Abstract
The 5′→ 3′ exoribonucleases (XRNs) have important functions in transcription, RNA metabolism, and RNA interference. The recent structure of Rat1 (Xrn2) showed that the two highly conserved regions of XRNs form a single, large domain, defining the active site of the enzyme. Xrn1 has a 510-residue segment following the conserved regions that is required for activity but is absent in Rat1. We report here the crystal structures at 2.9 Å resolution of Kluyveromyces lactis Xrn1 (residues 1–1245, E178Q mutant), alone and in complex with a Mn2+ ion in the active site. The 510-residue segment contains four domains (D1–D4), located far from the active site. Our mutagenesis and biochemical studies demonstrate that their functional importance is due to their stabilization of the conformation of the N-terminal segment of Xrn1. These domains may also constitute a platform for interacting with protein partners of Xrn1.
The 5′→ 3′ exoribonucleases (XRNs) belong to a family of conserved enzymes in eukaryotes and have important functions in transcription, RNA metabolism, and RNA interference. In fungi and animals, two XRNs have been identified. Xrn1 (175 kD) is primarily cytosolic and is involved in degradation of decapped mRNAs, nonsense mediated decay, microRNA decay and is essential for proper development 1–5. The Xrn1 homolog in Drosophila, known as Pacman, is required for male fertility 6.
Xrn2 (115 kD) is primarily nuclear, and its ortholog in S. cerevisiae, more commonly known as Rat1, was initially suggested to have roles in RNA trafficking from the nucleus to the cytoplasm 7,8 and in transcription of some genes by RNA polymerase III (Pol III) 9. More recent studies have identified Xrn2/Rat1 as the exoribonuclease that is crucial for the ‘torpedo’ model of transcription termination by Pol II 10–13 and Pol I 14,15. Rat1 also promotes telomere elongation 16 and (together with Xrn1) degradation of hypomodified mature tRNAs 17. The Rat1 ortholog in S. pombe is required for proper chromosome segregation 18. Of the three homologs of Xrn2 in the plant Arabidopsis, two function in the nucleus as RNA silencing suppressors 19, while the third functions in the cytoplasm in the ethylene response pathway 20,21 as well as in RNA interference 22.
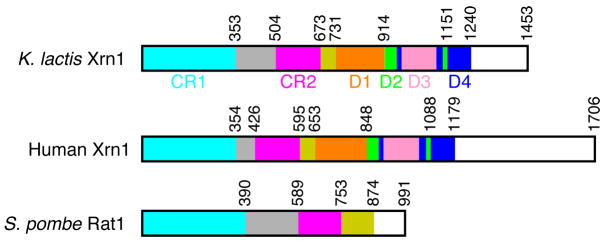
XRNs share two highly conserved regions (CR1 and CR2), corresponding to residues 1–354 and 426–595 of human Xrn1 (Fig. 1), with 58% sequence identity among the Xrn2/Rat1 proteins and 48% identity between Xrn1 and Xrn2 (Supplementary Fig. 1). A cluster of seven conserved acidic residues in CR1 23,24 is expected to coordinate two metal ions for catalysis 25. In comparison, conservation for residues outside of CR1 and CR2 is much weaker. However, the poorly conserved segment of about 120 residues just after CR2 is required for the function of Rat1 18.
Figure 1.
Domain organizations of Xrn1. Schematic drawing of the domain organization of K. lactis Xrn1, human Xrn1, and S. pombe Rat1. The various domains are labeled.
The larger size of Xrn1 compared to Xrn2/Rat1 is due to a much longer C-terminal segment, most of which is unique to Xrn1 (Fig. 1). Weak sequence conservation among Xrn1 proteins is observed up to residue 1180 of human Xrn1 or 1240 of S. cerevisiae and Kluyveromyces lactis (K. lactis) Xrn1 (Supplementary Fig. 1). This weakly conserved segment of approximately 570 residues is required for the nuclease activity of Xrn1. A C-terminal truncation mutant of S. cerevisiae Xrn1, removing residues 1206–1528 and therefore missing only the last 40 residues of this segment, could not complement growth defect in xrn1 null cells and had essentially no exonuclease activity in vitro 23.
We recently reported the crystal structure of S. pombe Rat1 in complex with its activating partner Rai1 26. The structure reveals that the two conserved regions of XRNs form a single, large domain. The poorly conserved segment directly following CR2 has tight interactions with CR1, and it also mediates recruitment of Rai1 26–28. However, it is not known whether Xrn1 has a structurally equivalent segment, and more importantly, why the 570-residue segment following CR2 in Xrn1 is required for activity.
We report here the crystal structures at 2.9 Å resolution of K. lactis Xrn1 (residues 1–1245, E178Q mutant), alone and in complex with a Mn2+ ion in the active site. The 570-residue segment following CR2 consists of two parts–a 60-residue segment directly following CR2 that has some similarity to the 120-residue C-terminal segment of Rat1, and a 510-residue segment that contains four additional domains (D1–D4). The four domains are located far from the active site, but may help to stabilize the N-terminal segment of Xrn1 for catalysis.
Results
Structure determination
After screening through Xrn1 proteins from various organisms, we purified and crystallized residues 1–1245 of K. lactis Xrn1, which cover the entire conserved region of Xrn1 (Fig. 1). The quality of these crystals was rather poor, with diffraction typically to about 3.2 Å resolution. We were able to locate four molecules of Xrn1 by the molecular replacement method, using the structure of S. pombe Rat1 as the search model 26. We also obtained primary phasing with the selenomethionyl single-wavelength anomalous diffraction method 29, and improved the phase information through four-fold non-crystallographic symmetry averaging. We discovered that the E178Q mutant produced better crystals, and by screening through a large number of them we were able to extend the resolution to 2.9 Å. This mutation is in the active site of Xrn1 and is expected to disrupt the nuclease activity 23. By co-crystallizing with MnCl2, we also obtained the structure at 2.9 Å resolution of this mutant in complex with a Mn2+ ion in the active site.
The refined structures have good agreement with the crystallographic data and the expected bond lengths, bond angles, and other geometric parameters (Table 1). The four molecules of Xrn1 have roughly the same conformation, with rms distance of 0.8 Å for equivalent Cα atoms between any pair of the molecules. The majority of the residues (82%) are in the most favored region of the Ramachandran plot, and 16% are in additionally allowed region. The current atomic model for K. lactis Xrn1 contains most of the residues of the recombinant protein. Residues 1–2, 47–53, 116–127, 230–238, 409–411, 463–468, 488–503, 520–525, 609–620, 628–640, 776–780, 983–1082, and 1241–1245 are omitted due to lack of electron density (Supplementary Fig. 1).
Table 1.
Summary of crystallographic information
| Structure | K. lactis Xrn1 E178Q mutant | E178Q mutant in complex with Mn2+ |
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 115.7, 132.3, 143.9 | 116.3, 132.6, 144.1 |
| α, β, γ(°) | 110.1, 105.7, 103.7 | 109.9, 105.8, 104.0 |
| Resolution (Å)* | 30–2.9 (3.0–2.9) | 30–2.9 (3.0–2.9) |
| Rmerge (%) | 4.5 (25.8) | 6.4 (45.8) |
| I/σI | 15.8 (2.7) | 8.8 (1.4) |
| Completeness (%) | 97 (96) | 96 (90) |
| Redundancy | 2.0 (2.0) | 2.0 (2.0) |
| Refinement | ||
| Resolution | 30–2.9 | 30–2.9 |
| No. of reflections | 146,829 | 143,344 |
| Rwork/Rfree (%) | 24.1/30.5 | 24.8/28.4 |
| No. atoms | ||
| Protein | 34,041 | 34,041 |
| Ligand/ion | 0 | 4 |
| Water | 0 | 0 |
| B-factors | ||
| Protein | 72 | 80 |
| Ligand/ion | – | 69 |
| Water | – | – |
| R.m.s. deviations | ||
| bond lengths (Å) | 0.016 | 0.017 |
| bond angles (°) | 1.7 | 1.8 |
The numbers in parentheses are for the highest resolution shell. One crystal was used for each data set.
Overall structure of Xrn1
Residues 3–1240 of K. lactis Xrn1 form a compact structure overall, in the shape of a rectangular box, with dimensions of 80Å × 60Å × 35Å (Fig. 2a). Only the linker segment (residues 354–503) between the two conserved regions and residues 979–1109 (domain D3, see below) are positioned outside this overall compact structure (Fig. 2b).
Figure 2.
Structure of K. lactis Xrn1. (a). Schematic drawing of the structure of K. lactis Xrn1. The domains are colored as in Fig. 1 and labeled. The Mn2+ ion in the active site is shown as a sphere in dark gray. (b). Structure of K. lactis Xrn1, viewed along the arrow indicated in panel a. (c). Overlay of the structures of K. lactis Xrn1 and S. pombe Rat1 26. For simplicity, the linker between CR1 and CR2 and domains D1–D4 of Xrn1 are not shown. All the structure figures were produced with PyMOL (www.pymol.org) unless stated otherwise.
Residues in CR1 (1–353) are located in the center of the structure of Xrn1 (Fig. 2a). CR2 (residues 505–673) surrounds one side of CR1, forming one edge of the box and helping to define the active site landscape (Fig. 2a). Residues 732–1240 at the C-terminus of the recombinant protein surrounds the other side of CR1. This 510-residue segment is unique to Xrn1 and has no equivalents in Rat1/Xrn2 (see below). On the other hand, residues 674–731 of Xrn1, directly following CR2 in the primary sequence, form a long, extended structure that has some similarity to the equivalent segment of Rat1 (see below).
The poorly conserved segment between CR1 and CR2 forms a distinct domain in this structure, connected to CR1 through a 13-turn, 70Å-long helix, which places the domain far away from the rest of the structure (Fig. 2b). The domain contains a three-stranded anti-parallel β-sheet with helices on one face. Many of the loops in this domain, as well as the loop connecting it to CR2, are disordered. This domain mediates the formation of a dimer in the crystal (Supplementary Fig. 2). This dimer is likely a crystal-packing artifact, as K. lactis Xrn1 is monomeric in solution based on gel-filtration data. Human Xrn1 contains only 70 residues for this segment, much shorter than that in K. lactis Xrn1 (150 residues, Fig. 1). Therefore, this domain is unlikely to be present in human Xrn1, and may instead be unique to the fungal Xrn1 proteins.
Comparison to the structure of Rat1
The overall structures of CR1 and CR2 of Xrn1 are similar to the equivalent regions of Rat1 (Fig. 2c), consistent with their high degree of sequence conservation (Supplementary Fig. 1). The rms distance for 437 equivalent Cα atoms of CR1 and CR2 between the two structures is 1.4 Å. Most of the central β-sheet and its surrounding helices of CR1 have the same conformation in the two structures. However, helices αB and αD, as well as several connecting loops (for example the β5 αG segment), have clear differences between the two structures (Fig. 2c). Another important structural difference concerns the tower domain (helix αD) in Rat1. The equivalent helix in Xrn1 is shorter, and Xrn1 does not have a pronounced structural feature in this region, consistent with earlier suggestions based on sequence analysis 26.
The 60-residue segment that directly follows CR2 in Xrn1 (residues 674–731) forms a structure that shows general similarity to that of the equivalent segment in Rat1 (Fig. 2c). The N-terminal part of this segment covers up one edge of CR1, and the C-terminal part traverses the bottom of CR1, although its exact location is noticeably different from that in Rat1 (Supplementary Fig. 3). This segment of Rat1 also mediates interactions with Rai1 26. The structural and amino acid sequence differences in this region of Xrn1 therefore define the molecular basis why it cannot interact with Rai1.
Unique structural features of Xrn1
In addition to the structural differences between Xrn1 and Rat1 described above, Xrn1 contains a 510-residue C-terminal extension that is absent in Rat1 (Fig. 1). Our structure shows that these residues form three distinct domains (D1, D2, D4), with possibly another domain (D3) that is mostly disordered in the current crystal (Fig. 2a). Residues in domains D1 and D4 cover up one side of CR1, but the four domains are located more than 20 Å away from the active site of Xrn1.
Unexpectedly, domains D1, D2 and D4 contain a similar central five-stranded βbarrel core, with a rms distance of 1.4 Å for approximately 40 equivalent Cα atoms (Supplementary Fig. 4). The sequence identity among these structurally equivalent residues is only about 15%. The backbone fold of this β-barrel core is related to that of tudor domains, chromo domains, and others 30, based on a DaliLite search through the PDB 31. Most of these domains are involved in protein-protein interactions, some of which are mediated through methylated lysines. However, the cluster of aromatic residues in tudor and chromo domains that is important for binding methylated lysines is absent in Xrn1, suggesting that these Xrn1 domains are unlikely to recognize methylated proteins.
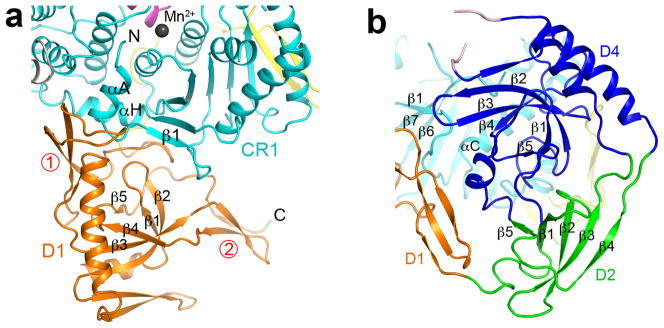
The structure of domain D1, formed by residues 732–914, also contains several additional features in the periphery, including two three-stranded β-sheets and two helices (Fig. 3a). The closest structural homolog for its central β-barrel core is the chromo domain of S. cerevisiae Eaf3 (Supplementary Fig. 4) 32,33. However, the Z score from DaliLite is 6.5 and the sequence identity for structurally equivalent residues is 12%, suggesting that this is a weak homology.
Figure 3.
Unique structural features in Xrn1. (a). Close up of the structure of domain D1, as well as its interactions with CR1. The two three-strand β-sheets in the periphery of the structure are labeled with the circled numbers in red. (b). Close up of the structure of domains D2 and D4, as well as the interactions between D4 and CR1.
Domain D2 is formed by two stretches of Xrn1, residues 915–960 and 1134–1151 (Fig. 3b), as there is an insert (residues 961–1133) between the third and fourth strands of the β-barrel. This domain has intimate interactions with domain D4, but does not contact CR1 directly. Domain D4 is formed by residues 1152–1240 as well as residues 961–978 and 1110–1133 at the two ends of the insert in D2 (Fig. 3b). The closest structural homologs of these two domains include a tudor domain in JMJD2A 34, a domain in the transcription anti-termination factor NusG 35,36, the N-terminal domain of type II restriction endonuclease Hpy99I 37, and the CPH domain of Cul7 and PARC 38 (Supplementary Fig. 5). The Z scores of these structural comparisons are approximately 6.5, with sequence identities of ~15%.
As domains D2 and D4 have strong interactions with each other, it is somewhat reminiscent of the double tudor domains that have been observed in some proteins, such as 53BP1 39,40 and Crb2 41. However, structural overlays show that the relative orientation and position of domains D2 and D4 in Xrn1 are different from those observed for these other double tudor domains.
Domains D1 and D4 have close contacts with CR1 (Fig. 2a), burying roughly 2,000 A2 of surface area in this interface (Supplementary Fig. 1). One of the three-stranded β-sheets in the periphery of domain D1 interacts with the N-terminal segment (helix αA and strand β1) and helix αH of CR1 (Fig. 3a). Domain D4 primarily interacts with the loop after β1, helix αC and the β6 β7 loop in CR1 (Fig. 3b).
The presence of another domain (D3) is suggested based on the current structure. This domain is formed by residues 979–1109, in the insert of domain D2 (Fig. 1). Two helical segments of this domain can be modeled in three of the four Xrn1 molecules. These two helices are positioned far away from the rest of the structure (Fig. 2b), and show large structural differences among the Xrn1 molecules. Residues in this putative domain show reasonable conservation between human and yeast Xrn1 (Supplementary Fig. 1), suggesting that it may also play a role in Xrn1 function (see below).
The active site of Xrn1
The active site of Xrn1 is located in a deep pocket near the bottom of the αD helix in CR1 (Fig. 4a). As observed in the structure of Rat1 26, CR2 encircles the active site in Xrn1, but makes few direct contributions to it (Fig. 4b). Nonetheless, this active site landscape probably explains why the XRNs are exoribonucleases. The location of the active site in a pocket is unlikely to be compatible with an endonuclease activity. Domains D1–D4 are located far from the active site. The amino acid sequences of these four domains contain an excess of basic residues (the pI of the entire 510-residue segment is about 10), as also indicated by the positively charged surface patches in domain D4 (Fig. 4a), suggesting that these domains might help in binding the RNA substrate. In comparison, the molecular surface of Rat1 is more electronegative in nature (Fig. 4b). The pI for the conserved regions of Xrn1 and Xrn2/Rat1 is about 5.5 23.
Figure 4.
Structure of the active site of K. lactis Xrn1. (a). Molecular surface of K. lactis Xrn1, colored by electrostatic potential. The active site region is indicated with the yellow star. Domain D3 and the linker between CR1 and CR2 are not shown. (b). Molecular surface of S. pombe Rat1–Rai1 complex 26, viewed in the same orientation as panel a. Panels a and b produced with the program Grasp 50. (c). Stereo drawing of the active site of K. lactis Xrn1. Side chains in the active site are shown as stick models and labeled. The bound Mn2+ ion is shown as a gray sphere. (d). Overlay of the structures of the seven conserved acidic residues in CR1 and the two Tyr residues in CR2 in the active sites of Xrn1 (in cyan for carbon atoms) and Rat1 (in gray). The liganding interactions of the Mn2+ ion are indicated with the dashed lines. Residues in Xrn1 are labeled in black, and those in Rat1 labeled in gray.
Almost all the residues in the active site region are highly conserved among the XRNs (Supplementary Fig. 1). The seven conserved acidic residues correspond to Asp35, Asp86, Glu176, Glu178, Asp206, Asp208 and Asp291 in K. lactis Xrn1 (Fig. 4c). The N-terminal end of helix αD provides several positive (Lys and Arg) and hydrophilic (Gln) residues to the active site. Helix αB also contributes several hydrophilic residues to the active site. Only three side chains in CR2, Tyr547, Tyr548 and Gln591, are located near the active site (Fig. 4c), and all three residues are conserved among the XRNs.
By co-crystallizing the E178Q mutant with 5 mM MnCl2 (Table 1), we were able to observe the binding of one Mn2+ ion in the active site (Fig. 4c). The metal ion is tightly coordinated by the side chains of three of the conserved acidic residues, Asp206, Asp208 and Asp291 (Fig. 4d), with distances of roughly 2.1 Å. There are no conformational changes in the enzyme upon the binding of this cation.
Domain D1 is required for nuclease activity
We next assessed the functional importance of the new domains in the structure of Xrn1. We created three internal deletion mutants, removing residues 355–492 (the linker between CR1 and CR2), 993–1098 (domain D3), 927–1145 (domains D2 and D3), as well as two C-terminal truncations, removing residues 752–1453 (from domain D1 onward) and 916–1453 (from domain D2 onward) of K. lactis Xrn1. Based on the structure, removing D3 or D2 and D3 is unlikely to have a large impact on the interactions of domains D1 and D4 with the rest of the structure (especially CR1) (Fig. 2a). All these mutants were expressed in and purified from E. coli. All gave clean gel-filtration profiles (Supplementary Fig. 6), suggesting that the deletions did not affect the overall folding of the protein. Exonuclease activity was measured using a 240-nt RNA substrate labeled at its 3′-end as well as a 55-nt ssDNA substrate labeled at its 5′-end. Wild-type K. lactis Xrn1 (residues 1–1245) demonstrated strong, time-dependent degradation of the RNA substrate (Fig. 5a). The activity appeared to be mostly processive, as there was little evidence for intermediates in the reaction.
Figure 5.
Biochemical studies on the nuclease activity of K. lactis Xrn1. (a). Time course showing the degradation of the 240-nt RNA substrate (indicated with the arrowhead, with 5′-end monophosphate and labeled at the 3′-end) by recombinant K. lactis Xrn1 (residues 1–1245). (b). Ribonuclease assays showing the effect of site-specific and deletion mutations on the activity of K. lactis Xrn1. All reactions contained 20 ng of enzyme, except for the 4 lanes at the right, which contained 50 ng of enzyme. (c). Nuclease assays showing the effect of C-terminal deletion mutations on the activity of K. lactis Xrn1 towards a 55-nt ssDNA substrate, labeled at the 5′-end. (d). Nuclease assays showing the effect of deleting the N-terminal four residues on the activity of K. lactis Xrn1 towards the 55-nt ssDNA substrate.
We next analyzed nuclease activity of the mutants. Deleting domain D3 or D2 and D3 essentially abolished nuclease activity towards the RNA substrate (Fig. 5b). On the other hand, the domain in the linker segment between CR1 and CR2 was not required for activity (Fig. 5b), consistent with its poor conservation among the XRNs. We also introduced a double mutation, K388A K389A, in the long helix in this region, and it had little impact on nuclease activity. The truncation mutant lacking all four domains (Δ752–C terminus) was inactive (Fig. 5b). In comparison, weak nuclease activity was observed for the truncation mutant lacking domains D2 D4 (Δ916–C terminus). To assess the nuclease activity of the proteins more carefully, we followed the reactions in a time course. The data confirmed that the mutant lacking the linker segment between CR1 and CR2 had slightly reduced activity compared to the wild-type enzyme, while the derivative lacking domains D2–D4 had much lower activity (Supplementary Fig. 7).
We further characterized the nuclease activity of wild-type Xrn1 and the deletion mutants using the ssDNA substrate. Earlier studies showed that yeast Xrn1 has nuclease activity towards ssDNA 23,24,28. Wild-type K. lactis Xrn1 readily degraded this ssDNA substrate, while the truncation mutant lacking all four domains (Δ752–C terminus) was essentially inactive (Fig. 5c). On the other hand, the truncation mutant lacking domains D2–D4 (Δ916–C terminus), as well as the two internal deletion mutants, showed weak activity.
We also assessed the functional importance of the highly conserved residues in the active site of Xrn1 (and Rat1). Many of them, especially those in CR1, have been examined earlier by mutagenesis studies 23,24. We selected several additional conserved residues in the active site for study, especially the three conserved residues in CR2 that make contributions to the active site, Tyr547, Tyr548 and Gln591 (Fig. 4c). The experiments confirmed that the E178Q mutant had essentially no nuclease activity (Fig. 5b). The D35A mutation also abolished activity. The R100A and Y547A Y548A mutants maintained some activity towards the RNA substrate, although they both showed the appearance of intermediates. Finally, the Q96A, N131A and Q591A mutations had only small effects on nuclease activity, which was confirmed by time course experiments (Supplementary Fig. 7).
Domain D1 may stabilize the N-terminal segment, which is important for nuclease activity
The crystal structure suggests a possible explanation for the functional importance of domain D1. It has close interactions with the N-terminal segment, and the N-terminus is located near the active site (Fig. 3a). Therefore, domain D1 may help to define and/or stabilize the conformation of the N-terminal segment, which may be important for Xrn1 catalysis.
To assess the functional importance of the N-terminal segment of Xrn1 for its nuclease activity, we created a deletion mutant that removed the first four residues (Δ1–4) (Fig. 3a and Supplementary Fig. 1). The mutant was purified following the same protocol as above and behaved the same on a gel-filtration column as wild-type Xrn1. However, this mutant had dramatically reduced nuclease activity towards the 55-nt ssDNA substrate (Fig. 5d). The experimental data are consistent with our model for why domain D1 is required for nuclease activity. Domains D2–D4 may help maintain domain D1 in the correct conformation, thereby indirectly stabilizing the conformation of the N-terminal segment.
We carried out additional experiments to characterize other possible molecular mechanism(s) for the reduced activity of the deletion mutants, including electrophoretic mobility shift assays and thermal shift experiments 42. The mutations weakened but did not disrupt binding of the 240-nt RNA (Fig. 6). The RNA substrate appeared to be able to bind multiple copies of Xrn1, resulting in a complex that failed to migrate into the gel for the wild-type enzyme at higher concentrations (Fig. 6). In comparison, several intermediate complexes were observed with the mutants, likely due to weaker affinity for the RNA. These data suggest that the four new domains contribute to RNA binding, which may be important for recruiting substrate and possibly for other functions.
Figure 6.

RNA binding activity of K. lactis Xrn1. Electrophoretic mobility shift assays for wild-type and various deletion mutants of Xrn1. The wild-type enzyme has high affinity for the 240-nt RNA, producing a large complex that did not migrate into the gel.
We also observed that the mutations reduced the thermal stability of the enzyme (Supplementary Fig. 8). Therefore, the new domains may also stabilize the overall structure of Xrn1, facilitating its catalysis. This is somewhat reminiscent of Rat1, where Rai1 can stimulate Rat1 nuclease activity by binding and stabilizing its overall structure 26,27. At the same time, the mutant lacking the CR1–CR2 linker also has reduced thermal stability but is nonetheless fully active (Fig. 5b), suggesting that there is not a direct correlation between stability and nuclease activity.
Discussion
We report the first crystal structure of an Xrn1 enzyme, which shows that the 510-residue segment unique to Xrn1 forms four distinct domains. Biochemical data show that these domains are important for the catalysis by Xrn1, even though they are located far from the active site and are only weakly conserved among the Xrn1 enzymes. Therefore, the highly conserved core of the XRNs (CR1 and CR2) is not sufficient for enzyme activity, and requires the help of less conserved segments (or another protein, such as Rai1 in the case of Rat1) for stability and/or activity.
The four new domains likely have other important functions besides catalysis. Xrn1 interacts with a large number of proteins during its functions in RNA metabolism 5, and the new domains may constitute a platform for the recruitment of these other proteins. Two weakly conserved surface patches can be identified in domains D1, D2, and D4 (Figs. 7a, 7b). The conserved surface patches may mediate interactions with these other proteins and/or the substrate of Xrn1.
Figure 7.
Conserved surface features in the structure of K. lactis Xrn1. (a). Molecular surface of K. lactis Xrn1 colored by sequence conservation among fungal and mammalian enzymes, analyzed by the program ConSurf 51. The colors vary from dark purple for highly conserved residues to cyan for residues with little conservation. A weakly conserved surface patch is highlighted with the red oval. (b). Surface conservation of Xrn1, viewed after a 90° rotation around the horizontal axis from panel a.
The position of the Mn2+ ion in Xrn1, and its coordination pattern, appears to be unique among enzymes with homologous structures (Supplementary Fig. 9). In fact, the observed positions of the metal ions are highly variable in these other enzymes 43, including bacteriophage T4 RNase H 44 and flap endonucleases 45–48. The distances between the metal ions and their ligands in these other structures are oftentimes 3 Å (or longer), too far for liganding interactions. Our inability to observe binding of a predicted second metal ion to the E178Q mutant suggests that the mutation may have disrupted the other binding site (Supplementary Fig. 9).
While most of the residues in the active sites of Xrn1 and Rat1 are conserved (Supplementary Fig. 1), the position and/or conformation of many of these residues show clear differences between the two structures. For example, the two conserved acidic residues that coordinate the Mn2+ ion in our structure, Asp206 and Asp208, have conformational differences in both their main-chain (β5–αG loop) and side-chain atoms as compared to Rat1 (Fig. 4d). As a consequence, the Mn2+ ion observed in the Xrn1 structure cannot maintain a similar binding mode in Rat1 unless there is a conformational change in the latter.
A collection of missense mutations in Xrn1 that had severely reduced nuclease activity was identified earlier from a genetic screen 23. The majority of these mutations are in CR1 and are located directly in the active site of Xrn1 (Fig. 4c). Three additional mutations, C201R, L592P and W798R, are located in the hydrophobic core of the structure. Therefore, all of these mutations either directly disrupt the active site, or indirectly disturb the overall structure of the enzyme.
Our biochemical data indicate that domain D1 is required for Xrn1 nuclease activity, while domains D2–D4 are important but may not be essential for catalysis. Earlier deletion studies with yeast Xrn1 were based on a truncation that started in the middle of domain D4 23, which very likely caused the unfolding of this domain and may have in turn had detrimental effects on the other domains.
The four new domains in Xrn1 cover up one side of the CR1 (Fig. 2a), while Rai1 is located at the bottom of CR1 in the complex with Rat1 (Fig. 4b) 26. Therefore, the relative positions of the four domains in Xrn1 are entirely different from that of Rai1. In addition, our biochemical and functional studies demonstrate that Rai1 possesses RNA 5′-end pyrophosphohydrolase and decapping activities, which are important for RNA 5′-end capping quality control 26,49. In comparison, the four domains in Xrn1 are unlikely to have any catalytic activity. On the other hand, Rai1 can indirectly enhance the activity of Rat1, possibly by stabilizing the overall structure of the enzyme 26,27. In that regard, it would be interesting to characterize whether the protein partners of Xrn1, possibly recruited through the four domains, can also regulate the exoribonuclease activity of this enzyme.
Methods
Protein expression and purification
Residues 1–1245 of K. lactis Xrn1 were subcloned into the pET26b vector (Novagen). The expression construct contained a C-terminal hexa-histidine tag, which was not removed for crystallization. The native protein was over-expressed overnight in E. coli BL21 Star (DE3) cells (Novagen) at 20°C in the presence of 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Gold Biotechnology, Inc). The soluble protein was eluted from nickel affinity beads (Qiagen) by a buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, 200 mM imidazole and 10 mM β-mercaptoethanol. The eluted protein was further purified by gel-filtration chromatography and then concentrated to 10 mg/mL in a buffer containing 20 mM Tris (pH 7.5), 200 mM NaCl, 2 mM DTT and 5 % (v/v) glycerol. Purified protein samples were flash frozen with liquid nitrogen and then stored at −80°C.
The selenomethionyl (SeMet) protein was produced in E. coli BL21 Star (DE3) cells grown in defined M9 media supplemented with selenomethionine and specific amino acids to block endogenous methionine biosynthesis 52. The SeMet protein was purified with the same protocol as the native protein, and the concentration of DTT was increased to 10 mM.
Protein crystallization
Crystals were obtained with the sitting-drop vapor diffusion method at 4 °C. The reservoir solution contained 100 mM Tris (pH 8.5), 0.9 M LiCl, 13.5% (w/v) PEG 6000. The protein solution was supplemented with 0.9 mM Triton X-100, 0.15 mM 1-s-nonyl-β-D-thioglucoside, and 60 mM Gly-Gly-Gly (Hampton Research) before crystallization. For the Mn2+ complex, 5 mM MnCl2 was included in the protein solution. The crystals were flash frozen in liquid nitrogen for diffraction analysis and data collection at 100 K.
Data collection and processing
Native and selenomethionyl single-wavelength anomalous diffraction data sets were collected on an ADSC Q315 CCD at the X29A beamline of the National Synchrontron Light Source (NSLS) at Brookhaven National Laboratory. The diffraction images were processed using the HKL package 53. The crystals belong to space group P1, and there are four molecules of Xrn1 in the crystallographic asymmetric unit. The data processing statistics are summarized in Table 1
Structure determination and refinement
The initial structure of Xrn1 was determined by the molecular replacement method with the program Phaser 54, using the structure of S. pombe Rat1 as the model (corresponding to the N-terminal 50% of XRN1) 26. The four molecules of XRN1 were located in the unit cell, although the subsequent electron density map was not of sufficient quality, especially for the C-terminal segment, which was not present in the Rat1 structure. Phase information from the atomic model was combined with the seleno-methionyl SAD data set to produce better phases with the program Phaser, which were further improved by four-fold NCS averaging with the program DM in the CCP4 package 55. The atomic model was built with the programs O 56 and Coot 57. The structure refinement was carried out with the programs CNS 58 and Refmac 59. The statistics on the structure refinement are summarized in Table 1.
Exoribonuclease assay
Site-specific mutations were created with the QuikChange kit (Stratagene), and deletion mutations were created by PCR. The mutants were sequenced to confirm correct incorporation of the mutations, and purified following the same protocol as the wild-type protein.
To prepare the RNA substrate for exoribonuclease assays, the plasmid pG3SVL-A, containing the SV40 late (SVL) polyadenylation site, was linearized with DraI and transcribed with Sp6 RNA polymerase (Promega) for 2 h at 37 °C. The 240-nt RNA transcripts were gel purified using standard procedures 60. 5′ termini were dephosphorylated with calf intestinal phosphatase (New England Biolabs) for 2 h at 37 °C and the RNAs were subsequently labeled at their 3′-ends with 32P Cp and T4 RNA ligase (Promega) overnight at 4 °C. 5′ termini were mono-phosphorylated by T4 polynucleotide kinase (New England Biolabs) and 1 mM ATP at 37 °C for 2 h.
To prepare the DNA substrate, a 55-nt ssDNA (5′-CTTAAACAGCAAGAAGATCGTCGTAACGAAAATACTGATACAGTCCGCCTGTATG-3′) was purchased from Operon. The 5′-end was labeled with [γ-32P]ATP and T4 polynucleotide kinase for 2h at 37 °C.
Exoribonuclease assays were performed at 37 °C for 30 min or as indicated. Reaction mixtures (20 μl volume) contained either 3′-end labeled RNA (~100 counts per second) or 5′-end labeled ssDNA (100 ng) as a substrate, 50 mM NaCl, 10 mM MgCl2,20 mM Tris (pH 8.0), 0.5 mM DTT, 50μg/ml BSA, 1U RNasin and 20 or 50 ng of recombinant Xrn1. RNA products were isolated by phenol/chloroform extraction and ethanol precipitation and fractionated by PAGE using a gel with 10% acrylamide and 7 M urea. The data were analyzed by PhosphorImager. Assays were repeated several times to ensure reproducibility.
Electrophoretic mobility shift assay (EMSA)
Reaction mixtures (10 μl) containing the 3′-end labeled 240-nt RNA (~100 count/second) in binding buffer (25 mM Tris (pH 7.5), 200 mM NaCl, 2 mM DTT, 0.5 mM EDTA) and indicated amount of protein were incubated at room temperature for 30 min and then fractionated using a 6% native polyacrylamide gel at 4 °C. The data were visualized by PhosphorImager.
Thermal shift assay
Thermal stabilities of wild-type and truncated mutant Xrn1 proteins were analyzed at various temperatures by the Mx3005P Real-Time PCR system (Stratagene). Each protein (4–10 μM) was mixed with fluorescence dye, SYPRO orange (Invitrogen), for monitoring conformational changes in 20 mM Tris (pH 7.5), 200 mM NaCl, and 2 mM DTT. The temperature was increased from 25 to 75 °C over a 50 min period.
Supplementary Material
Acknowledgments
We thank Neil Whalen and Stuart Myers for setting up the X29A beamline at the NSLS. This research is supported in part by grants from the NIH to LT (GM077175) and JLM (GM028983).
Footnotes
Accession codes. The atomic coordinates have been deposited at the Protein Data Bank, with accession codes 3PIE and 3PIF.
Author Contributions. J.H.C. and S.X. carried out protein expression, purification and crystallization experiments. J.H.C., S.X. and L.T. carried out crystallographic data collection, structure determination and refinement. J.H.C. carried out mutagenesis, nuclease assays and thermal shift experiments. K.X. helped with the preparation of the nuclease substrate. J.L.M. designed experiments and analyzed the data. All authors commented on the manuscript. L.T. designed the experiments, analyzed the data and wrote the paper.
References
- 1.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Newbury SF. Control of mRNA stability in eukaryotes. Biochem Soc Trans. 2006;34:30–34. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- 3.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 4.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 5.Coller J, Parker R. Eukaryotic mRNA decapping. Ann Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 6.Zabolotskaya MV, Grima DP, Lin MD, Chou TB, Newbury SF. The 5′–3′ exoribonuclease Pacman is required for normal male fertility and is dynamically loalized in cytoplasmic particles in Drosophila testis cells. Biochem J. 2008;416:327–335. doi: 10.1042/BJ20071720. [DOI] [PubMed] [Google Scholar]
- 7.Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharymyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Develop. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 8.Kenna M, Stevens A, McCammon M, Douglas MG. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.di Segni G, McConaughy BL, Shapiro RA, Aldrich TL, Hall BD. TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol Cell Biol. 1993;13:3424–3433. doi: 10.1128/mcb.13.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 11.West S, Gromak N, Proudfoot NJ. Human 5′->3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 12.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Develop. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Develop. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Develop. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechnisms of transcriptional termination. Genes Develop. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luke B, et al. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharocmyces cerevisiae is mediated by Met22 and the 5′ –3′ exonucleases Rat1 and Xrn1. Genes Develop. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shobuike T, Tatebayashi K, Tani T, Sugano S, Ikeda H. The dhp1+ gene, encoding a putative nuclear 5′->3′ exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucl Acid Res. 2001;29:1326–1333. doi: 10.1093/nar/29.6.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gy I, et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olmedo G, et al. Ethylene-insensitive5 encodes a 5′->3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potuschak T, et al. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 23.Page AM, Davis K, Molineux C, Kolodner RD, Johnson AW. Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucl Acid Res. 1998;26:3707–3716. doi: 10.1093/nar/26.16.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solinger JA, Pascolini D, Heyer WD. Active-site mutations in the Xrn1p exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis. Mol Cell Biol. 1999;19:5930–5942. doi: 10.1128/mcb.19.9.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: Two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Xiang S, et al. Structure and function of the 5′->3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue Y, et al. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens A, Poole TL. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J Biol Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 29.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 30.Maurer-Stroh S, et al. The tudor domain ‘royal family’: tudor, plant Agenet, chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v. 3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun B, et al. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 35.Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burmann BM, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 37.Sokolowska M, Czapinska H, Bochtler M. Crystal structure of the bba-Me type II restriction endonuclease Hpy99I with target DNA. Nucl Acid Res. 2009;37:3799–3810. doi: 10.1093/nar/gkp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaustov L, et al. The conserved CPH domains of Cul7 and PARC are protein-protein interaction modules that bind the tetramerization domain of p53. J Biol Chem. 2007;282:11300–11307. doi: 10.1074/jbc.M611297200. [DOI] [PubMed] [Google Scholar]
- 39.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 40.Charier G, et al. The tudor tandem of 53BP1: A new structural motif involved in DNA and RG-rich peptide binding. Structure. 2004;12:1551–1562. doi: 10.1016/j.str.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vedadi M, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Syson K, et al. Three metal ions participate in the reaction catalyzed by T5 Flap endonuclease. J Biol Chem. 2008;283:28741–28746. doi: 10.1074/jbc.M801264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueser TC, Nossal NG, Hyde CC. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 45.Hwang KY, Baek K, Kim HY, Cho Y. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat Struct Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- 46.Feng M, et al. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nature Struct Mol Biol. 2004;11:450–456. doi: 10.1038/nsmb754. [DOI] [PubMed] [Google Scholar]
- 47.Sakurai S, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dore AS, et al. Structure of an archael PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucl Acid Res. 2006;34:4515–4526. doi: 10.1093/nar/gkl623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao X, et al. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 51.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 52.Doublie S, et al. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384:219–221. doi: 10.1016/0014-5793(96)00316-x. [DOI] [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.CCP4. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 56.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 60.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a Poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.