1987). This mechanism is distinct from that of the LDL receptor pathway, where LDL is internalized and the particle is degraded in the endosomal/lysosomal pathway (Brown and Goldstein, 1986). SR-BI also mediates the bidirectional flux of FC between HDL and cells with the direction of cholesterol movement determined by the cholesterol concentration gradient between cells and HDL (Ji et al., 1997; de la Llera-Moya et al., 1999; de la Llera-Moya et al., 2001). By accelerating the transfer of HDL lipids, SR-BI provides a conduit for the rapid mass movement of CE and FC between cells and HDL. Very little is known about the plasma membrane locale where SR-BI-mediated cholesterol trafficking occurs. In steroidogenic cells in vivo, SR-BI is found in an elaborate cell surface compartment of HDL-filled microvillar channels that form by the stacking of microvilli or by juxtaposition of microvilli against the cell surface (Reaven et al., 1989; Reaven et al., 1998). In cultured cells SR-BI has been reported to be present in caveolae, which act as sites of HDL cholesterol trafficking (Babitt et al., 1997; Graf et al., 1999). In the present study we have used SR-BI-specific localization methods at the light and electron microscopic levels to define the cell surface location of the active SR-BI receptor in a variety of cultured cells. The results show that the vast majority of SR-BI is present in patches or clusters that are predominantly on microvillar extensions of the plasma membrane. Detergent solubilization experiments suggest that SR-BI is not in a classical raft or ordered membrane domain. These findings suggest that the microvillar extensions containing clusters of SR-BI are a membrane domain that serves as a way station for cholesterol trafficking between cells and HDL.
MATERIALS AND METHODS
Materials
Monomeric avidin columns and sulfo-NHS-LC-LC-biotin were from Pierce (Rockford, IL). Rabbit polyclonal antibody against the c-terminal portion of SR-BI was from Novus Biologicals (Littleton, CO). Rabbit polyclonal antibody against the extracellular portion of SR-BI has been described (no. 356; Temel et al., 1997). Rabbit polyclonal antibody against caveolin-1 (cav-1) was from Transduction Laboratories (Lexington, KY). Mouse monoclonal antibodies against transferrin receptor 1 (TfR-1) was from Alpha Diagnostic (San Antonio, TX). All fluorescently tagged secondary antibodies and the Alexa-568 protein-labeling kit were from Molecular Probes (Eugene, OR). All HRP-conjugated secondary antibodies were from Amersham (Piscataway, NJ). The 10-nm gold-conjugated goat antibiotin secondary antibody was from BB International (Cardiff, United Kingdom). The 15-nm gold-conjugated goat anti-rabbit secondary antibody was from Amersham. Phosphatidylethanolamine labeled with a 1.4-nm nanogold cluster and gold enhancement kits were from Nanoprobes (Yaphank, NY). Lubrol WX was purchased from Serva (Paramus, NJ), Triton X-100 from Sigma (St. Louis, MO), and phospholipids from Avanti Polar Lipids (Birmingham, AL). CNBr-activated Sepharose 4B was obtained from Sigma.
HDL Labeling
Human HDL3 (1.125 g/ml < ρ < 1.210 g/ml; hereafter referred to as HDL) was prepared by ultracentrifugation (Havel et al., 1955) and labeled with [125I]dilactitol tyramine and [3H]cholesteryl oleoyl ether (125I/[3H]HDL) for HDL CE selective uptake experiments as described (Connelly et al., 1999). For measurements of HDL CE selective uptake, WI38[SR-BI] (de la Llera-Moya et al., 2001) or adrenocorticotropic hormone (ACTH)-treated Y1-BS1 (Temel et al., 1997) cells were incubated with 10 μg/ml 125I/[3H]HDL for 60 min at 37°C, washed, and processed to determine radiolabel incorporation (Connelly et al., 1999). The values for 125I radioactivity in acid-insoluble and -soluble fractions and the value for organic solvent extractable 3H radioactivity were used as described (Connelly et al., 1999) to determine the amount of HDL CE that was cell surface associated, endocytosed, and taken up by selective uptake.
For fluorescent staining experiments, HDL was labeled with Alexa-568 protein labeling kit using the company protocol. For electron microscopy, biotinylated HDL was prepared by incubating HDL (13 mg/ml, 150 μl) with sulfo-NHS-LC-LC-biotin (5 mg/ml, 20 μl) for 2 h at room temperature. Biotinylated HDL was purified by binding and elution from a monomeric avidin column. Reconsituted 11 nm discoidal HDL were prepared from purified apoA-I and palmitoyloleolylphosphatidylcholine as described (Sparks et al., 1992) except that nanogold-phosphatidylethanolamine was included at 1% (mole/mol) of phospholipid. The reconstituted gold HDL was purified from unincorporated nanogold-phosphatidylethanolamine by chromatography on a Superose 6 column (Pharmacia) and collection of the HDL region of the profile. Stoichiometry of gold incorporation into the HDL was determined by the absorbances at 420 nm (gold) and 280 nm (apoA-I) and ranged from 0.5 to 0.8 nanogold complex/HDL particle.
Cell Culture and Transfection
All cell lines were grown in T-175 flasks and maintained at 37°C in a humidified incubator under 5% CO2. COS-7 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, San Diego, CA) supplemented with 10% calf serum, 1% glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1% sodium pyruvate (Invitrogen). LdlA7[SR-BI] cells were maintained in Ham's F12 complete media, 5% heat-inactivated fetal bovine serum, 1% glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, with 0.25 mg/ml G418. WI38-VA13 cells (human lung fibroblast) stably transfected with SR-BI (WI38[SR-BI]) or parent WI38 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, with or without 0.8 mg/ml G418, respectively. Y1-BS1 cells (murine adrenocortical cells) were grown in F10-HAM media (Sigma) supplemented with 2.5% fetal bovine serum, 12.5% horse serum, 1% glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. FRT cells were maintained in Ham's F12 medium with Coon's modification containing 10% FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin. Before transfection, cells were grown in six-well plates for 24 h. All cell lines were transfected using Fugene (Roche Molecular Biochemicals, Indianapolis, IN) based on the company's protocol.
Fluorescence Staining
Cells were grown on coverslips for 48 h before staining. Transiently transfected cells were replated onto coverslips 24 h after transfection, and cells were processed 24 h after replating. Y1-BS1 cells were treated with 100 nM ACTH for 24 h before staining. For fluorescent HDL labeling experiments, cells were incubated with Alexa-568 HDL (20 μg/ml) for 30 min on ice and then washed with PBS and fixed for 1 h with 4% (wt/vol) paraformaldehyde in 77 mM PIPES, pH 7.5. Cells were washed in PBS and mounted using ProLong antifade mounting media.
For immunofluorescence experiments, cells were fixed with 4% paraformaldehyde for 1 h, permeabilized with 0.3% Triton X-100 for 15 min, blocked with 3% BSA, 10 mM glycine in PBS for 1 h, and then incubated with anti-cav-1 antibody (0.5 μg/ml). After a 1-h incubation, cells were washed with PBS and treated with secondary antibody for 30 min. Cells were washed and mounted as above. Cells were examined using a Nikon Eclipse E800 microscope (Garden City, NJ) and confocal images were collected with a Bio-Rad Radiance 2000 system (Cambridge, MA).
Electronmicroscopy
Cells were grown on ACLAR (Ted Pella, Redding, CA) for 48 h before experiments. For experiment using biotinylated HDL, cells were incubated with labeled HDL (40 μg/ml) at 4°C for 90 min. Cells were fixed with 2% glutaraldehyde/1% paraformaldehyde in PBS for 30 min, blocked with 10% normal goat serum in PBS for 15 min, and incubated with goat antibiotin gold secondary antibody for 1 h.
For experiments using antibodies raised against the extracelluar portion of SR-BI, cells were fixed with 4% paraformaldehyde/0.25% glutaraldehyde in PBS for 1 h and blocked with 3% BSA/10 mM glycine in PBS for 1 h. Cells were incubated with primary antibody, 5 μg/ml, for 1 h and gold-conjugated secondary antibody for 30 min. Cells were then postfixed with 2% glutaraldehyde in PBS. For these experiments the anti-SR-BI 356 antibody was processed to remove reactivity to glutathione-S-transferase by overnight incubation with glutathione-S-transferase coupled to CNBr-activated Sepharose 4B in 150 mM NaCl, 20 mM Tris, pH 7.4, containing 0.1% Triton X-100. The nonabsorbed fraction was then bound to the glutathione-S-transferase/SR-BI-extracellular domain fusion protein similarly coupled to CNBr-activated Sepharose 4B by overnight incubation at 4°C in 150 mM NaCl, 20 mM Tris, pH 7.4, containing 0.1% Triton X-100. After washing, the resin was eluted with 5 M MgCl2, 0.1% Triton X-100, 18 mM Tris-HCl, pH 5.5, dialyzed against 150 mM NaCl, 50 mM Tris, pH 8.0, 0.1% Triton X-100, and concentrated using a Centricon-50 centrifugal concentrator (Millepore, Bedford, MA).
For experiment using gold HDL, cells were incubated with gold HDL (10 μg/ml) at 4°C for 90 min. Cells were fixed with 2% glutaraldehyde/1% paraformaldehyde in PBS for 30 min and then gold-enhanced according to company protocol. Samples were postfixed with reduced osmium tetroxide (2% OsO4/3% potassium ferrocyanide) for 90 min, rinsed three times for 10 min in 0.1 M phosphate buffer, pH 7.4, three times for 10 min in deionized water, and en bloc stained with aqueous 1% uranyl acetate for 1 h. Samples were rinsed three times for 10 min in deionized water, dehydrated through an ascending series of ethanol, and embedded in Durcupan (Fluka, Buchs, Switzerland) between sheets of Aclar. Ultrathin sections (60-90 nm) were cut on a Reichert Ultracut E ultramicrotome, picked up on formvar-coated nickel slot grids, and poststained with 1% methanolic uranyl acetate and 0.3% aqueous lead citrate. The grids were then examined with a JEOL 1200 EX electron microscope (JEOL, Peabody, MA).
For postembedding staining of cav-1, cells were fixed in 1% paraformaldeyde, 0.5% glutaraldehyde, postfixed in 1% osmium tetroxide, and embedded as described above. Thin sections (60-90 nm, pale gold) were cut, picked up on formvar-coated nickel slot grids, and treated in a saturated aqueous solution of sodium metaperiodate for 10 min (Rajamannan et al., 2002). Grids were rinsed four times for 5 min in dH2O, three times for 5 min in PBS + 0.05% Tween-20, blocked 15 min in PBS + 0.05% Tween-20 + 1% glycine + 2% normal goat serum, and incubated with anti-cav-1 antibody in blocking buffer overnight at room temperature. Grids were rinsed four times for 5 min in PBS + 0.05% Tween-20, incubated 1 h with 15-nm gold-labeled goat anti-rabbit (Amersham) in PBS + 0.05% Tween-20, rinsed three times for 5 min in PBS + 0.05% Tween-20, three times for 5 min in dH2O, postfixed 10 min in 2% glutaraldehyde, and rinsed two times for 5 min in dH2O. Grids were then poststained with uranyl acetate and lead citrate and examined with a JEOL 1200 EX electron microscope.
Sucrose Gradient Fractionation
Cells were grown to confluency in 35-mm dishes and lysed in 0.5 ml TNE buffer (25 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM EDTA) containing 1% Triton X-100 (vol/vol) or Lubrol WX (wt/vol) by passage through a 26-gauge needle. The cell lysates were incubated on ice for 30 min, mixed with 0.5 ml 80% (wt/vol) sucrose in TNE buffer, and then transferred to SW60 centrifuge tubes. A step sucrose gradient was formed by layering of 2.5 ml of 38% (wt/vol) and 1 ml of 5% (wt/vol) sucrose in TNE buffer. To decide the amount of time needed to form stable gradients, samples were centrifuged at 38,000 rpm (148,305 × g) for a series of time points. Stable gradients were formed after 3 h of centrifugation, and further spinning did not change the protein distribution pattern (personal communication from Dr. Deborah A. Brown). On the basis of the results from this pilot experiment, we decided to centrifuge all samples for 3 h. After centrifugation, 12 fractions were collected from each sample from top to bottom. The distribution patterns of SR-BI, cav-1, and transferrin receptor 1 (TfR1) were analyzed by immunoblotting. TfR1 was used as a marker for nonraft membranes. In Triton X-100 extracts TfR1 was found only in high-density fractions, indicating detergent solubilization was complete.
RESULTS
Cell Surface Distribution of SR-BI
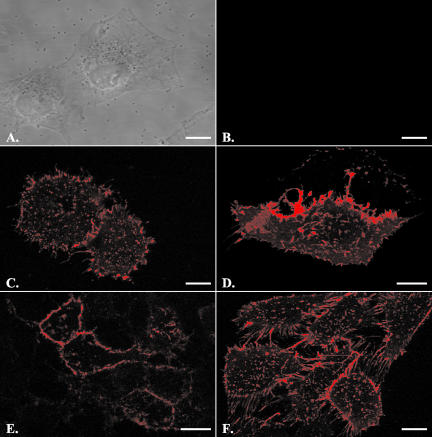
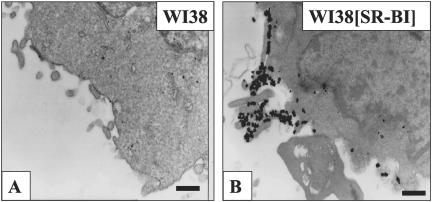
To specifically monitor SR-BI on the cell surface, cells were incubated at 4°C with HDL that was labeled on the protein moiety with Alexa-568. As shown in Figure 1, A and B, the WI38 human lung fibroblast cell line shows no Alexa-568 HDL binding in the absence of SR-BI expression. In contrast, WI38[SR-BI] cells stably expressing SR-BI show prominent cell surface Alexa-568 HDL binding in variable-sized patches, on short cell surface extensions, and on larger membrane extensions that extend between adjacent cells (Figure 1C). WI38[SR-BI] cells express levels of SR-BI similar to those seen in adrenal or liver cells naturally expressing SR-BI (unpublished data). Thus, this pattern of SR-BI distribution is not the result of vast over expression. Additionally, WI38[SR-BI] cells exhibit the various activities of SR-BI on cholesterol transport as well as its effects on membrane cholesterol distribution as observed in other cell types (de la Llera-Moya et al., 2001). A similar distribution of cell surface SR-BI was seen on transiently transfected COS-7 cells (Figure 1D), murine Y1 adrenocortical cells (Figure 1E), and ldlA7[SR-BI] cells (Figure 1F), a Chinese hamster ovary cell line lacking the LDL receptor and stably expressing murine SR-BI (Acton et al., 1996).
Figure 1.
Cells expressing SR-BI showed prominent cell surface HDL binding. Cells were incubated with fluorescently labeled HDL (20 μg/ml) for 30 min on ice and then washed with PBS and fixed with 4% paraformaldehyde. Scale bars, 10 μm. The five cell lines used were WI38 cells (A and B), stably transfected WI38[SR-BI] cells (C), transiently transfected COS-7 cells (D), ACTH treated Y1-BS1 cells (E), and stably transfected ldlA7 cells (F). A and B demonstrate that in the absence of SR-BI expression, no HDL staining was observed. Cells from all SR-BI-transfected cell lines showed prominent cell surface HDL binding in irregularly shaped patches with varying sizes. They also showed heavy staining on cell processes and on extensions connecting the cells.
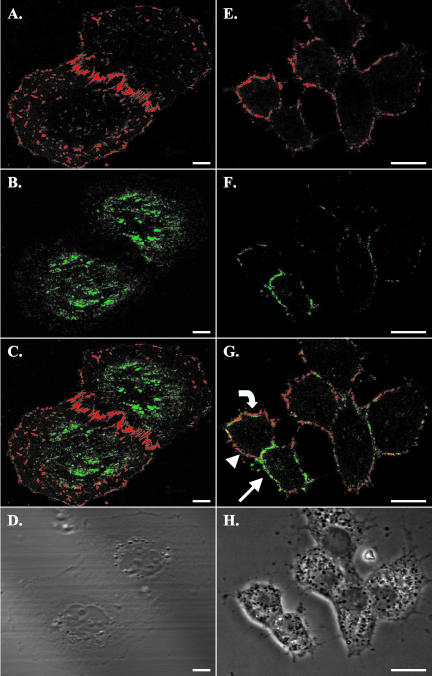
The relationship between cell surface SR-BI and cav-1 was determined by first incubating cells with Alexa-568 HDL, followed by fixation, permeabilization with detergent, incubation with polyclonal anti-cav-1 antibody, and detection with Alexa-488-labeled secondary antibody. Figure 2A (phase image in Figure 2D) shows a confocal section of WI38[SR-BI] cells near the coverslip and illustrates cell surface SR-BI in numerous patches and on cell surface extensions. Cav-1 staining (Figure 2B) was interspersed, but not coincident, with SR-BI on the surface of the cell (merged image, Figure 2C). Cav-1 staining was stronger in the central regions of the cells where it was interspersed with lighter patches of SR-BI. In a few places, colocalization of the labels was seen.
Figure 2.
Costaining for SR-BI and caveolin in stably transfected WI38[SR-BI] and ACTH-treated murine adrenal Y1-BS1 cells showed little colocalization of the two proteins. Cells were first incubated with fluorescently labeled HDL as described in Figure 1. Cells were fixed with 4% paraformaldehyde, permeabilized with detergent, incubated with polyclonal antibody against cav-1, and stained with Alexa-488-tagged secondary antibody. Scale bars, 10 μm. (A-D) WI38[SR-BI] cells; (E-H) Y1-BS1 cells. (A and E) SR-BI staining is shown; (B and F) Cav-1 staining is shown. (C and G) The merged images. (D and H) The phase contrast images. Y1-BS1 cells were treated with ACTH for 24 h before staining to maximize SR-BI expression. The images for WI38[SR-BI] cells were taken near the coverslips, whereas the images for the Y1-BS1 cells were taken from the middle of the cells. HDL staining (A and E) showed a patched distribution pattern of SR-BI on the cell membrane, as observed in other cell types. In WI38[SR-BI] cells, cav-1 staining was prominent under the nucleus but was also seen throughout the flattened regions of the cells. In Y1-BS1 cells cav-1 staining was clear on the cell edge in patches (B and F, respectively). In the merged images, some colocalization was observed (C and G, yellow), but the primary impression is the lack of colocalization. There were several prominent features in Y1-BS1 cell staining. Some regions of membrane showed only SR-BI staining (G, arrowhead), or only cav-1 staining (G, arrow), but not both. There were also patches where cav-1 staining appeared to be internal to SR-BI staining (G, curved arrow).
Figure 2E (phase image in Figure 2H) shows a confocal section of ACTH-treated Y1-BS1 adrenal cells through the central region of the cells after incubation with Alexa-568 HDL. Staining was seen in patches along the plasma membrane. Cav-1 staining was also largely along the plasma membrane (Figure 2F), but as illustrated in the merged image (Figure 2G), most of the cav-1 and SR-BI staining were not coincident. Although minor colocalization of SR-BI and cav-1 was seen (yellow in Figure 2G), two prominent features characterize their distributions. First, large regions of the plasma membrane show either cav-1 stain (Figure 2G, arrow) or SR-BI stain (Figure 2G, arrowhead), but not both. Second, in some regions (curved arrow) cav-1 stain appears slightly internal to the SR-BI stain. These features were also observed in WI38[SR-BI] cells with confocal sections through the central region of the cell (unpublished data). Similar patterns of SR-BI and cav-1 staining were seen in transiently transfected COS-7 cells and in ldlA7[SR-BI] cells (unpublished data). These results indicate that SR-BI is localized in patches on the cell surface that show little overlap with cav-1-containing structures.
SR-BI Localization at the Electron Microscopic Level
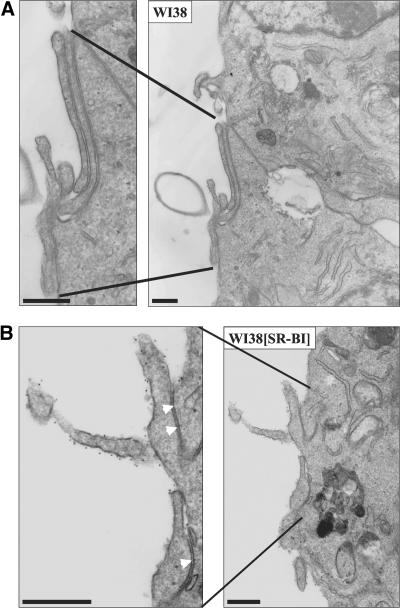
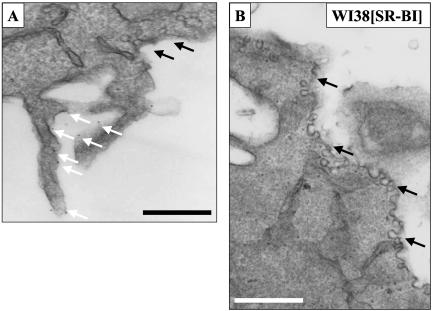
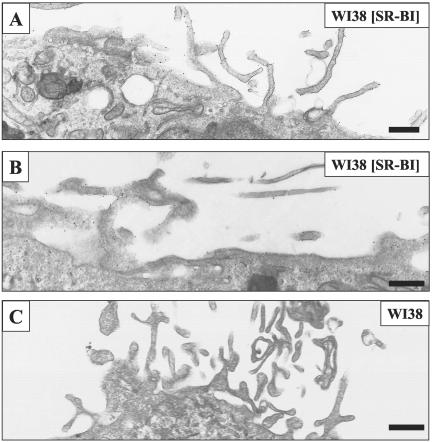
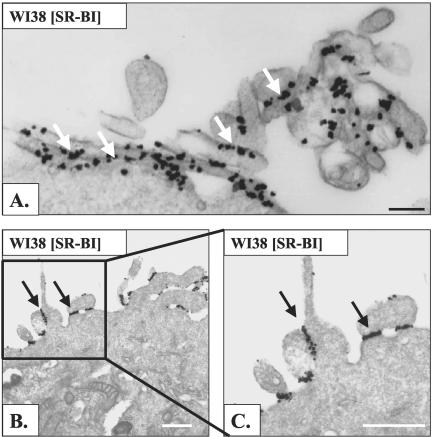
Three techniques were used to visualize cell surface SR-BI at the electron microscopic level. In the first, cells were incubated with biotinylated HDL, fixed with glutaraldehyde, incubated with colloidal gold-labeled antibiotin antibody, and prepared for standard transmission electron microscopy. Figure 3A shows no gold on the surface of a WI38 cell not expressing SR-BI. In contrast, Figure 3B shows a WI38[SR-BI] cell with numerous gold particles decorating microvillar extensions from the cell surface. These data indicate that staining with biotinylated HDL is highly specific for cells expressing SR-BI. Some microvilli extend from the cell and others are snuggled against each other or up against the cell surface to form channels (arrowheads) similar to those observed on steroidogenic cells in vivo (Reaven et al., 1989). Figure 4A shows biotinylated HDL labeling on microvillar extensions (white arrows) adjacent to a group of caveolae (black arrows) that show no labeling. Similarly, Figure 4B shows a large region of plasma membrane replete with caveolae that is devoid of HDL labeling.
Figure 3.
Biotinylated HDL specifically labeled SR-BI on microvillar extensions of WI38[SR-BI] cells. Cells were incubated with biotinylated HDL (40 μg/ml) at 4°C for 90 min. Cells were fixed with 2% glutaraldehyde/1% paraformaldehyde and incubated with 10-nm gold-conjugated goat antibiotin secondary antibody, before preparation for electron microscopy. Scale bar, 500 nm. WI38 cells without SR-BI expression did not show biotinylated HDL binding, even when observed under higher magnification (A). In WI38 [SR-BI] cells, biotinylated HDL dotted the surface of microvillar extensions (B), indicating SR-BI was concentrated on these extensions. However, gold was not observed between juxtaposed microvillar extensions, as indicated by the arrowheads in the enlarged portion of B.
Figure 4.
Biotinylated HDL labeled SR-BI located close to, but not within, morphologically distinct caveolae. Cells were processed as described in Figure 3. Scale bar, 500 nm. In some WI38 [SR-BI] cells labeled with biotinylated HDL, gold (white arrows) was observed on microvillar extensions that were in close proximity to caveolae. However, caveolae (black arrows) did not contain gold label (A). (B) A region of plasma membrane replete with caveolae but lacking gold label.
Very similar results were obtained with antibody directed against the extracellular domain of SR-BI. In this case, cells were incubated with affinity-purified antibody that was detected with colloidal gold-labeled secondary antibody. As shown in Figure 5, A and B, WI38[SR-BI] cells show gold decorating microvillar extensions from the cell surface and apparently undifferentiated regions of membrane. In contrast, WI38 cells lacking SR-BI expression show no gold labeling (Figure 5C). As was the case with biotinylated HDL, only occasional labeling of caveolae was observed with anti-SR-BI antibody (unpublished data).
Figure 5.
Antibody staining confirmed SR-BI localization on microvillar extensions in WI38[SR-BI] cells. The affinity-purified antibody used in this experiment was raised against the extracellular portion of SR-BI. Cells were fixed and incubated with primary antibody for 1 h and gold-conjugated secondary antibody for 30 min. Cells were then postfixed with 2% glutaraldehyde in PBS and processed for electron microscopy. Scale bar, 500 nm. (A and B) WI38[SR-BI] cells with gold particles mostly decorating microvillar extensions and some on nonmicrovillar regions of membrane. (C) A WI38 cell not expressing SR-BI.
With biotinylated HDL, gold labeling was seen primarily on exposed regions of microvillar extensions from the plasma membrane. No labeling was seen in channels formed between juxtaposed microvilli (see arrowheads in Figure 3B), as might be anticipated by analogy to the localization of SR-BI and HDL in microvillar channels of steroidogenic cells in vivo (Reaven et al., 1989, 1998, 2000). A potential explanation for the absence of labeling within the channels is that the colloidal gold-labeled secondary antibodies are too large to enter the channels that already are occupied with biotinylated HDL. To examine this point, we prepared reconstituted 11-nm-diameter HDL particles into which was incorporated a single dipalmitoylphosphatidylethanolamine that was covalently attached to a 1.4-nm nanogold cluster. The nanogold cluster on this HDL is ∼13% the diameter of the HDL particle and should not hinder penetration into the channels formed by juxtaposed microvillar extensions. Cells were incubated with the nanogold-HDL and fixed in glutaraldehyde, the nanogold complex was enlarged by a chemical enhancement procedure, and the samples were prepared for electron microscopy.
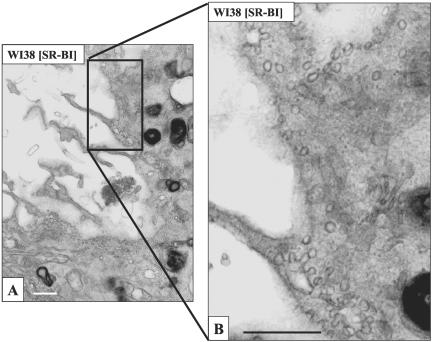
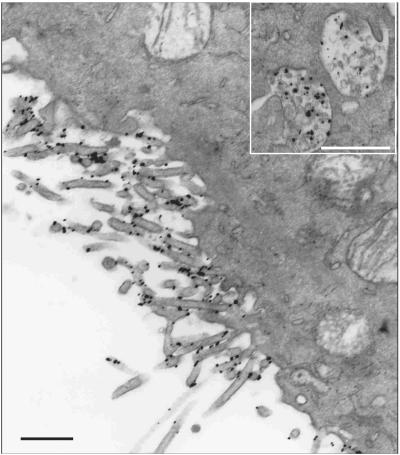
Figure 6 shows that when incubated with gold-HDL, WI38 cells lacking SR-BI were nearly devoid of gold (Figure 6A), whereas WI38[SR-BI] cells showed heavy labeling on cell surface microvillar extensions and in the regions between juxtaposed microvillar extensions (Figure 6B). Labeling with gold HDL between stacked microvillar extensions and between the cell surface and juxtaposed microvillar extensions are further illustrated in Figure 7. Serial sections of these samples showed that the clusters of bound HDL occur in patches of 100-500-nm dimensions. Furthermore, it was clear that gold HDL was heavily clustered in some groups of microvillar extensions, whereas other microvillar extensions had little or no label. These data confirm the localization of SR-BI to microvillar extensions, as noted with antibody and biotinylated HDL staining, and show further that SR-BI is prominent in patches between juxtaposed microvilli and in regions where microvilli snuggle against the plasma membranes to form microvillar channels. As was the case with antibody and biotinylated HDL staining of WI38[SR-BI] cells, gold HDL did not localize in substantial amounts in caveolae (Figure 8). Table 1 shows quantitative data on the labeling of WI38 and WI38[SR-BI] cells with the three methods for detecting SR-BI and also shows the distribution of gold label between microvillar extensions plus undifferentiated plasma membrane vs. caveolae plus small vesicles located near the cell surface. Note that with SR-BI antibody and gold HDL ∼80% of cell surface staining was on microvillar extensions, whereas this was ∼50% with the biotinylated HDL (Table 1, legend).
Figure 6.
Gold HDL binds in patches on WI38 [SR-BI] cell membranes. Cells were incubated with gold HDL (10 μg/ml) at 4°C for 90 min, fixed, and gold-enhanced. Cells were then processed for electron microscopy. WI38 cells were devoid of gold particles (A). WI38 [SR-BI] cells showed extensive gold staining on the cell surface (B). As evident from B, the gold particles did not uniformly stain the cell membrane, but were concentrated on and between microvillar extensions. Scale bars, 500 nm.
Figure 7.
Gold HDL binds on stacked microvillar extensions and between the cell surface and juxtaposed microvillar extensions in WI38 [SR-BI] cells. Cells were stained with gold HDL as in Figure 6. Scale bars, (A) 200 nm; (B and C) 500 nm. Gold HDL is located on and between stacked or juxtaposed microvillar extensions (arrows) where it is often seen in patches of 100-500-nm dimensions.
Figure 8.
Gold HDL staining was not seen in caveolae of WI38[SR-BI]. Cells were stained with gold HDL as in Figure 6. (A) A membrane region replete with caveolae and showing some typical rosettes of caveolae; (B) higher magnification view of A. None of the caveolae was stained, even though other regions of the same cell were heavily stained with gold HDL (unpublished data). Scale bar, 500 nm.
Table 1.
Distribution of gold particles with different labeling methods
| Percent distributionc
|
|||||
|---|---|---|---|---|---|
| Cella | Method | Gold particles | Printsb | MVE | Caveolae |
| WI38 | bHDL | 0 | 10 | ndd | nd |
| WI38[SR-BI] | bHDL | 417 | 10 | 100 | 0 |
| WI38 | α-SR-BI | 29 | 43 | nd | nd |
| WI38[SR-BI] | α-SR-BI | 843 | 43 | 99.9 | 0.1 |
| WI38 | gHDL | 5 | 17 | nd | nd |
| WI38[SR-BI] | gHDL | 1543 | 17 | 98.5 | 1.5 |
Cells were stained by the indicated method to detect SR-BI as described in MATERIALS AND METHODS. bHDL, biotinylated HDL; α-SR-BI, antibody directed against the extracellular domain of SR-BI; gHDL, HDL containing DPPE-nanogold
Number of prints examined for comparison between WI38 and WI38[SR-BI]. Micrographs were taken at 12,500× and enlarged 3× upon printing
Values show the percent distribution of gold particles between MVE and caveolae. MVE values include the sum of gold on cell surface microvillar extensions plus gold on undifferentiated membrane. For WI38[SR-BI] cells the distributions of gold particles between microvillar extensions and undifferentiated membrane regions for the three methods were, respectively, (in percent of total): bHDL, 47.5%, 52.5%; α-SR-BI, 78.3, 21.6%; gHDL, 83.5%, 15%. Caveolae included morphologically identifiable caveolae plus small vesicles near the cell surface
nd, not determined
Cell Type Distribution of Gold HDL Binding by Electron Microscopy
Three additional cell types were examined to test the generality of SR-BI localization to clusters on microvillar extensions of the plasma membrane. Cell lines examined were the ldlA7[SR-BI] Chinese hamster ovary cell stably expressing murine SR-BI (Acton et al., 1996), the Y1-BS1 murine adrenocortical cell in which SR-BI is expressed naturally and is inducible by ACTH treatment (Rigotti et al., 1996), and the Fisher rat thyroid cell (FRT) that expressed SR-BI via transient transfection. The FRT cell does not express cav-1 or SR-BI and has been shown to exhibit SR-BI-mediated free cholesterol efflux and HDL CE selective uptake upon SR-BI expression (Wang et al., 2003). Each of these cell lines showed a cell surface pattern of gold HDL binding that was similar to that in the WI38[SR-BI] cell (unpublished data). Most of the HDL was bound on microvillar extensions and apparently undifferentiated regions of membrane with little localization to caveolae or small vesicles near the cell surface. These data are summarized in Table 2. The absence of cav-1 expression in FRT cells had little or no effect on the localization of gold HDL to microvillar extensions. Note also that 70-94% of the gold HDL was localized to microvillar extensions with 6-30% on undifferentiated membrane regions, depending on the cell type (Table 2, legend). Additionally, we carried out immunostaining for cav-1 at the electron microscopic level in WI38[SR-BI] cells as described in MATERIALS AND METHODS. These experiments detected cav-1 in caveolae and small vesicles near the cell surface, but detected no cav-1 in microvillar extensions (unpublished data).
Table 2.
Cell type distribution of gold HDLa
| Percent distribution of gold HDL
|
||
|---|---|---|
| Cell type | Microvilli + cell surface | Caveolae + small vesicles |
| WI38[SR-BI] | 96.2 | 3.8 |
| IdlA7[SR-BI] | 96.7 | 3.3 |
| ACTH-Y1-BS1 | 99.4 | 0.6 |
| FRT/SRB1 | 93.6 | 6.4 |
Cells were incubated with 10 μg/ml gold HDL for 90 min on ice, washed, fixed, subjected to gold enhancement, and processed for electron microscopy as indicated in MATERIALS AND METHODS. Total number of gold particles counted were as follows: WI38[SR-BI], 2658; ldlA7[SR-BI], 729; Y1-BS1, 516; FRT/SR-BI, 1612. The distributions of gold particles between microvillar extensions and undifferentiated membrane regions for the four cell types were, respectively (in percent of total): WI38[SR-BI], 77.9%, 18.3%; ldlA7[SR-BI], 88.3%, 8.4%; ACTH-Y1-BS1, 93.6%, 5.8%; FRT/SRB1 70%, 23.6%. Micrographs were taken at 12,000× and enlarged 3× upon printing
Distribution of Gold HDL at 37°C
To determine whether the interaction of HDL with SR-BI was altered at 37°C, WI38[SR-BI] cells were incubated with gold HDL for 5, 15, or 60 min before analysis by electron microscopy. The basic pattern of gold HDL labeling was largely unchanged compared with incubation on ice, with most of the HDL remaining on the cell surface (Table 3A). Additionally, there was no transfer of HDL to cavolae or cell surface vesicles. By 60 min at 37°C, a small amount of gold HDL was found intracellularly, and this was exclusively in multivesicular bodies (MVB), suggestive of a minor amount of endocytic uptake. These data agree well with biochemical measurements of HDL CE binding and uptake in WI38[SR-BI] cells using HDL particles labeled with 3H cholesteryl oleolyl ether to trace HDL CE and 125I-dilactitoltyramine to label HDL proteins as previously described (Connelly et al., 1999). With WI38[SR-BI] cells incubated with HDL for 60 min at 37°C, the distribution of HDL CE was (ng HDL CE/mg cell protein): cell surface, 17.6; endocytosed, 1; selective uptake, 333 (unpublished data). Thus, the distribution between cell surface (17.6 = 94.6%) and endocytosed (1 = 5.4%) HDL with radiolabeled HDL is similar to the cell surface (91.9%) vs. MVB (7.8%) distribution observed with gold HDL (Table 3A).
Table 3.
Distribution of gold HDL at 37°Ca
| Percent distribution of gold HDL
|
|||
|---|---|---|---|
| Time at 37°C (min) | Microvilli + cell surface | Caveolae + small vesicles | MVB |
| A. WI38[SR-BI] | |||
| 5 | 99.8 | 0.2 | 0 |
| 15 | 96.9 | 3.1 | 0 |
| 60 | 91.9 | 0.3 | 7.8 |
| B. Y1-BS1 | |||
| 5 | 99 | 1 | 0 |
| 15 | 94.6 | 1.5 | 3.9 |
| 60 | 73.4 | 0.6 | 26 |
Cells were incubated with 10 μg/ml gold HDL for the indicated times, washed, fixed, subjected to gold enhancement, and processed for electron microscopy. Total number of gold particles counted for the 5-, 15-, and 60-min time points were, respectively: WI38[SR-BI] 518, 1040, 1137; Y1-BS1 393, 613, 1445. The distributions of gold particles between microvillar extensions and undifferentiated cell surface membrane regions for the three time points were, respectively (in percent of total): WI38[SR-BI] (5 min) 66.4%, 33.4%; (15 min) 66.1%, 30.8%; 60 min) 77.1%, 14.8%; Y1-BS1 (5 min) 89%, 10%; (15 min) 88.7%, 5.9% (60 min) 57.9%, 15.5%. Micrographs were taken at 12,000× and enlarged 3× upon printing. MVB, multivesicular body
Experiments with ACTH-treated Y1-BS1 cells showed a similar pattern with gold HDL remaining largely on the cell surface even after 60 min at 37°C (Figure 9), with little or no accumulation of gold HDL in caveolae (Table 3B). Compared with WI38[SR-BI] cells, Y1-BS1 cells showed greater accumulation of gold HDL in MVBs at 60 min (Figure 9, inset). In this case the percent distribution of gold between the cell surface and MVBs was 73.4% vs. 26%. These data also agree well with biochemical measurements of HDL CE binding and uptake for 60 min at 37°C using doubly radiolabeled HDL as noted above. In this case the distribution of HDL CE was (ng HDL CE/mg cell protein): cell surface), 70.8; endocytosed, 34.3; selective uptake, 175 (unpublished data). Thus, the distribution between cell surface (70.8 = 67.4%) and endocytosed (34.3 = 32.6%) HDL with radiolabeled HDL is similar to the cell surface (73.4%) vs. MVB (26%) distribution observed with gold HDL (Table 3B).
Figure 9.
Gold HDL localized on the cell surface and in multivesicular bodies in Y1-BS1 cells at 37°C. ACTH-treated Y1-BS1 cells were incubated with gold HDL (10 μg/ml) for 60 min at 37°C, fixed, and goldenhanced. Cells were then processed for electron microscopy. Most of the gold label was found on microvillar extensions but some was seen in MVBs within the cells (inset). Scale bars, 500 nm.
SR-BI Membrane Localization Examined by Detergent Extraction and Gradient Floatation
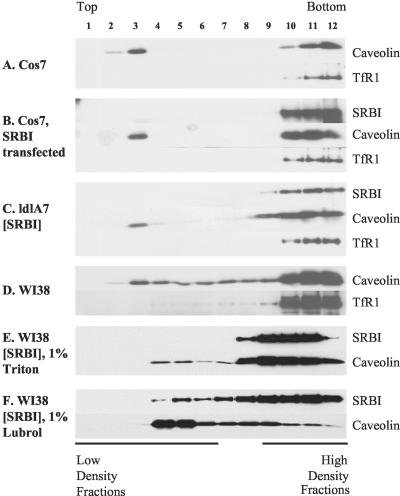
By confocal microscopy and electron microscopy, SR-BI was found in irregularly shaped patches and clusters of microvillar extensions on the cell membrane, but was not found in caveolae. To test whether SR-BI might be located in rafts, or detergent resistant membranes (DRM), sucrose gradient fractionation was used on several cell lines to further examine the membrane localization of SR-BI. Cells were solubilized with 1% Triton X-100, floated in sucrose step gradients, and analyzed by Western blotting to determine the distribution between DRM (low-density fractions) and non-DRM (high-density fractions; Figure 10).
Figure 10.
SR-BI was not shown to be tightly associated with detergent-resistant microdomains. Cells were grown to confluency in 35-mm dishes, lysed with 0.5 ml TNE buffer containing 1% Triton, and incubated on ice for 30 min. Step sucrose gradients were centrifuged at 38,000 rpm (148,305 × g) for 3 h. Twelve fractions were collected from top to bottom as noted in the figure, and distributions of SR-BI, cav-1, and TfR1 were analyzed by immunoblotting. The various cell lines examined are indicated on the figure. (F) The sucrose gradient distribution of SR-BI and cav-1 in WI38[SR-BI] cells solubilized in 1% Lubrol WX.
There have been many studies conducted to evaluate the effectiveness of the DRM isolation method. It has been reported that the detergent: protein ratio is one important factor determining the effectiveness of DRM isolation (Ostermeyer et al., 1999), and variation in the ratio could produce markedly varied results. To obtain consistent results, we kept the detergent: protein ratio at 10:1, as suggested (Ostermeyer et al., 1999). We also used nonraft protein TfR1 as a negative control (Schuck et al., 2003; Roper et al., 2003). For all Triton X-100 experiments, TfR1 was found only in high-density fractions, indicating that detergent solubilization of nonraft membrane was complete (Figure 10).
In COS-7 cells, cav-1 was separated into DRM and non-DRM, with slightly more protein in the non-DRM in both control (Figure 10A) and SR-BI-expressing cells (Figure 10B). In contrast, in COS-7 cells transiently transfected with SR-BI, almost all SR-BI was in non-DRM, with only a very faint band appearing at fraction 3 (Figure 10B).
We also used a cell line stably transfected with SR-BI, ldlA7[SR-BI], to examine the detergent solubility of SR-BI in comparison to cav-1 (Figure 10C). Cav-1 was also separated into DRM and non-DRM fractions, with more protein in the non-DRM. In this cell type as well, SR-BI was completely solubilized, detectable only in non-DRM fractions (Figure 10C). Data from these two cell lines suggest that even though SR-BI is located in clusters on the cell membrane, it is not stably associated with conventional Triton X-100 DRMs.
Different results were obtained from WI38 cells, a human lung fibroblast cell line. In both WI38 cells and WI38[SR-BI], clean separation of cav-1 between DRM and non-DRM was not observed (Figure 10, D and E); rather the cav-1 profile was smeared across the gradient from fraction 3 to fraction 12. In WI38[SR-BI] cells, SR-BI was largely solubilized (Figure 10E, fractions 8-12), although in some experiments SR-BI also was seen to some extent in fractions 3-7 (unpublished data). The variability of the SR-BI gradient profile in WI38[SR-BI] cells prompted us to test another detergent. Simons and colleagues recently demonstrated that cell types may differ in resistance to various detergents used to isolate DRMs (Schuck et al., 2003). Similarly, prominin, a pentaspan plasma-membrane protein located in microvilli of epithelial cells, is Triton soluble but Lubrol insoluble (Roper et al., 2003). On the basis of these findings, we also used the weaker detergent Lubrol WX for DRM isolation. As illustrated in Figure 10F, SR-BI was partially solubilized by Lubrol with most of the protein distributed toward the high-density region of the gradient. In contrast, much less cav-1 was solubilized by Lubrol with most of the cav-1 distributed toward the low-density region of the gradient. Thus, as in the other cell types, SR-BI in WI38[SR-BI] cells is less sensitive to detergent solubilzation than cav-1. Note that Lubrol also incompletely solubilized TfR1 in these experiments (unpublished data).
DISCUSSION
The present study provides new insight into the mechanisms by which SR-BI mediates cholesterol trafficking between cells and lipoproteins. The major finding is that SR-BI in the plasma membrane is localized primarily in clusters on microvillar extensions of the plasma membrane. This pattern was invariant among four cell types and was the same at 4 and 37°C. It was also the same in transfected cells stably expressing SR-BI, in transiently transfected cells, and in adrenocortical cells expressing the endogenous SR-BI gene. At the light microscopic level SR-BI showed little colocalization with cav-1, and at the electron microscopic level was nearly absent from caveolae. This was strikingly clear in the WI38[SR-BI] cell that is replete with caveolae and expresses high levels of cav-1. The clustering of SR-BI on microvillar extensions was also seen in FRT cells that lack cav-1 expression and that show SR-BI-mediated HDL CE selective uptake and FC efflux to HDL (Wang et al., 2003). These data indicate that SR-BI-mediated cholesterol trafficking between cells and HDL occurs primarily in clusters of SR-BI on microvillar extensions of the plasma membrane independently of caveolae or cav-1. Clustering of SR-BI on microvillar extensions was not uniform; some groups of microvilli were heavily labeled, whereas others were largely free of SR-BI. This lack of uniformity suggests that SR-BI is somehow targeted to and clustered in some membrane regions but not others.
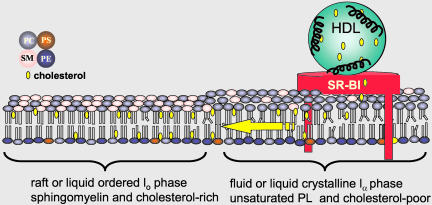
Analysis of DRMs via Triton X-100 solubilization showed no DRM association of SR-BI in COS-7 or ldlA7 cells. In WI38[SR-BI] cells, results with Triton X-100 were variable as to the DRM distribution of SR-BI. Extraction with Lubrol WX showed a very different gradient distribution of SR-BI and cav-1 between low- and high-density fractions indicating greater solubilization of SR-BI with the weaker detergent. These cell type differences may reflect differences in SR-BI association with membrane domains of differing lipid composition (Schuck et al., 2003). Overall, the detergent solubilization experiments indicate that SR-BI is not strongly associated with DRMs. This observation, the near absence of SR-BI from caveolae, and the localization of plasma membrane SR-BI in clusters on microvillar extensions raises the important issue of the nature of the lipids in these membrane domains where SR-BI mediates HDL CE selective uptake and the bidirectional flux of FC between the membrane and HDL. DRMs are believed to represent liquid-ordered domains enriched in saturated phospholipids in which cholesterol is tightly packed with the phospholipid acyl tails (Brown and London, 1998; Rietveld and Simons, 1998; Xu and London, 2000). The lack of association with DRMs suggests, but does not establish, that SR-BI is in a more fluid membrane domain that contains a greater fraction of unsaturated phospholipids with cholesterol less tightly packed than in DRMs. We speculate that SR-BI-mediated cholesterol trafficking into and out of such fluid domains, compared with liquid-ordered domains, would be energetically favorable because of reduced interactions of cholesterol with saturated acyl tails (Figure 11). Such domains could serve as a way station for SR-BI-mediated cholesterol movement between HDL and other domains of the plasma membrane or internal membranes.
Figure 11.
Model of HDL cholesterol trafficking via SR-BI in microvillar membrane domains. The model shows SR-BI localized in a more fluid membrane domain rich in unsaturated phospholipids and poor in sphingolipids and cholesterol (yellow ovals) adjacent to a membrane raft or more liquid-ordered domain rich in sphingolipids and cholesterol. The speculation is that the localization of SR-BI in a more fluid or disordered domain will be energetically favorable for the rapid trafficking of cholesterol between HDL and the membrane and the subsequent redistribution of cholesterol to other membrane domains where cholesterol is sequestered via interactions with saturated acyl tails of sphingolipids. PC, phosphatidylcholine; PS, phosphtidylserine; PE, phosphatidylethanolamine; SM, sphingomyelin.
In addition to mediating cholesterol movement between HDL and cells, several lines of evidence indicate that clusters of SR-BI on microvillar extensions impart unique properties to the plasma membrane. SR-BI-expressing cells show an increase in the fast kinetic pool for cholesterol efflux to cyclodextrin acceptors and an increase in the membrane cholesterol pool that is susceptible to exogenous cholesterol oxidase (de la Llera-Moya et al., 1999; Kellner-Weibel et al., 2000). These data suggest that cholesterol more readily desorbs from SR-BI-containing membranes. Importantly, these features occur in the absence of HDL, indicating that these effects on the membrane are due to SR-BI and not to HDL interactions with SR-BI. The latter point is supported by separation of function mutations in the extracellular domain of SR-BI that eliminate the cholesterol oxidase-sensitive membrane cholesterol pool without disrupting cholesterol flux between HDL and cells (Connelly et al., 2003).
In vivo, SR-BI is most highly expressed in steroidogenic cells (Acton et al., 1996; Landschulz et al., 1996; Rigotti et al., 1996) where it is found predominantly on cell surface microvillar channels that fill with HDL (Reaven et al., 1989, 1998). Microvillar channels are an elaborate network of juxtaposed microvilli that act as the site of SR-BI-mediated HDL CE selective uptake. The organization of SR-BI in clusters on and between microvillar extensions in cultured cells (Figure 7) appears to be similar to but less elaborate than the organization seen in vivo. SR-BI expression in insect cells elicits the formation of double membrane structures resembling the microvillar channels of steroidogenic cells (Reaven et al., 2001). SR-BI also appears to have substantial effects on membrane properties in vivo. Comparison of adrenal gland ultrastructure in wild-type and SR-BI knockout mice revealed that SR-BI is absolutely required for the formation of microvillar channels (Williams et al., 2002). Additionally, the microvillar membrane is 17% thinner in Srb1-/- mice, indicative of SR-BI effects on membrane structure. Taken together with the cell culture data, these findings suggest that SR-BI has a preferential association with specific membrane domains and may play a role in the organization of the domains.
A previous report suggested that SR-BI was localized in caveolae in ldlA7[SR-BI] and Y1-BS1 cells based on partial colocalization with cav-1 by immunostaining and cofractionation of SR-BI and cav-1 by a biochemical isolation procedure (Babitt et al., 1997). In the present study, we examined SR-BI localization at both light and electron microscopic levels, using antibodies and labeled HDL particles. The results indicate that SR-BI is predominantly located in clusters on microvillar extensions with very little in caveolae. This difference with the immunostaining results in the previous study may reflect the use of detergent permeabilization in that study as was necessary with an antibody directed against an intracellular domain of SR-BI. In our experience it is difficult to distinguish between cell surface and intracellular SR-BI and to determine authentic cav-1 colocalization in permeabilized and antibody-stained cells due to the numerous microvillar extensions and the convoluted cell surface. Coisolation of cav-1 and SR-BI in light membrane fractions using a nondetergent protocol (Babitt et al., 1997) may reflect similar lipid to protein ratios, difficulty in separating nearby membrane domains that contain SR-BI or cav-1, or disruption and reorganization of some plasma membrane components during sonication. As noted in Figures 2 and 4, regions of the membrane containing cav-1 or SR-BI are often intermingled. Additionally, the staining for SR-BI that appears slightly outside the cav-1 staining (Figure 2G, curved arrow) is consistent with the pattern at the electron microscopic level showing much of the SR-BI on microvillar extensions extending out from the membrane regions that contain caveolae. Additionally, electron microscopic observation of SR-BI clustered in some groups of microvilli but not others is consistent with the fluorescence microscopy, which showed irregularly sized patches of SR-BI on the membrane and on membrane extensions.
In summary, the present study shows that SR-BI is localized in clusters on plasma membrane microvillar extensions. The finding that most of the cell surface SR-BI is organized in this manner suggests that this microvillar domain serves as a way station for cholesterol trafficking between HDL and cells. The nature of the lipids in this domain is unknown, but detergent solubilization experiments suggest that SR-BI is not strongly associated with classical membrane rafts rich in detergent-resistant saturated phospholipids. We speculate that the presence of SR-BI in a more fluid membrane domain will favor rapid cholesterol flux between the membrane and HDL (Figure 11). Similarly, the movement of HDL cholesterol into such a domain may facilitate rapid cholesterol distribution to other membrane domains, including caveolae, and other cellular compartments.
Acknowledgments
We thank Ryan Temel for purification of the anti-SR-BI antibody and Ryan Temel and Seth Klein for performing the HDL CE selective uptake experiments. We thank Deborah Brown for advice with the DRM isolation experiments and for critical comments. This work was supported by National Institutes of Health Grants HL63768 and HL22633.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-06-0445. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0445.
Abbreviations used: LDL, low-density lipoprotein; SR-BI, scavenger receptor class B, type I; HDL, high-density lipoprotein; CE, cholesteryl ester; FC, free cholesterol; cav-1, caveolin-1; MVB, multivesicular body; DRM, detergent-resistant membrane.
References
- Acton, S., Rigotti, A., Landschulz, K.T., Xu, S., Hobbs, H.H., and Krieger, M. (1996). Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271, 518-520. [DOI] [PubMed] [Google Scholar]
- Arai, T., Wang, N., Bezouevski, M., Welch, C., and Tall, A.R. (1999). Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor BI transgene. J. Biol. Chem. 274, 2366-2371. [DOI] [PubMed] [Google Scholar]
- Babitt, J., Trigatti, B., Rigotti, A., Smart, E., Anderson, R.G.W., Xu, S., and Krieger, M. (1997). Murine SR-BI, a high density lipoprotein receptor that mediates selective lipid uptake, is N-glycosylated and fatty acylated and colocalizes with plasma membrane caveolae. J. Biol. Chem. 272, 13242-13249. [DOI] [PubMed] [Google Scholar]
- Braun, A., Trigatti, B.L., Post, M.J., Sato, K., Simons, M., Edelberg, J.M., Rosenberg, R.D., Schrenzel, M., and Krieger, M. (2002). Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ. Res. 90, 270-276. [DOI] [PubMed] [Google Scholar]
- Brown, D., and London, E. (1998). Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164, 103-114. [DOI] [PubMed] [Google Scholar]
- Brown, M.S., and Goldstein, J.L. (1986). A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34-47. [DOI] [PubMed] [Google Scholar]
- Connelly, M.A., de la Llera-Moya, M., Peng, Y., Drazul-Schrader, D., Rothblat, G.H., and Williams, D.L. (2003). Separation of lipid transport functions by mutations in the extracellular domain of scavenger receptor class B, type I (SR-BI). J. Biol. Chem. 278, 25773-25782. [DOI] [PubMed] [Google Scholar]
- Connelly, M.A., Klein, S.M., Azhar, S., Abumrad, N.A., and Williams, D.L. (1999). Comparison of class B scavenger receptors, CD36 and SR-BI, shows that both receptors mediate HDL-cholesteryl ester selective uptake but SR-BI exhibits a unique enhancement of cholesteryl ester uptake. J. Biol. Chem. 274, 41-47. [DOI] [PubMed] [Google Scholar]
- de la Llera-Moya, M., Connelly, M.A., Drazul, D., Klein, S.M., Favari, E., Yancey, P.G., Williams, D.L., and Rothblat, G.H. (2001). Scavenger receptor, class B, type I, (SR-BI) affects cholesterol homeostasis by magnifying cholesterol flux between cells and HDL. J. Lipid Res. 42, 1969-1978. [PubMed] [Google Scholar]
- de la Llera-Moya, M., Rothblat, G.H., Connelly, M.A., Kellner-Weibel, G., Sakar, S.W., Phillips, M.C., and Williams, D.L. (1999). Scavenger receptor BI (SR-BI) mediates free cholesterol flux independently of HDL tethering to the cell surface. J. Lipid Res. 40, 575-580. [PubMed] [Google Scholar]
- Glass, C., Pittman, R.C., Civen, M., and Steinberg, D. (1985). Uptake of high-density lipoprotein-associated apoprotein A-I and cholesterol esters by 16 tissues of the rat in vivo and by adrenal cells and hepatocytes in vitro. J. Biol. Chem. 260, 744-750. [PubMed] [Google Scholar]
- Glass, C., Pittman, R.C., Weinstein, D.B., and Steinberg, D. (1983). Dissociation of tissue uptake of cholesterol ester from that of apoprotein A-I of rat plasma high density lipoprotein: selective delivery of cholesterol ester to liver, adrenal, and gonad. Proc. Natl. Acad. Sci. USA 80, 5435-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, G.A., Connell, P.M., van der Westhuyzen, D.R., and Smart, E.J. (1999). The class B, type I scavenger receptor promotes the selective uptake of high density lipoprotein cholesteryl ethers into caveolae. J. Biol. Chem. 274, 12043-12048. [DOI] [PubMed] [Google Scholar]
- Gwynne, J.T., and Hess, B. (1980). The role of high density lipoproteins in rat adrenal cholesterol metabolism and steroidogenesis. J. Biol. Chem. 255, 10875-10883. [PubMed] [Google Scholar]
- Havel, R.J., Eder, H.A., and Bragdon, J.H. (1955). The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34, 1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y., Jian, B., Wang, N., Sun, Y., de la Llera Moya, M., Phillips, M.C., Rothblat, G.H., Swaney, J.B., and Tall, A.R. (1997). Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272, 20982-20985. [DOI] [PubMed] [Google Scholar]
- Kellner-Weibel, G., de la Llera-Moya, M., Connelly, M.A., Stoudt, G., Christian, A.E., Haynes, M.P., Williams, D.L., and Rothblat, G.H. (2000). Expression of scavenger receptor B.I in C.O.S-7 cells alters cholesterol content and distribution. Biochemistry 39, 221-229. [DOI] [PubMed] [Google Scholar]
- Kozarsky, K.F., Donahee, M.H., Glick, J.M., Krieger, M., and Rader, D.J. (2000). Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arteriosclerosis Thromb. Vasc. Biol. 20, 721-727. [DOI] [PubMed] [Google Scholar]
- Kozarsky, K.F., Donahee, M.H., Rigotti, A., Iqbal, S.N., Edelman, E.R., and Krieger, M. (1997). Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature 387, 414-417. [DOI] [PubMed] [Google Scholar]
- Krieger, M. (1999). Charting the fate of the “good cholesterol”: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu. Rev. Biochem. 68, 523-558. [DOI] [PubMed] [Google Scholar]
- Krieger, M., and Herz, J. (1994). Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu. Rev. Biochem. 63, 601-637. [DOI] [PubMed] [Google Scholar]
- Landschulz, K.T., Pathak, R.K., Rigotti, A., Krieger, M., and Hobbs, H.H. (1996). Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J. Clin. Invest. 98, 984-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeyer, A.G., Beckrich, B.T., Ivarson, K.A., Grove, K.E., and Brown, D.A. (1999). Glycosphingolipids are not essential for formation of detergent-resistant membrane rafts in melanoma cells. J. Biol. Chem. 274, 34459. [DOI] [PubMed] [Google Scholar]
- Pittman, R.C., Knecht, T.P., Rosenbaum, M.S., and Taylor, C.A., Jr. (1987). A nonendocytotic mechanism for the selective uptake of high density lipoprotein-associated cholesterol esters. J. Biol. Chem. 262, 2443-2450. [PubMed] [Google Scholar]
- Rajamannan, N.M., Springett, M.J., Pederson, L.G., and Carmichael, S.W. (2002). Localization of caveolin 1 in aortic valve endothelial cells using antigen retrieval. J. Histochem. Cytochem. 50, 617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven, E., Chen, Y.-D.I., Spicher, M., and Azhar, S. (1984). Morphological evidence that high density lipoproteins are not internalized by steroid-producing cells during in situ organ perfusion. J. Clin. Invest. 74, 1384-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven, E., Leers-Sucheta, S., Nomoto, A., and Azhar, S. (2001). Expression of scavenger receptor class B type I (SR-BI) promotes microvillar channel formation and selective cholesteryl ester transport in a heterologous reconstituted system. Proc. Natl. Acad. Sci. USA 98, 1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven, E., Nomoto, A., Leers-Sucheta, S., Temel, R., Williams, D.L., and Azhar, S. (1998). Expression and microvillar localization of scavenger receptor, class B, type I (a high density lipoprotein receptor) in luteinized and hormone-desensitized rat ovarian models. Endocrinology 139, 2847-2856. [DOI] [PubMed] [Google Scholar]
- Reaven, E., Spicher, M., and Azhar, S. (1989). Microvillar channels: a unique plasma membrane compartment for concentrating lipoproteins on the surface of rat adrenal cortical cells. J. Lipid Res. 30, 1551-1560. [PubMed] [Google Scholar]
- Reaven, E., Zhan, L., Nomoto, A., Leers-Sucheta, S., and Azhar, S. (2000). Expression and microvillar localization of scavenger receptor class B, type I and selective cholesteryl ester uptake in Leydig cells from rat testis. J. Lipid Res. 41, 343-356. [PubMed] [Google Scholar]
- Rietveld, A., and Simons, K. (1998). The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim. Biophys. Acta 1376, 467-479. [DOI] [PubMed] [Google Scholar]
- Rigotti, A., Edelman, E.R., Seifert, P., Iqbal, S.N., DeMattos, R.B., Temel, R.E., Krieger, M., and Williams, D.L. (1996). Regulation by adrenocorticotropic hormone of the in vivo expression of scavenger receptor class B type I (SR-B1), a high density lipoprotein receptor, in steroidogenic cells of the murine adrenal gland. J. Biol. Chem. 271, 33545-33549. [DOI] [PubMed] [Google Scholar]
- Rigotti, A., Trigatti, B.L., Penman, M., Rayburn, H., Herz, J., and Krieger, M. (1997). A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. USA 94, 12610-12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, K., Corbeil, D., and Huttner, W.B. (2003). Retention of prominin in microvilli reveals distinct cholesterol-based lipid micro-domains in the apical plasma membrane. Nat. Cell Biol. 2, 582-592. [DOI] [PubMed] [Google Scholar]
- Schuck, S., Honsho, M., Ekroos, K., Shevchenko, A., and Simons, K. (2003). Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. USA 100, 5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, D.L., and Tall, A.R. (2001). The cellular biology of scavenger receptor class B type I. Curr. Opin. Lipidol. 12, 497-504. [DOI] [PubMed] [Google Scholar]
- Sparks, D.L., Phillips, M.C., and Lund-Katz, S. (1992). The conformation of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particles. 13C NMR studies of lysine ionization behavior. J. Biol. Chem. 267, 25830-25838. [PubMed] [Google Scholar]
- Stein, Y., Dabach, Y., Hollander, G., Halperin, G., and Stein, O. (1983). Metabolism of HDL-cholesteryl ester in the rat, studied with a nonhydrolyzable analog, cholesteryl linoleyl ether. Biochim. Biophys. Acta 752, 98-105. [DOI] [PubMed] [Google Scholar]
- Temel, R.E., Trigatti, B., DeMattos, R.B., Azhar, S., Krieger, M., and Williams, D.L. (1997). Scavenger receptor B, type I (SR-BI) is the major route for the delivery of high density lipoprotein cholesterol to the steroidogenic pathway in cultured mouse adrenocortical cells. Proc. Natl. Acad. Sci. USA 94, 13600-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigatti, B. et al. (1999). Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proc. Natl. Acad. Sci. USA 96, 9322-9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, Y., Gong, E., Royer, L., Cooper, P.N., Francone, O.L., and Rubin, E.M. (2000). Relationship between expression levels and atherogenesis in scavenger receptor class B, type I transgenics. J. Biol. Chem. 275, 20368-20373. [DOI] [PubMed] [Google Scholar]
- Ueda, Y., Royer, L., Gong, E., Zhang, J., Cooper, P.N., Francone, O., and Rubin, E.M. (1999). Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J. Biol. Chem. 274, 7165-7171. [DOI] [PubMed] [Google Scholar]
- Varban, M.L. et al. (1998). Targeted mutation reveals a central role of SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc. Natl. Acad. Sci. USA 95, 4619-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Connelly, M.A., Chen, H., Williams, D.L., and Brown, D.A. (2003). Caveolin-1 does not affect SR-BI-mediated cholesterol efflux or selective uptake of cholesteryl ester in two cell lines. J. Lipid Res. 44, 807-815. [DOI] [PubMed] [Google Scholar]
- Wang, N., Arai, T., Ji, Y., Rinninger, F., and Tall, A.R. (1998). Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein apoB, low density lipoprotein apoB, and high density lipoprotein in transgenic mice. J. Biol. Chem. 273, 32920-32926. [DOI] [PubMed] [Google Scholar]
- Williams, D.L., Connelly, M.A., Temel, R.E., Swarnakar, S., Phillips, M.C., de la Llera-Moya, M., and Rothblat, G.H. (1999). Scavenger receptor BI (SR-BI) and cholesterol trafficking. Curr. Opin. Lipidol. 10, 329-339. [DOI] [PubMed] [Google Scholar]
- Williams, D.L., Wong, J.S., and Hamilton, R.L. (2002). SR-BI is required for microvillar channel formation and the localization of HDL particles to the surface of adrenocortical cells in vivo. J. Lipid Res. 43, 544-549. [PubMed] [Google Scholar]
- Xu, X., and London, E. (2000). The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry 39, 843-849. [DOI] [PubMed] [Google Scholar]