Abstract
The characteristics of epigenetic control including the potential for long lasting, stable effects on gene expression that outlive an initial transient signal, could be of singular importance for post-mitotic neurons, which are subject to changes with short to long lasting influence on their activity and connectivity. Persistent changes in chromatin structure are thought to contribute to mechanisms of epigenetic inheritance. Recent advances in chromatin biology offer new avenues to investigate regulatory mechanisms underlying long-lasting changes in neurons, with direct implications for the study of brain function, behavior and diseases.
Introduction
One of the most intriguing and fundamental properties of brain function is the ability to sustain long-term changes in patterns of neuronal activity, a phenomenon broadly defined as memory. Memory lasts minutes to years 1 underscoring the existence of multiple strategies that afford neurons with short- to long-lasting functional changes. Precise mechanisms underlying memory formation and associated plasticity of neuronal function have been subject to intense investigation at the molecular, cellular and neuronal network levels, and are likely to involve all, or combination of changes in protein synthesis, gene expression, and cellular and anatomical structure.
Recent years have seen an extensive search for gene regulatory mechanisms that respond on the short time scale associated with memory formation, while persisting over the long time scale over which memory can last. This has prompted a singular interest for the process of epigenetic inheritance. Epigenetic changes are defined as alterations in gene expression that are self-perpetuating in the absence of the original signal that caused them 2,3. The idea of persistent changes in gene expression triggered by transient events is intuitively parallel to the long term effects believed to be involved in memory.
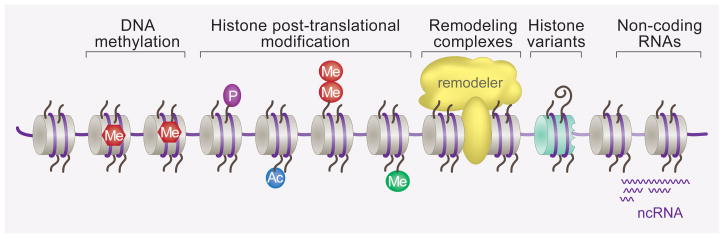
A major class of epigenetic mechanisms is thought to involve persistent changes in chromatin structure 2 (Figure 1). Most, if not all, transcriptional regulatory events cause changes to chromatin structure and composition, due to the recruitment of chromatin modifying enzymes by transcription factors and by the transcriptional machinery itself. Less is known about whether, or under which circumstances, chromatin modifications can be stably maintained or propagated. Nevertheless, the recent realization that most genes associated with mental retardation affect chromatin-remodeling processes 4,5, together with the identification of chromatin alterations in the process of neuronal plasticity and long-lasting changes in brain function, have recently brought chromatin biology to the forefront of molecular neuroscience and neuropathology. A key question is whether the seemingly specific requirement of chromatin modifiers in neuronal processes merely reflects the complexity of transcriptional regulation in the nervous system, or indicates a special function of chromatin related mechanisms in memory and behavioral control.
Figure 1. Mechanisms involved in chromatin modifications.
Five broad and interrelated mechanisms are known to affect chromatin structure: DNA methylation, histone modification, insertion of histone variants, remodeling complexes, and non-coding RNAs. All five have been shown to be essential contributors to the development and cell fate determination of tissues including in the nervous system, while histone modifications and DNA methylation have so far been more extensively investigated in the context of adult brain function.
This essay will use representative examples in the recent literature to assess the contribution of various chromatin remodeling events to long-lasting changes in brain function. In order to investigate how, and when specific chromatin modifications impact brain function and behavior, the contribution of chromatin alterations to changes in brain function will be discussed according to their timing and duration, from the most to the least transient, throughout the life of the organism, and possibly across generations (Figure 2).
Figure 2. Contribution of various chromatin remodeling events throughout life of an organism.
Chromatin modifications occurring at different time points during the life of an organism have been associated with various short to long-lasting regulatory events that affect the development and the function of the brain and other tissues.
Sustained changes in neuronal activity affect the chromatin
Neuronal activity induces changes in gene expression that are essential to the establishment and maintenance of long-term neuronal plasticity in the adult brain 6. Consequently, and perhaps not unexpectedly, promoter regions of genes involved in neuronal plasticity show alterations in chromatin composition, and a growing number of reports have described changes in chromatin states, particularly in DNA methylation and histone marks, associated with long-term plasticity.
1 DNA methylation and brain activity
DNA methylation of cytosine residues into 5-methyl cytosine, which in mammalian cells is mainly confined to CpG dinucleotides, is viewed as the most stable and long-lasting chromatin modification. Although the role of DNA methylation in constitutive silenceing of chromatin regions, X-inactivation, parental allele imprinting, retroviral and individual gene silencing is established, the precise mechanisms by which DNA methyl marks are set, maintained and erased are the topic of much debate (Box 1). The importance of DNA methylation in assisting essential gene regulation events associated with brain function and disease was revealed by the identification of Mecp2, a known methyl-CpG-binding domain protein (MBD) protein, as the target of mutations causing Rett syndrome 7. Rett syndrome is a severe X-linked mental retardation disorder characterized by late onset neurological defects in affected girls. Although Mecp2 is ubiquitously expressed in the mouse, the conditional knockout of Mecp2 in the mouse brain recapitulates the entire phenotype of the Mecp2-null 8,9, while rescue of expression of Mecp2 in postmitotic neurons prevents the emergence of phenotype in the mouse 10. Mecp2 is highly expressed by post-mitotic neurons, and the neurotrophin BDNF, a key player in neuronal plasticity events, has been identified as one of the main target genes of Mecp2 repression following neuronal activity 11,12. Recent analysis in mouse mutant lines that lack or over-express Mecp2 has pointed to additional candidate target genes of Mecp2 function in the hypothalamus 13. Interestingly, changes in gene expression observed in the mutant lines, though relatively modest, as well as demonstration of the direct binding of Mecp2 to promoter regions of candidate target genes, suggest a role of Mecp2 in direct activation as well as repression of transcription. Some of the transcriptional activation by Mecp2 was shown to involve CREB1, a major transcriptional activator and essential component of signaling pathways underlying neuronal plasticity.
BOX 1. DNA methylation and demethylation.
The DNA can be covalently modified by methylation of the cytosine residue into 5-methyl cytosine, which in mammalian cells is mainly confined to CpG dinucleotides. The presence of another methyl cytosine modification, 5-hydroxymethyl-2′-deoxycytidine (hmdC) was recently reported in the adult mouse brain 22, and its functional significance is yet to be determined.
CpG methylation has been involved in X-inactivation, genomic imprinting, suppression of transposable elements, and is required for proper embryonic development. The locations of CpG-rich regions of the genome, also called CpG islands, are often correlated with gene promoter regions, and changes in the methylation status of key developmental genes has been proposed to participate in restriction of pluripotency and lineage commitment. The role and mechanisms of DNA methylation in ensuring tissue-specific gene expression are not entirely clear. Moreover, the extent and underlying mechanisms of changes in methyl marks are highly debated questions that are of particular relevance for the study of long lasting transcriptional changes in the brain.
To directly visualize changes in methylation marks associated with the establishment of lineage- and pluripotency-specific transcriptional programs, large scale and genome-wide analysis of DNA methylation has been recently performed, documenting differences in patterns of methylation according to the developmental stage or cell type analyzed 84,85,86,87. Surprisingly, significant differences were also uncovered in gene regions outside CpG islands, and for ES cell outside of CG context, underscoring the still poorly understood complexity of transcriptional control in promoter regions as well as gene-bodies. Widespread differences in composition and pattern of cytosine methylation were observed in different cell types 85,86, although the number of promoters that display either loss or gain of methylation between ES cells and terminally differentiated neurons appears rather modest 41
Microarray analysis of the methylation status of 15,000 promoters in ES cells and terminally differentiated pyramidal neurons in vitro reported a gain of DNA methylation on only 343 (2.3%), and an even less frequent loss of DNA methylation, on 22 (0.1%) of the tested promoters during neuronal differentiation. Strikingly, analysis at an intermediate developmental stage, that of neuronal progenitors, shows that most changes occur at the transition from ES cells to progenitor state, suggesting that alterations in DNA methylation correlate more strongly with fate commitment and loss of pluripotency, rather than with terminal neuronal differentiation 41. Interestingly, many promoters bearing the Polycomb-mediated histone H3 methylation (H3K27me3) in ESCs acquired DNA methyl marks during differentation, suggesting interrelated processes.
The mechanisms by which differential methylation patterns are established in mammals remain highly debated. While enzymes carrying DNA methylation are shared between plants and mammals, and their mechanisms of action well understood 88, a lot of uncertainty is left regarding mechanisms of DNA de-methylation 89. DNA demethylation can result from passive demethylation in absence of maintenance methylation following DNA replication, or from an active process of enzymatic removal of the 5-methylcytosine mark. Active genome-wide demethylation are thought to occur at two times of development (figure 2), in the male pronucleus of the zygote, right after fertilization, and in primordial germ cells of E11.5-12.5 embryos. Unfortunately none of the plant enzymes involved in the active process appear conserved in mammals, and there is still some debate as to how these events occur. The DNMTs, which are expressed in both fetal and adult tissues may be involved, while other studies, including in the adult brain, have proposed a very different mechanism in which 5-methylcytosine is removed from DNA in a deamination and base excision/repair process 17,18,21,90,91,92. Two recent reports in mouse primordial germ cells and somatic cells induced to pluripotency have given additional credence to the deamination-repair dependent DNA demethylation process 93,94. How more targeted promoter demethylation process may occur is unknown, although some reports have shown interesting switch of DNA methylation and demethylation in hormone-induced transcriptional control 82.
The major defects in brain function and the late onset of the phenotype observed in Rett syndrome and related mouse models, together with cognitive impairments and defects in neuronal differentiation found in mutant lines for MBD1 14 suggest an important role for DNA-methylation marks in assisting transcriptional networks mediating normal neuronal homeostasis 5.
Yet, perturbations in gene transcription in mutants for methyl-CpG-binding domain proteins (MBD) such as Mecp2 are perhaps not so surprising, as the role of CpG methylation in gene silencing is established, although several MBD proteins appear dispensable for embryonic development in the mouse 15. A slightly different set of questions concerns the extent to which DNA methyl marks can be modified in the adult brain, and whether these changes can affect neuronal function. Studies along these lines in the nervous system are still in their infancy, and results quite controversial. In part, there is still major uncertainty over key mechanisms underlying the establishment and erasure of methyl marks in early embryonic and germ cell precursors, where robust and widespread changes in methylation and demethylation are known to occur (box 1). Thus, extreme caution should be used in interpreting data in the brain, where such changes, if any, may be rather modest and affect only a few genes.
Both maintenance and de novo DNA methyltransferases DNMT1, and DNMT3a respectively, are found expressed at high levels in the developing and adult nervous system 16. The expression of these enzymes in post-mitotic neurons is rather intriguing but does not necessarily imply a function in active methylation in these cells, as de novo and maintenance methylation in germ cell development and embryogenesis occurs during DNA replication. Further, injection of DNMT inhibitors reportedly leads to defect in memory-associated neuronal plasticity. However, the requirement of DNA synthesis for the activity of the drugs, together with the toxicity and lack of specificity of the inhibitors employed make the interpretation of the results difficult 16. CNS-specific conditional DNMT knockouts affecting DNA methylation in dividing neuronal precursors lead to profound neuronal defects, suggesting a role of DNA methylation in neuronal development. These results, however, provide little information about changes in DNA methylation in postmitotic neurons.
Surprisingly, a recent report described a significant, though rather modest reduction in DNA methylation at specific promoter regions of BDNF and FGF1 in the adult dentate gyrus of the mouse following electroconvulsive treatment 17. Through loss of function experiments, the authors invoked the participation of Gadd45b (growth arrest and DNA damage-inducible protein 45 beta) in this phenomenon, a member of a family of molecules that has been shown in some systems, though refuted in others, to act as cofactors promoting DNA demethylation through DNA repair 18,19,20,21. The functional impact on gene transcription of rather weak incremental reductions rather than genuine loss of promoter methylation is unclear. Further, as the precise mechanistic links between Gadd45 activation, DNA demethylation and DNA repair remain to be clarified (Box 1), the simple interpretation of these results as an indication that active DNA demethylation is detected in postmitotic neurons will await further supporting evidence.
As will be detailed in a later part of this review, the methylation status of a number of genes involved in behavioral control has similarly been reported to vary according to early postnatal environmental conditions, raising particularly intriguing questions about the ability of the environment to affect the DNA methylation status of neural genes. Interestingly, 5-hydroxymethyl-2′-deoxycytidine (hmdC) was recently identified as an abundant nucleotide in many regions of the adult brain, including the cortex, brain stem and in cerebellar Purkinje neurons 22. This is an intriguing discovery, as hmdC may represent an intermediate for oxidative demethylation, or an end product that could modulate the binding of proteins that normally recognize 5-methyl cytosine.
2 Histone modification and neuronal plasticity
Molecular analysis of signaling pathways underlying neuronal plasticity has identified alterations of histone marks, particularly histone acetylation, in transcriptional units induced by neuronal activity, and has implicated histone modifying enzymatic complexes in memory formation (see background information on histone modifications in Box 2). These findings have raised interesting mechanistic questions, as well as new ideas for the design of drugs aimed at memory impairment.
BOX 2. Histone modifications.
Histones, particularly H3 and H4, are subject to extensive covalent post-translational modifications (PTM) that include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, biotinylation, ADP-ribosylation, and likely more to be discovered, each occurring at specific sites and residues 95. Some histone modifications act in cis to directly alter the local chromatin structure, while others act in trans to influence the recruitment of chromatin-modifying factors. In trans, histone modifications enable specific binding partners to dock, often as part of larger multi-molecular complexes that generate further chromatin remodeling. Acetylated histone residues are recognized by bromodomains, often associated with histone acetyltransferases (HAT), thus leading to spread of the histone modification. Similarly methylated lysine residues are recognized by chromodomain- containing proteins. This recognition is highly dependent on the chromatin context, such that a given chromodomain- or bromodomain-containing protein may only bind to a given set of methylated, or acetylated histone residues, respectively, and only in the presence of other defined chromatin effectors 96.
Histone modifications do not occur in isolation, but often as combinations of marks. The understanding of the regulation and physiologically relevant substrate specificity of these enzyme complexes remains a challenge. Moreover, many histone-modifying enzymes also target non-histone substrates, underscoring the complexity of chromatin dynamic and associated cellular processes.
Concerted efforts have been made to establish clear functional links between histone modifications and changes in transcriptional activity, leading to the enticing and highly debated hypothesis of a “histone code” with predictive value on the transcriptional status of genes 97,98. For example silenced chromatin typically displays low levels of histone acetylation, together with high levels of H4K20me3 and H3K27me3, while hyperacetylation, H3K4me3 and H3K36me3 are recognizable marks of active transcription. Faced with the ever growing number and complexity of chromatin modifications within a given transcriptional unit, it is however becoming clear that a single histone mark, or defined combination of, are not be simply predictive of a given transcriptional outcome: H3K9me2/3 and H3K4me2/3 for example are found enriched on silenced and actively transcribed genes, respectively, but are also present in the reciprocal state 99. The possible combinatorial effect of multiple histone modifications has recently been addressed by a genome-wide analysis of PTMs occurring on single nucleosomes in correlation with transcription levels 96,100. The data largely confirm known bias towards certain combination of histone marks at promoters, transcription sites, gene bodies and 5’ and 3’ UTRs, which had previously been associated with active or repressive chromatin states. However, discrete PTM combinations rarely appear repeated within the genome, with most patterns detected on single promoters. Thus, instead of revealing a simple predictive “code” shared by many genes, in depth observation of histone modification patterns highlights instead the unique complexity of each transcriptional unit and associated transcriptional regulatory machinery to ensuring proper response to cellular signals.
Sensory experience and resulting neuronal activation leads to depolarization and calcium influx into the postsynaptic cell, which in turn triggers signals orchestrating short- and long-term changes in synaptic strength. The induction of specific activity-dependant transcriptional programs has been shown to play a key role in experience-dependent long-term neural plasticity 6. In depth studies have led to the characterization of a prototypical signaling pathway that is evolutionally conserved in Aplysia, Drosophila, and mouse, and by which extracellular stimuli are transformed into changes in activity-dependent gene expression 23. Gene regulation by the cyclic AMP response element binding protein (CREB), originally identified as binding to the cAMP- and Ca-dependent response elements of the somatostatin and cfos genes, respectively, as well as mediating long term synaptic potentiation in Aplysia 24, is particularly central to the expression of many forms of long-term memory. Following postsynaptic depolarization and Ca entry, activated CREB binds to the cAMP response element (CRE) in the promoter region of activity-induced genes such as the immediate early gene cfos, and the neurotrophin BDNF, and, in conjunction with different combinations of other factors 23 orchestrates long term activity-induced changes in gene expression.
Changes in histone post-translational modifications in general, and in histone acetylation in particular, have been extensively documented at promoters of genes induced by sustained neuronal activity. For example repeated electroconvulsive treatment, which induces long-term changes in neuronal activity that are beneficial for treatment of depression, triggers histone modifications at promoters of genes such as CREB, BDNF, c-fos that display sustained changes in transcription, but not at neuronal genes with unchanged expression 25. Similarly, an increasingly large number of paradigms have documented alterations in histone post-translational modifications in activity-dependent neuronal plasticity, addiction and long-term memory formation 6,26,27,28,29. Furthermore, pharmacological alteration of histone deacetylase (HDAC) activity significantly affects the process of memory formation, although the poor specificity of currently available reagents clearly limits the interpretation of the results 30,31,32.
At the mechanistic level, CREB-associated transcriptional regulation has been shown to involve the recruitment of multi-component regulator complexes as well as the initiation of chromatin remodeling events. Activated CREB recruits CREB-binding protein (CBP), or its paralog p300, which functions both as a scaffolding protein and a histone acetyltransferase (HAT) 6. CBP recruitment in turn stimulates histone acetylation and transcriptional complex formation at the promoters, leading to transcriptional activation of many CREB-target genes. Mutations in the CBP/p300 gene are responsible for the mental retardation syndrome Rubinstein–Taybi 33 the phenotype of which may result from impairment in either or both of CREB-dependent and -independent functions of CBP. The essential role of HAT activity in CBP-mediated neuronal plasticity has been genetically demonstrated by the selective long-term memory defects of a transgenic mouse line carrying a dominant negative CBP that blocks the HAT activity of the endogenous protein 34.
Similarly, histone deacetylase (HDAC) activity has been associated with repression of neuronal activity-dependant gene transcription. HDAC2 has recently been identified by chromatin immunoprecipitation (ChIP) at the promoter regions of a large number of genes involved in synaptic plasticity or activity-dependent processes, such as Bdnf, Egr1, Fos, Cpg15, Camk2a, Creb1, Crebbp, NRXN3 and the NMDA receptor subunits, and appears to associate with known neuronal transcriptional co-repressors such as mSin3, MTA2 and CoREST 31. This has led to the suggestion that a balance between histone acetylation, leading to transcriptional activation, and histone de-acetylation with subsequent gene repression is as an essential component of the long-term regulation of activity-dependent genes in the brain.
The study of genetically modified mouse strains, in which the function of specific histone-modifying enzymes has been altered in the brain has further revealed the fundamental contribution of chromatin remodeling to long-term neuronal plasticity and addiction. Cocaine induces HDAC5 phosphorylation and nuclear export in the Nucleus Accumbens (NuAc), and viral and genetic manipulations of HDAC5 expression in the NuAc significantly alter the response to chronic, but not acute cocaine and stress exposure 29. In another study, overexpression of HDAC2 but not HDAC1 leads to impairment in synapse formation and plasticity and in hippocampus-dependent long term, but not short-term memory formation, while conditional neuron-specific HDAC2 knockout leads to increased synapse formation and memory facilitation 31.
Postnatal, forebrain-specific deficiency of the histone methyltransferase complex GLP/G9 leads to a drastic reduction in neuronal euchromatic H3K9me2 levels 35. Genetically modified animals display complex behavioral abnormalities, including defects in learning, motivation, and environmental adaptation but no apparent structural abnormality. Importantly the behavioral phenotypes are distinct from those found in mice with forebrain ablation of another histone lysine methyltransferase, Ezh2 which is essential for H3K27 methylation, and has been shown to play an important role in lineage specification and neuronal and astrocyte differentiation 36,37. Interestingly upregulation of neuronal progenitor and non-neuronal genes was identified in GLP/G9 deficient mice, suggesting an essential role of GLP/G9 in maintaining neuron-specific transcriptional homeostasis and in protecting adult neurons from expression of non-neuronal and neuronal progenitor genes.
In another recent study, repeated cocaine administration was shown to induce repression of G9a and H3K9me2 and to promote cocaine preference, in part through the transcriptional activation of numerous genes known to regulate aberrant forms of dendritic plasticity. Thus H3K9 dimethylation appears essential to ensure the stability of proper neuronal gene expression programs 38.
3. Debating the role of histone modifications in plasticity
The studies reported here document a fascinating new side of the control of neuronal plasticity. However, one must exercise extreme caution in interpreting the role of histone modifications in this process, as histone marks are an extension and reflection of the underlying transcriptional network, and cannot therefore be interpreted alone (Box 2). The highly transient nature of histone marks must also be kept in mind. Histone acetylation for example was shown in some experimental systems to display a half-life in the order of minutes 39. Similarly a histone methyl mark such as H3K4me3, although more stable, has been shown in yeast not to be maintained after removal of the gene activating stimulus, even a loci known to be subject to epigenetic regulation 40. In a study looking at neuronal differentiation in vitro 41 H3K27me3 marks associated with polycomb-mediated repression emerge in progenitor cells as in anticipation of neuronal differentiation, but not in embryonic stem cells despite the fact that the corresponding genes are silenced in these cells as well. Thus, the histone mark does not indicate silencing in absolute term, but instead the ability for dynamic regulation and recruitment of sequence- and context- specific transcription factors. These illuminating studies in simpler experimental systems emphasize the need for more detailed mechanistic studies of transcriptional events related to neuronal plasticity.
What is the adult brain inheriting from the neonatal epigenome?
The origin of behavioral diversity within individuals of a given species constitutes one of the most fundamental questions in behavioral neuroscience, and decades of research has tried to determine the respective roles of genetic and early environmental influences in shaping adult behavioral patterns. The term of epigenetics is increasingly invoked to interpret studies from rodents to non-human primates and humans, in which stochastic developmental events and environmental information appear to stably sculpt physiological, behavioral traits and disease susceptibility from the early perinatal period into adulthood.
Studies of human monozygotic twins raised together, compared to monozygotic twins raised apart reveal significant discordance in behavioral and physiological phenotypes as well as in disease susceptibility that cannot be accounted for by simple Mendelian inheritance of genetic traits, nor by identifiable environmental differences 42. These results have been confirmed by analysis of genetically identical inbred rodents and cattle raised in tightly controlled versus variable pre- and postnatal environments 43, and more recently by observation of cloned animals 44. This paradox led to the early hypothesis of a “third component …. effective at or before fertilization” 43, the basis of which was proposed by some authors to rest in differential and heritable chromatin remodeling events that occur during cell differentiation and embryonic morphogenesis 42. Consistent with this hypothesis is the example of monozygotic twins discordant for Beckwith–Wiedemann syndrome (BWS) 45, which is thought to result from unequal distribution among twins of DNA methylation enzymes in the inner mass stage, leading to a defect in maintenance of imprinting at KCNQ1OT1. However, variability in chromatin modifications cannot be interpreted in abstract, outside the context of specific gene transcriptional regulation. Sophisticated analyses of stochastic variation in eukaryotic gene expression, from yeast to metazoans, suggest that fluctuation in chromatin-mediated events may indeed participate in gene expression variability, and that the range of variability is tightly linked to the degree of connectivity of genes within a transcriptional network, such that highly interconnected developmental networks are better able to buffer stochastic variability 46,47.
Thus, a certain range of stochastic variability in the epigenetic inheritance of neuronal progenitors may underlie stable differences in brain function, behavior and neurological disease susceptibility among individuals sharing similar genomes and environmental conditions. However the precise mechanisms involved are far from being elucidated, and in depth studies of stochastic variability during brain development, such as those performed in simpler experimental systems, are lacking.
Perhaps even more striking than a rather limited stochastic variability, are published reports suggesting that the early perinatal environment may actively and durably shape the neural, behavioral, and pathological state of individuals. Examples in the literature are numerous, affecting both neural and non-neural functions, and although the underlying mechanisms are still largely undefined, the influence of the environment on the chromatin configuration of certain genes has been put forward as a leading hypothesis for these stable changes.
The ability of the pre- and early post-natal environment in establishing distinct behavioral traits among genetically identical animals was directly demonstrated by combining embryo transfer and cross-fostering among the two inbred mouse strains C57BL/6J and BALB/cj 48. Differences between the C57BL/6J and BALB/cj strains have been well documented in exploratory and anxiety-related behaviors, watermaze performance and sensory motor gating, and were widely assumed to result from genetic factors. Surprisingly, C57BL/6J mice developing in a BALB/cj uterus and reared by a BALB/cj mother showed 3 out of 4 tested behaviors identical to that of BALB/cj mice and significantly different from other C57BL/6J. Thus a combination of pre- and early post-natal maternal environment is able to significantly shape the development of adult behavior.
In studies with far reaching impact on human health, worldwide epidemiological analyses in humans as well as direct experimentation in animal models indicate that defects in maternal and early postnatal nutrition influence a number of health risks factors in adult life, mainly cardiovascular and metabolic diseases such as hypertension, insulin resistance and obesity, a phenomenon commonly named metabolic syndrome 49. Nutritional deficiency, and restriction or excess in the maternal and post-natal diet during critical developmental time-windows results in permanent alterations in the adult function of peripheral organs, such as the liver, kidney, heart, adipocyte, and of the hypothalamo-pituitary-adrenal (HPA) axis. In addition to a direct perturbation of developmental events underlying organogenesis, the influence of early nutrition on the establishment and maintenance of cytosine methylation, including the methylation of retrotransposons 50,51 and of imprinted genes 52 points to chromatin remodeling as a potential target of early environmental influence.
More generally, the HPA axis and the organization of peripheral and central stress responses have emerged as a main target of long-lasting perinatal environmental influences. Seminal work with rodent neonates showed that manipulations of the mother–infant relationship have long-term consequences on neuroendocrine and behavioral responses later in life, and that maternal handling exerts a strong inhibitory effect on the HPA function of adult offspring, resulting in lower stress and fear responses 53,54. A series of recent studies from rodents to non-human primates and humans have investigated the molecular mechanisms by which maternal-infant relationship may exert such lasting changes on HPA function.
In the rat, variation in the amount of maternal grooming, licking and associated somatosensory stimulation of the pups lead to differences in fear responses and HPA function of the adult offspring. In a remarkable non-genetic transmission of behavior traits, daughters raised or cross-fostered by poorly grooming mothers, become highly fearful and stressed adults, and in turn are poorly grooming dams 55. A signaling pathway linking maternal care to the stress response of the offspring has been proposed. High maternal somatosensory stimulation increases 5HT signaling in the hippocampus of the pups, and activates a cAMP-dependant protein kinase signaling pathway, which in turn leads to increase in the expression of the transcription factor nerve growth factor induced protein A (NGFI-A). NGFI-A binds to and regulates the activation of the exon 17 promoter of the glucocorticoid receptor (GR) promoter, leading to increase in transcription of specific isoforms of the GR. High level of GR expression in offspring of high grooming moms is stably maintained into adulthood, well after maternal stimulation has ceased, suggesting a mechanism to permanently increase GR transcription. Interestingly, differences in GR expression between the offspring of high- and low-grooming females correlate with differential levels of DNA methylation and histone acetylation at the exon 17 GR promoter that display NGFI-A recognition sites, resulting in alterations in NGFI-A binding 56. Low DNA methylation, high histone acetylation of exon 17, and resulting high NGFI-A binding and GR expression correlated with high level of maternal care are established within the first week of postnatal life, and maintained in the adult. Notwithstanding the caveats related to the low target specificity of the drug, and its rather indirect effect, brain infusion of the adult offspring with the HDAC inhibitor trichostatin A (TSA) is reported to eliminate the maternal effect on GR expression 56. In another report of the effect of early life stress on chromatin remodeling of genes with important behavioral functions, mild though significant hypomethylation of the arginine vasopressin (AVP) promoter was recently described in the mouse hypothalamus following experimental mom-infant separation 57.
These still highly correlative data have led to the suggestion of a mechanistic link between life-long changes in behavioral traits, and the establishment of chromatin modifications of key genes in a critical perinatal period. Identifying a clear causal mechanistic relationship between these events will require an in depth understanding of the players and mechanisms involved: What are the neuronal types involved across the brain? What broader changes may be taking place in chromatin and in transcriptional networks? In a system as complex as the brain and underlying behavioral circuits, obtaining specific genetic and pharmacological tools will significantly enhance the ability to answer these questions. Chromatin remodeling is only one part of a larger puzzle of how behavioral traits are generated and maintained, and the rather modest changes in methylation levels observed in only a few genes seem unlikely to underlie such profound behavioral differences across the population. Other mechanisms proposed to mediate stable behavioral differences are equally attractive. The maternal-infant relationship in non-human primates for example has been shown to affect brain function through neurotrophin action 58, which may or may not include any chromatin related events.
The influence of mom and dad on the brain of offspring
Genomic imprinting
Genomic imprinting is a set of epigenetic modifications unique to placental mammals and flowering plants that is established in the parental germ lines and somatically maintained, and that results in the preferential expression of the maternal or the paternal allele of certain genes. The monoallelic expression makes these loci especially vulnerable to mutations and deregulation, and they often contribute to diseases and disorders 59. The evolutionary pressures that lead to imprinting of specific loci are matters of great debate 60,61,62. The first imprinted genes discovered: Igf2 63 and Igf2r 64 are paternally and maternally expressed, respectively, and have opposing effects on embryonic growth. Since these landmark papers, the vast majority of imprinting studies have focused on embryonic growth and development 65,66. In rodents and humans, nearly 100 imprinted genes have been identified, which are often organized in clusters in the genome. A bioinformatic approach estimated the existence of 600 imprinted genes in the mouse genome 67, although this study failed to predict any imprinted genes on the X-chromosome, which are now known to exist 68,69.
Strikingly, many imprinted genes have been found to be expressed in the brain, where they serve unknown functions, and genetic analysis in mice has identified behavioral and neurological function as the second most frequent function affected in mouse mutants for imprinted genes, right behind embryonic growth 70. A handful of studies have demonstrated roles for some imprinted genes in the regulation of homeostatic brain functions such as thermoregulation, maintenance of circadian rhythm, feeding behavior, as well as maternal and mating behaviors 70. Further, it has been proposed that imprinted genes regulate a broad spectrum of social behaviors, including mother-infant bonding, kin recognition, risk taking behavior, the sharing of resources within social groups, and sexually dimorphic behaviors 70 Clinical studies of patients with neurological disorders related to imprinting, such as Angelman Syndrome, Prader-Willi Syndrome and Turner Syndrome, have also demonstrated clear roles for imprinted genes in the regulation of human social behaviors 71.
Work on imprinted loci demonstrates that the imprinting status of some genes can be both temporally and spatially regulated. These data on only a small number genes so far provide additional complexity to the long held view that imprinting is stably established in parental gametes and early embryonic stages, and suggest instead that mechanisms may exist throughout adulthood to dynamically modulate the outcome of parental chromatin marks. Interestingly, as parental marks are established in primordial germ cells of offspring’s, environmental factors in the prenatal maternal environment will affect genomic imprinting of F2s (Figure 2).
Trans-generational inheritance
As shown in many of the examples cited in this review, a growing body of evidence indicates that the chromatin state can be influenced by environmental conditions. This, in turn, opens the door for a mechanistic underpinning of the so-called “soft inheritance” according to which specific environmental conditions may lead to a non-Mendelian transgenerational inheritance of certain traits, a phenomenon widely reported in plants, and increasingly discussed in animals as well 72. For example in utero alterations of DNA methylation affecting F1s and F2s have been reported as a result of a maternal diet that affects single carbon metabolism, or that contains endocrine disrupting compounds 72,73,74,75. Although no direct evidence yet links imprinting, perturbed chromatin states and nutritional environment changes, the hypothesis that imprinted genes may play a role in the trans-generational effects of the maternal diet on the physiology of offspring has increasingly been suggested 73. More generally, environmental effects on genomic imprinting during pregnancy appears as an attractive mechanism to explain trans-generational effects, which in mammals has yet to be observed beyond F2s 72,76
Outlook on chromatin remodeling processes in the adult brain
The increasingly large number of experimental data associating long-term changes in brain activity with alterations in chromatin raises several fundamental questions. Histone posttranslational modifications and other chromatin remodeling events are expected mechanisms of gene regulation in any cellular system undergoing long-lasting changes. Nothing unique to the brain in the chromatin remodeling events has been reported so far, and it is unclear that these provide anything but a transient contribution to the underlying transcriptional network that may not be sustained in the absence of the network itself. Finally, the predictive value of the observed changes, a “code” of modification associated with specific changes in brain activity that in turn may be exploited for further experimental or clinical intervention is far from established.
Chromatin remodeling events described so far in the context of long-term changes in brain function are merely part of a larger and much more complex transcriptional control pathway. The level of mechanistic detail achieved in understanding chromatin remodeling within the context of transcriptional control in simpler or reconstituted experimental systems 77 is far from being reached in the nervous system. Only a few histone PTMs or other chromatin remodeling events have so far been investigated. Moreover, little mechanistic insight has been gained to date, that may underlie the reported specificity of chromatin remodeling events to the subsets of genes affected by neuronal activity. The interesting examples of ncRNA-mediated targeted DNA methylation identified in plants 78, the transcriptional silencing of distant chromosome domains by long non-coding RNAs 79 represent new mechanisms providing specificity to chromatin remodeling events that may be worth investigating in the context of the brain development and function. Finally, chromatin remodeling events are not intrinsically long lasting, and in fact, with perhaps the exception of Polycomb group proteins 80, the inheritance of chromatin marks through DNA replication is still an open question. Thus, chromatin components, as well as the associated transcriptional regulatory machinery may be required to determine the stability or dynamic state of chromatin changes.
A complicating and often neglected factor in the interpretation of histone modifications in neuronal plasticity results from the fact that what so-called “histone-modifying enzymes” play in fact other roles in the cell beyond histone modifications, such as scaffolding and modifying non-histone substrates. The Elongator complex, for example, plays an essential role in the migration and differentiation of cortical neurons 81. Although Elongator is a known histone H3 acetylase in the nucleus, it also targets cytoplasmic proteins such as a-tubulin and other unknown substrates. The reduction of a-tubulin acetylation via expression of a nonacetylatable a-tubulin mutant leads to defects in neuronal branching of cortical neurons that are similar to mutations in the acetylation subunit of Elongator, demonstrating that a-tubulin is in fact the key target of this complex. Moreover, Elongator acetylation of a-tubulin in vitro is counteracted by HDAC6-mediated deacetylation, illustrating a delicate balance in acetylation-deacetylation of substrates distinct from histones in the process of neuronal maturation.
In another study, HDAC6, in addition to its well-described histone-modifying activity, was shown to biochemically and functionally interact with Cdc20 and to stimulates Cdc20-APC activity through polyubiquitination 82 in a process essential to dendrite morphogenesis in post-mitotic neurons.
In conclusion, despite the clear involvement of chromatin modifications demonstrated in many paradigms of long term changes in brain function, the relative lack of mechanistic insights beyond correlative observations with a handful of changes such as few of the known histone PTMs, does not yet permit one to draw a precise picture of the impact of chromatin remodeling on changes in neuronal activity. The transcriptional machinery itself is not invariant, and in addition to specific transcription factors, core components of the transcription machinery could also vary among different cell types. For example, during skeletal myogenesis, cells no longer use the canonical TFIID complex but instead use a specialized complex, generating a customized pre-initiation machinery for this cell type 83. Similarly, different effectors may interact with different chromatin marks according to the biological context. Clearly, the complexity in histone PTMs and chromatin remodeling has not yet been approached in the context of neuronal function, and genetic dissection of essential substrates and enzymes, together with precise reconstitution experiments will be critical to gain insights into the chromatin machinery that orchestrates stable changes in brain processes.
Acknowledgments
We wish to thank Renate Hellmiss for illustrations and Frances Meale for help with the manuscript. The author wishes to thank Nicole Francis, Dirk Schubeler, and members of the lab for helpful discussions. The author is supported by the Howard Hughes Medical Institute, the Klarman Foundation, and the National Institute of Health.
References
- 1.Squire LR. Memory and brain. Oxford University Press; 1987. [Google Scholar]
- *2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *3.Ptashne M. On the use of the word 'epigenetic'. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. Two interesting reviews on the meaning and prospective mechanisms of epigenetic regulation. [DOI] [PubMed] [Google Scholar]
- 4.Kramer JM, van Bokhoven H. Genetic and epigenetic defects in mental retardation. Int J Biochem Cell Biol. 2009;41:96–107. doi: 10.1016/j.biocel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Guan Z, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. An early mechanistic analysis of memory formation and associated chromatin modifications. [DOI] [PubMed] [Google Scholar]
- 7.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 8.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 9.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 10.Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci U S A. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 12.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- *13.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. An interesting mechanistic analysis of Mecp2 function in the hypothalamus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin Caballero I, Hansen J, Leaford D, Pollard S, Hendrich BD. The Methyl-CpG Binding Proteins Mecp2, Mbd2 and Kaiso Are Dispensable for Mouse Embryogenesis, but Play a Redundant Function in Neural Differentiation. PLoS One. 2009;4:e4315. doi: 10.1371/journal.pone.0004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 17.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 19.Jin YH, et al. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 2008;4:e1000053. doi: 10.1371/journal.pgen.1000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. An intriguing new nucleotide modification identified in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 25.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 2004;24:5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koshibu K, et al. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Levenson JM, et al. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 29.Renthal W, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 30.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 31.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- 33.Alarcon JM, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katan-Khaykovich Y, Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–752. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radman-Livaja M, Liu CL, Friedman N, Schreiber SL, Rando OJ. Replication and active demethylation represent partially overlapping mechanisms for erasure of H3K4me3 in budding yeast. PLoS Genet. 6:e1000837. doi: 10.1371/journal.pgen.1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. Insightful analysis of changes in DNA and histone H3 methylation at promoter regions during fate determination and early differentiation of neuronal progenitors. [DOI] [PubMed] [Google Scholar]
- 42.Wong AH, Gottesman II, Petronis A. Phenotypic differences in genetically identical organisms: the epigenetic perspective. Hum Mol Genet. 2005;14(Spec No 1):R11–18. doi: 10.1093/hmg/ddi116. [DOI] [PubMed] [Google Scholar]
- 43.Gartner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratoryanimals? Lab Anim. 1990;24:71–77. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- 44.Tamashiro KL, et al. Phenotype of cloned mice: development, behavior, and physiology. Exp Biol Med (Maywood) 2003;228:1193–1200. doi: 10.1177/153537020322801015. [DOI] [PubMed] [Google Scholar]
- 45.Weksberg R, et al. Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum Mol Genet. 2002;11:1317–1325. doi: 10.1093/hmg/11.11.1317. [DOI] [PubMed] [Google Scholar]
- 46.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 463:913–918. doi: 10.1038/nature08781. Interesting analysis of variability in regulatory networks of genetically identical animals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *48.Francis DD, Szegda K, Campbell G, Martin WD, Insel TR. Epigenetic sources of behavioral differences in mice. Nat Neurosci. 2003;6:445–446. doi: 10.1038/nn1038. A short but remarkable demonstration of the effect of perinatal environment on behavioral traits. [DOI] [PubMed] [Google Scholar]
- 49.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 50.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 52.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Levine S. Infantile experience and resistance to physiological stress. Science. 1957;126:405. doi: 10.1126/science.126.3270.405. [DOI] [PubMed] [Google Scholar]
- 54.Denenberg VH, Brumaghim JT, Haltmeyer GC, Zarrow MX. Increased adrenocortical activity in the neonatal rat following handling. Endocrinology. 1967;81 :1047–1052. doi: 10.1210/endo-81-5-1047. [DOI] [PubMed] [Google Scholar]
- 55.Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 56.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 57.Murgatroyd C, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 58.Cirulli F, et al. Early life stress as a risk factor for mental health: role of neurotrophins from rodents to non-human primates. Neurosci Biobehav Rev. 2009;33 :573–585. doi: 10.1016/j.neubiorev.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keverne EB. Genomic imprinting and the evolution of sex differences in mammalian reproductive strategies. Adv Genet. 2007;59:217–243. doi: 10.1016/S0065-2660(07)59008-5. [DOI] [PubMed] [Google Scholar]
- 61.Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 62.Wolf JB, Hager R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006;4:e380. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 64.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 65.Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006;65 (Suppl 3):50–58. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- 66.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 67.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies W, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 69.Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 70.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 71.Kozlov SV, et al. The imprinted gene Magel2 regulates normal circadian output. Nat Genet. 2007;39:1266–1272. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- 72.Richards EJ. Inherited epigenetic variation--revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 73.Lim AL, Ferguson-Smith AC. Genomic imprinting effects in a compromised in utero environment: Implications for a healthy pregnancy. Semin Cell Dev Biol. 2009 doi: 10.1016/j.semcdb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 74.Patisaul HB, Adewale HB. Long-term effects of environmental endocrine disruptors on reproductive physiology and behavior. Front Behav Neurosci. 2009;3:10. doi: 10.3389/neuro.08.010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *76.Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. A insightful review of the phenomenon of transgenerational inheritance. [DOI] [PubMed] [Google Scholar]
- 77.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 78.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harb Symp Quant Biol. 2006;71:449–459. doi: 10.1101/sqb.2006.71.028. [DOI] [PubMed] [Google Scholar]
- 79.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *80.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. A unique demonstration of the retention of a chromatin modification through cell division. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Creppe C, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 82.Kim AH, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Bustos C, et al. Tissue-specific variation in DNA methylation levels along human chromosome 1. Epigenetics Chromatin. 2009;2:7. doi: 10.1186/1756-8935-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 89.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 90.Jost JP, et al. 5-Methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res. 2001;29:4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kangaspeska S, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 92.Metivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 93.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allis CD, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- *96.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- *97.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. Two insightful reviews on the functional significance of individual or patterns of histone modifications with a debate on an updated histone code hypothesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 99.Barski A, et al. High–resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 100.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]