Abstract
d-Aspartate (d-Asp) is found in specific neurons, transported to neuronal terminals and released in a stimulation-dependent manner. Because d-Asp formation is not well understood, determining its function has proved challenging. Significant levels of d-Asp are present in the cerebral ganglion of the F- and C-clusters of the invertebrate Aplysia californica, and d-Asp appears to be involved in cell-cell communication in this system. Here, we describe a novel protein, DAR1, from A. californica that can convert aspartate and serine to their other chiral form in a pyridoxal 5′-phosphate (PLP)-dependent manner. DAR1 has a predicted length of 325 amino acids and is 55% identical to the bivalve aspartate racemase, EC 5.1.1.13, and 41% identical to the mammalian serine racemase, EC 5.1.1.18. However, it is only 14% identical to the recently reported mammalian aspartate racemase, DR, which is closely related to glutamate-oxaloacetate transaminase, EC 2.6.1.1. Using whole-mount immunohistochemistry staining of the A. californica central nervous system, we localized DAR1-like immunoreactivity to the medial region of the cerebral ganglion where the F- and C-clusters are situated. The biochemical and functional similarities between DAR1 and other animal serine and aspartate racemases make it valuable for examining PLP-dependent racemases, promising to increase our knowledge of enzyme regulation and ultimately, d-serine and d-Asp signaling pathways.
Keywords: Amino Acid, Enzymes, Neurobiology, Neurochemistry, Neurotransmitters, Aplysia, Racemase
Introduction
d-Serine (d-Ser) and d-aspartate (d-Asp) are endogenous molecules with known or putative cell-to-cell signaling activities in the CNS. d-Ser is predominantly found in forebrain structures (1), and acts as a co-agonist of the NMDA receptor, regulating activity at its glycine modulation site (2). Converted from l-serine (l-Ser) by the pyridoxal 5′-phosphate (PLP)2-dependent serine racemase (SerR) in astrocytes (3), d-Ser can be degraded by d-amino acid oxidase (DAO) (4) and acts as a novel glial neurotransmitter/neuromodulator (5, 6). Abnormal levels of SerR and DAO have been observed in schizophrenia patients (7–9), suggesting a connection between d-Ser levels and brain function, including mental illness.
In contrast to d-Ser, d-Asp is widely distributed in animals. It has been found in the brain, retina, and endocrine and exocrine tissues of both vertebrates and invertebrates (10). d-Asp displays a temporal pattern during animal development, beginning with transient high levels between embryonic and early postnatal stages, and rapidly declining to trace levels in most tissues in young adults (4, 11). Interestingly however, as animals mature, d-Asp is found at increasing levels in endocrine tissues (12). The enzyme required for the biosynthesis of d-Asp in the brain was not identified until recently, when the first mammalian aspartate racemase, DR, was cloned and characterized (13), although, how much this enzyme accounts for overall d-Asp synthesis is unclear. While the mechanism by which d-Asp forms remains under investigation, its catabolism is well understood and occurs through the enzyme d-aspartate oxidase (14, 15).
Efforts to understand d-Asp function are also ongoing. Indirect evidence points to hormonal roles (10), whereas other studies have shown its involvement in vision (16), embryonic development (17, 18), learning and memory (19), and neurogenesis (13). Several groups, including ours, have proposed that d-Asp can act as a neurotransmitter (12, 20–22). Further research is required to elucidate the physiological role of this intriguing signaling molecule.
Clearly, many questions remain about d-Asp formation, regulation and ultimately, its function; uncovering the answers will require having knowledge of enzymatic d-Asp biosynthesis. Although the d-Ser biosynthesis pathway was described in 1999 (3), as noted, DR, the first d-Asp racemase enzyme from the animal brain, was just recently reported (13). DR converts l-Asp to d-Asp, and its knockdown reduces newborn neuron survival and dendritic arborization in adult mouse hippocampus. In invertebrates, significant quantities of d-Asp are also found in nervous and endocrine tissues, indicating a potentially conserved d-Asp signaling pathway among Metazoan. However, the origin of the d-Asp in these tissues has not been clarified, nor has a DR-like enzyme been reported in A. californica. So far, the only invertebrate Asp racemase described, SbAspR, was isolated from the foot muscle of the bivalve Scapharca broughtonii (23). But it is unknown if the enzyme is also present in bivalve nervous and endocrine systems. In a prior study (24), we detected high levels of d-Asp (%d/l + d) in the F-, C-, and G-clusters of the cerebral ganglion of the A. californica CNS, with the highest d-Asp content (85%) found in the insulin-producing F-cluster cells. We also observed enzyme activity in the cerebral ganglion that could transfer [14C] from l-Asp to d-Asp in radioisotope pulse-and-chase experiments.
In this report, we describe the cloning of an Asp racemase gene, dar1, from the A. californica CNS, the characterization of a DAR1 protein by enzyme assay, and examination of CNS enzyme localization. Racemase assays were performed using small-volume capillary electrophoresis (CE) with laser-induced fluorescence (LIF) detection, selected because the methodology is well suited for separating chiral amino acids from small-volume samples (25, 26). We characterized the enzyme activity and show that DAR1 converts l-Asp to d-Asp, and vice versa, with a much smaller Km for l-Asp substrate than for d-Asp substrate. The enzyme also exhibits Ser racemase activity with a similar Km for both l-Ser and d-Ser substrates. The distribution of DAR1 in A. californica indicates it is found in tissues previously shown to have significant levels of d-Asp. Based on this dual racemase activity, the same tissues have been characterized for d-Ser, and both d-Ser and d-Asp are shown to co-localize.
EXPERIMENTAL PROCEDURES
Animals
Adult A. californica (125–250 g) were purchased from the Aplysia Research Facility in Miami, FL. Animals were kept in an aquarium of artificial sea water (460 mm NaCl, 10 mm KCl, 10 mm CaCl2, 22 mm MgCl2, 6 mm MgSO4, and 10 mm HEPES, pH 7.8) with constant aeration at 14–15 °C and were used within 2 weeks of arrival. For dissection, animals were anesthetized with an intraperitoneal injection of MgCl2 solution (390 mm) at 30–50% of body weight (v/w), and the interconnected CNS ganglia removed with surgical tools. The ganglia were either immediately processed for RNA isolation, total protein extraction or amino acid extraction; or treated with 1% protease Type IX (Sigma-Aldrich) in artificial sea water containing 100 units/ml penicillin G, 100 μg/ml streptomycin, and 100 μg/ml gentamicin for 1 h at 34 °C to aid in the removal of connective tissues prior to CNS immunohistochemistry staining.
cDNA Cloning of the dar1 Gene
The RNA preparation and DNA cloning reagents were purchased from Invitrogen unless otherwise specified. Chemicals were purchased from Sigma-Aldrich and the DNA oligos from IDT and the W. M. Keck Center for Comparative and Functional Genomics of the University of Illinois at Urbana-Champaign. A. californica genomic searches for potential amino acid racemase genes were conducted at the NCBI website using the SbAspR protein sequence (GenBankTM accession: BAE78960.1) as the search query. The target gene found from the BLAST search was named dar1. The cDNA sequence of the dar1 gene was cloned from A. californica CNS ganglion poly(A)+ RNA templates prepared by TRIzol Reagent and Dynabeads by conventional RT-PCR, 5′-RACE and 3′-RACE cloning techniques using the GeneRacerTM SuperScript III module kit and following product instructions, with the thermocycler details optimized for each cloning reaction. Gene-specific primers (GSPs) for RT and PCR reactions were created from the dar1 gene and high fidelity DNA polymerases were used for the PCR reactions. PCR products of interest were cloned into pCRTM4-TOPO®, transformed into TOP10 cells and sequenced. Three independent clones were obtained which covered the complete cDNA sequence of the dar1 gene. Clone 3 contained almost the complete dar1 protein coding sequence except for the last 15 nucleotides (nt). Clone 23 was produced by 5′-RACE and contained the 5′-UTR and a partial coding sequence overlapping with the 5′-end of clone 3 by 882 nt. Clone 108 was produced by 3′-RACE and contained the 3′-UTR and a partial protein coding sequence overlapping the 3′-end of clone 3 by 282 nt. From the sequence information obtained from these three clones, two GSP primers were designed and used to clone the entire dar1 ORF by standard RT-PCR from the Aplysia CNS ganglion poly(A)+ with Platinum® Pfx DNA Polymerase. The final ORF clone was named T90.
The primers used for the RT and PCR reactions include the following. For clone 3: GeneRacerTM Oligo dT for RT; GSP 5′-GTGACACACTCCAGTGGGAAC-3′ (GSP forward) and 5′-CTCAATGTCCAGATTCCCACC-3′ (GSP reverse). For clone 23: 5′-CTCAATGTCCAGATTCCCACC-3′ (GSP reverse) for RT, and 5′-AGCAGCGGCAACCGACGCCCC-3′ (GSP reverse) and GeneRacerTM 5′ Primer (forward). For clone 108: GeneRacerTM Oligo dT for RT; 5′-ATGGCAGCTTCGTGTGGAGTAACTT-3′ (GSP forward) and GeneRacerTM 3′ Primer (reverse) for primary PCR; 5′-CCGGCCCAGTACCTGGACACC-3′ (GSP forward) and GeneRacerTM 3′ Nested Primer (reverse) for nested PCR. For T90 cloning: GeneRacerTM Oligo dT for RT; forward GSP primer 5′-GGCATATGGCAGCTTCGTGTGGA-3′ (the Ndel site is underlined) and reverse GSP primer 5′-CCCTCGAGTCAAAAAGGCAAATTCTCAATGTC-3′ (the Xhol site is underlined) for full-length ORF PCR.
Construction of the dar1 Protein Expression Vector
The clone T90 plasmid and pET15b bacterial expression vector (EMD Chemicals) were double digested with NdeI and XhoI restriction enzymes (New England BioLabs, Inc.). The dar1 fragment and cleaved vector were gel-purified, ligated, and transformed into TOP10 cells. Transformants were mini-screened and sequenced. A sequence-confirmed clone was transformed into BL21 (DE3) bacterial cells (EMD Chemicals) to generate the bacterial clone 15bT90-31.
DAR1 Expression and Purification
Protein expression and purification reagents were purchased from EMD Chemicals unless specified otherwise. Chemicals were purchased from Sigma-Aldrich. 15bT90-31 bacteria were grown in 250 ml of Luria-Bertani broth supplemented with 100 μg/ml ampicillin at 37 °C until the optical density 600 reached 0.4–0.5. The dar1 gene expression was then induced by addition of 0.5 mm of isopropyl β-d-1-thiogalactopyranoside (IPTG) and the culture continued at 17 °C for 16 h. Soluble His tag protein purification from the bacterial cells was performed by following the BugBuster Mix lysis and nickel-nitrilotriacetic acid (NiNTA) purification product instructions with some modifications. All purification procedures were performed at 4 °C. Briefly, bacterial cells were lysed in the presence of 1× protease inhibitor mixture and DTT. Cleared lysate was incubated with bulk NiNTA resin for 1 h. The resin was then packed into a 24-ml Econo column (Bio-Rad Laboratories) by gravity. The column was washed and protein eluted with a NiNTA wash and elution buffer. The eluted protein was concentrated down to 2 ml and subjected to gel filtration chromatography with a HiPrep Sepharcryl 200 HR 16/60 column (GE Healthcare Life Sciences) using 50 mm Tris-HCl buffer, pH 8.0, and 2 mm DTT following product instructions. Protein fractions with enzyme activity were concentrated and stored in a solution of 50 mm NaPO4, 150 mm NaCl, pH 8.0, 20% glycerol, and 1 mm DTT. Protein concentration was determined with a BCA kit (ThermoFisher Scientific) and the protein molecular size and purity were estimated by SDS-PAGE with 12% Tris-glycine gel with Coomassie staining. Enzyme activity was determined by serine racemase assay as described below.
Aplysia Tissue Protein Extraction and Western Blot
Four adult animals were anesthetized and tissues (CNS ganglia, liver, buccal muscle, ovotestis, atrial gland) were dissected and pooled. CNS ganglia were desheathed to remove the fibrous connective tissues, as described above, before protein extraction. The desheathed CNS and other tissues were quickly frozen in liquid nitrogen and then homogenized with TissueRuptor in Qproteome protein preparation lysis buffer (Qiagen). Total protein was extracted from the homogenized lysates by following the Qproteome kit instructions. Protein concentration was determined by BCA assay. Proteins were boiled in 1× Lamini buffer containing 10 mm β-mercaptoethanol and loaded to 12% Tris-glycine gel and subjected to reducing SDS-PAGE. Resolved proteins were transferred to HybondTM-P (GE Healthcare) PVDF membrane, blocked with 5% nonfat Carnation milk (Nestlé) in Tris-buffered saline with Tween (50 mm Tris, 150 mm NaCl, pH 7.6, 0.05% Tween 20), stained with anti-DAR1 rabbit serum (Immunological Resource Center, University of Illinois at Urbana-Champaign) and goat anti-rabbit HRP secondary antibody (Jackson ImmunoResearch). Antibody-treated membrane was reacted with ImmobilonTM Western chemiluminescent HRP substrate (Millipore) and exposed to Amersham Biosciences HyperfilmTM (GE Healthcare Life Sciences).
CE-LIF Instrumentation and Sample Analysis
Separations were performed using an automated P/ACE MDQ CE system equipped with LIF detection (Beckman Coulter). The CE system was coupled to an external diode laser 56ICS426 (Melles Griot) emitting 440 ± 8 nm, with the coupling performed via a fiber optic cable (OZ Optics). The laser power was adjusted to 3 milliwatt at the output terminus of the optic cable to avoid damage to the photomultiplier tube (PMT). A bandpass filter of 490 ± 15 nm (Omega Optical), located prior to the PMT, selects the appropriate fluorescence emission band for detection. For separations, uncoated fused-silica capillaries (Polymicro Technologies) were used. Capillaries were injected with 0.1 m NaOH at a pressure of 30 psi for 1 min and allowed to be primed in NaOH for 25 min before their initial use. Samples were introduced into the capillaries by pressure injection of 0.5 psi for 5 s. The separation buffer for the Asp and glutamate (Glu) enantiomers contained 200 mm borate, pH 9.5, with 60 mm sodium deoxycholate (SDC) and 40 mm β-cyclodextrin (β-CD) (27). The Ser enantiomer separation buffer contained 75 mm borate, pH 10.5, with 50 mm SDS and 10 mm γ-cyclodextrin (28). For chiral alanine (Ala) separation, the inlet buffer consisted of 25 mm phosphate, pH 2.2, containing 4% (w/v) sulfated β-CD, and the outlet buffer of 50 mm phosphate, pH 2.2 (29). For Asp, Glu, and Ser chiral separations, capillaries of 75 μm inner diameter (ID) and 360 μm outer diameter (OD) were used. Capillary total length was 80 cm with an effective length of 70 cm to the detection window. Separations were carried out by applying 27 kV of normal polarity. Between two runs, the capillaries were rinsed with methanol (30 psi, 0.5 min), 0.1 m NaOH (30 psi, 1 min), and separation buffer (30 psi, 1 min). For Ala enantiomer separations, capillaries of 50 μm ID and 360 μm OD were used with 60 cm total length and 50 cm effective length. A reverse polarity of 20 kV was applied for separations. After each run, the capillaries were rinsed first with methanol (30 psi, 0.5 min), followed by the inlet separation buffer (30 psi, 1 min). CE-LIF results were displayed as electropherograms, which show analyte peak fluorescence intensities against analyte migration time from the capillary injection site to detection window. For quantitative analysis, the peak areas of amino acid analytes and internal control glycine from the CE electropherograms were obtained using Origin 8 software (Origin Lab Corp.). To obtain analyte concentrations, analyte peak areas were divided by glycine peak areas to generate normalized analyte peak area values, and converted to concentrations using working curves generated with known concentrations under the same experimental conditions.
DAR1 Racemase Assay
Chemicals and reagents for the enzyme assays were of the highest grade available and purchased from Sigma-Aldrich unless specified otherwise. Naphthalene-2,3-dicarboxaldehyde (NDA) was purchased from Invitrogen. Water was deionized and purified using a Barnstead E-PURE purification system (ThermoFisher Scientific) and sterilized by passing through 0.22 μm filters. Procedures were performed at room temperature unless otherwise noted. The racemase assays contained a cofactor mix composed of 20 μm PLP, 4 mm ATP, 2 mm DTT, and 2 mm MgCl2 in Tris-HCl buffer of various concentrations and pH depending on the specific experiment. To perform each assay, l/d-Asp, l/d-Ser, l-Glu, or l-Ala (2.5–200 mm) substrates were incubated with 1–10 μg of purified DAR1 in 100–150 μl Tris-HCl buffer (25–50 mm, pH 7.0–8.5) and cofactor mix at 30 °C for 2–4 h. Negative controls contained heat-inactivated enzyme or no substrates. At the end of the incubation period, glycine (3–5 mm) was added to each reaction and used as an internal control for subsequent sample treatments and analyses. Enzyme was removed by either TCA precipitation or the alternative filtration method described below, before amino acid derivatization for CE analysis. To remove the enzyme, 5% cold TCA was added to the reaction samples and incubated for 10 min on ice followed by centrifugation at 10,000 × g for 10 min. TCA was then removed from the samples by two extractions with equal volume of water/saturated ether. Enzyme can also be removed from an assay sample by centrifugation in a Pierce® Concentrator 7 ml/9K molecular-weight cutoff (ThermoFisher Scientific) for 5 min to collect protein-free filtrate. Protein-free samples were diluted in 50 mm borate buffer, pH 9.4, to appropriate volumes so that amino acids were derivatized with an 8× excess of NDA and 20× excess of potassium cyanide (KCN) in 50 mm borate buffer, pH 9.4, for 45 min. The reaction was stopped by diluting 1:10 with 50 mm borate buffer, pH 8.5, and immediately frozen on dry ice. Samples were then subjected to CE-LIF analysis within 24 h.
Enzyme Kinetics Assay
A series of concentrations (2.5–80 mm) of Ser or Asp substrates were incubated with 1–2 μg of purified enzyme in 100-μl reactions in 50 mm Tris-HCl buffer, pH 8.5, for Ser reactions, or pH 8.0 for Asp reactions, containing cofactor mix at 30 °C for 70–150 min. At the end of the incubations, 5 mm glycine was added to each reaction as an internal control. In addition, l-Ala was added to the Ser samples and l-Glu was added to the Asp samples so that the total amino acid concentrations in all reactions were equal to the highest substrate concentrations. The enzymes were immediately removed by centrifugal filtration as described above. The protein-free samples were then diluted to appropriate concentrations with 50 mm borate buffer, pH 9.4, so that 800 μm of the total amino acids were derivatized with 6.4 mm NDA and 16 mm KCN in a 10-μl reaction for 60 min. The reactions were stopped by 1:10 dilution with 50 mm borate buffer, pH 8.5, and quickly frozen on dry ice until CE-LIF analysis. The quantity of amino acid products was calculated from the linear function produced by a standard amino acid calibration curve by co-assaying standard amino acids without enzyme with samples containing enzyme. The enzyme catalysis velocity is defined as a μmol product formed by 1 mg of enzyme during a 1 h reaction time at 30 °C under the described assay conditions. The inverse enzyme velocities were plotted against inverse substrate concentrations and fitted to a linear model using Microsoft Excel to generate Lineweaver-Burk Plots. The Km and Vmax values were calculated from the inverse x- and y-intercepts of the plots.
Aplysia CNS Whole-mount Immunohistochemistry
A rabbit polyclonal antibody against purified full-length DAR1 was generated by the Immunological Resource Center, University of Illinois at Urbana-Champaign. All animal work was performed in compliance with federal, state and university guidelines for animal care and usage. Normal goat serum and rhodamine Red-X-conjugated goat anti-rabbit IgG (H+L) secondary antibody were purchased from Jackson ImmunoResearch. Paraformaldehyde was purchased from Electron Microscopy Sciences. The periesophageal ring and abdominal ganglia without connective tissue (or desheathed) were stretched onto Sylgard in 35 mm Petri dishes and fixed with paraformaldehyde as described by Llewellyn-Smith et al. (30). The tissues were rinsed with PBS to remove the fixative, blocked overnight with 10% goat serum, then incubated with the rabbit antibody (1:1000 dilution) for 7 days. The samples were washed with 10 mm phosphate buffer, pH 7.5, containing 154 mm NaCl, 1% BSA, 0.25 mm thimerosal, and 2% Triton X-100 for 3 days, incubated with the secondary antibody (1:500 dilution) for 3 days in the dark, and washed with the phosphate buffer without Triton X-100 for 3 days. The post-staining preparation for light microscopy visualization was performed as described by Fujisawa et al. (31). Immunohistochemistry control samples were stained with either primary antibody or secondary antibody alone. The samples were kept in the dark and photographed within a week with a Zeiss Axiovert 200 m inverted research-grade microscope equipped with a Roper Scientific Cascade 512B EMCCD camera and Zeiss Axiovision software. The camera obtained black/white and color fluorescence images with appropriate filters; the software was used to perform Z-stacking, time-lapse and tiling. The rhodamine images were obtained via 560 mm excitation and 620 nm emission.
Detection of d-Asp and d-Ser in Aplysia CNS Ganglion Clusters
A. californica cerebral ganglion clusters were dissected from anesthetized animals and immediately placed into 100–200 μl of 0.1 n HCl. The cells were homogenized manually with a glass homogenizer followed by sonication for 15 min in a waterbath sonicator. The materials were then centrifuged at 10,000 × g for 10 min at 4 °C and the supernatants dried down in a microcentrifuge under vacuum. The dried materials were reconstituted with 10 μl of 100 mm borate buffer, pH 9.4. Samples (0.5 μl) were then derivatized with 1 μl of 20 mm NDA and 1 μl of 20 mm KCN for 1 h. The derivitized samples were diluted 5–20 fold with 75 mm borate buffer, pH 10.5, for Ser separation, or 200 mm borate buffer, pH 9.5, for Asp separation. The diluted samples were injected into capillary by pressure at 0.5 psi for 5 s and separated under 27 kV with normal polarity.
RESULTS
Cloning of the dar1 Full-length cDNA
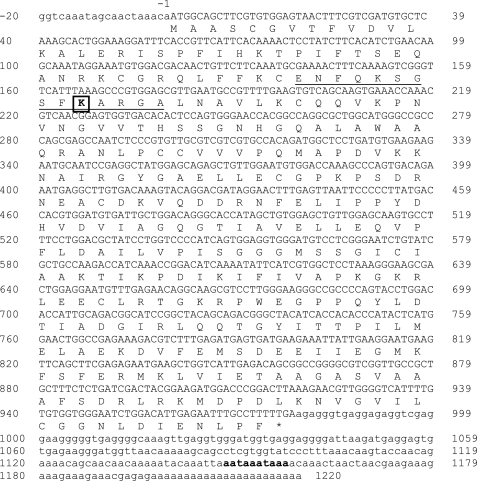
To locate an aspartate racemase in the A. californica nervous system, we searched the online A. californica genome data base and discovered a candidate gene (gb_AASC02056815.1), naming it dar1. We cloned the full-length cDNA of the dar1 gene, including the protein coding sequence (deposited in GenBankTM HM776055), and the 5′-UTR and 3′-UTR from poly(A)+ RNA, by using conventional RT-PCR, 5′-RACE and 3′-RACE cloning techniques. The cDNA sequence and its protein translation are shown in Fig. 1. The cDNA includes a 20 nt 5′-UTR, 978 nt coding sequence, and 241 nt 3′-UTR. The 3′-UTR contains a poly(A) tail of 23 adenines and two overlapping AATAAA polyadenylation signals. In total, the cDNA contains 1220 nt and codes for a 325 amino acid-long protein. At the protein level, DAR1 shares a 55% sequence identity to the bivalve aspartate racemase, and more than 40% to human, rat, and mouse brain serine racemases. A multiple sequence alignment of these five related proteins is shown in supplemental Fig. S1. The proteins have similar lengths, ranging from 325 to 340 amino acids, and share multiple conserved regions, including a putative PLP-binding motif near the N terminus around the DAR1 PLP-binding Lys-56 (32) and the DAR1 PLP-binding stabilizing clusters, 81Ser-Ser-Gly-Asn-His and 183Ser-Gly-Gly-Gly (33).
FIGURE 1.
Complete cDNA sequence and predicted protein sequence of dar1. 5′-UTR: nt −20 to nt −1 in lowercase letters; amino acid coding sequence: uppercase single letters; predicted amino acid sequence: single letter codes in uppercase; *: stop codon TGA; 3′-UTR: nt 979 to nt 1220 in lowercase letters; polyadenylation signals: bolded lowercase letters (aataaa). The underlined region is the predicted PLP-binding domain with a PLP-binding residue Lys-56 (boxed). GenBankTM accession number for the complete CDS: HM776055.
Microbial Expression of the dar1 Coding Sequence and Protein Purification
We cloned the dar1 ORF into a microbial expression vector, over-expressed the gene as a soluble recombinant protein with an N-terminal His6 tag in E. coli, and purified it for functional studies. The recombinant protein was only expressed under ITPG induction and purified from bacterial cell lysate to near homogeneity using NiNTA affinity purification and gel filtration chromatography (see supplemental Fig. S2). The recombinant DAR1 exhibited an electrophoresis mobility of 37 kDa in a reducing SDS-gel, a result consistent with the predicted molecular weight of the recombinant protein, but it had a molecular mass of 57 kDa as determined by a sizing column. The considerably larger molecular mass than molecular weight indicates that DAR1 molecules form homodimers in solution. Dimerization seems to be essential for DAR1 activity because the protein fraction of 30–35 kDa off the sizing column had little enzyme activity (data not shown).
DARI Expression in A. californica Tissues
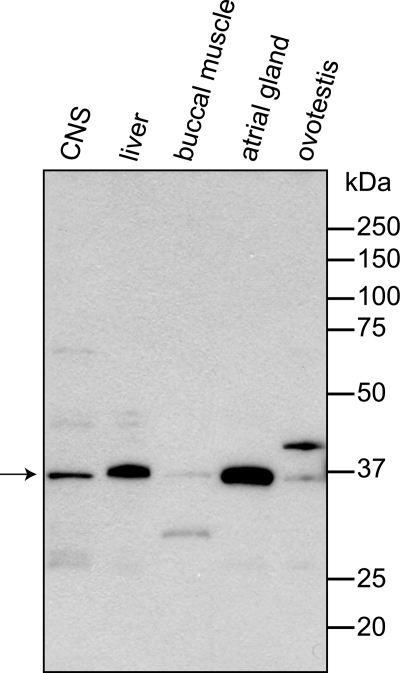
We raised a rabbit polyclonal antibody against purified recombinant DAR1 and examined dar1 expression in A. californica CNS neurons, and liver, buccal muscle, ovotestis, and atrial gland tissues by Western blot. As shown in Fig. 2, dar1 was expressed as a ∼36 kDa protein (predicted molecular mass is 35.4 kDa) in the CNS ganglion lysate. The protein was also strongly expressed in atrial gland, moderately in liver, and minimally in buccal muscle and ovotestis when an equal amount of total protein was loaded for each tissue.
FIGURE 2.
Western analysis of A. californica tissue expression of DAR1. Tissue protein lysates of CNS, liver, buccal muscle, atrial gland, and ovotestis tissues were subjected to reducing SDS-PAGE. Ten micrograms of each sample were co-run with Precision Plus Protein Standards in 12% Tris-glycine gel. The transfer blot was stained with rabbit anti-DAR1 serum (1:30,000) and goat anti-rabbit HRP conjugate (1:20,000). Film was exposed for 5 min. The predicted native DAR1 protein is 35.4 kDa.
DAR1 Racemase Activity and Characterization
We examined DAR1 PLP-dependent racemase activity toward l/d-Asp, l/d-Ser, l-Glu, and l-Ala substrates, and observed enzyme behaviors under different temperatures and buffer pH levels, and the effects of ATP/MgCl2 on the enzyme activity. These measurements were accomplished with CE-LIF, an approach well suited for chiral amino acid separation and detection. Our assay results demonstrated that DAR1 was active toward both Asp and Ser substrates and converted these chiral enantiomers in both directions (Fig. 3). However, DAR1 had no detectable activity when 10 μg of purified enzyme was incubated with 100 mm l-Glu or 100 mm l-Ala (d-Glu and d-Ala were not tested) in a 150 μl reaction containing 50 mm Tris-HCl, pH 8.0, with cofactors at 30 °C for 3 h, a condition where Asp and Ser conversion were easily observed.
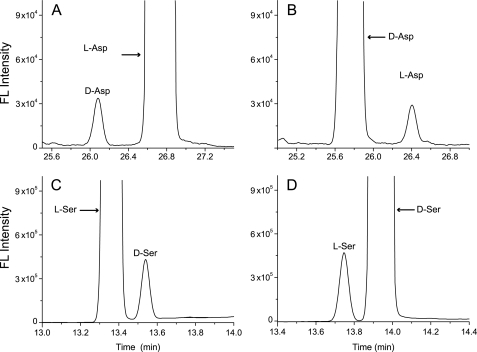
FIGURE 3.
DAR1 Asp and Ser racemase activities. CE-LIF electropherograms of enzyme reactions with four substrates (A) l-Asp, (B) d-Asp, (C) l-Ser, and (D) d-Ser. l-Asp/d-Asp (80 mm), or l-Ser/d-Ser (10 mm), were incubated with 1 μg of enzyme and cofactors in a 150-μl reaction buffer of 50 mm Tris-HCl, pH 8.0 (Asp reactions), or in 25 mm Tris-HCl, pH 9.0 (Ser reactions), at 30 °C for 2.5 h. A substrate starting concentration of 12 μm l-Asp, 4 μm d-Asp, 20 μm l-Ser, or 20 μm d-Ser was injected into the capillary. Migration time: time for analytes moving from the capillary injection site to detection window.
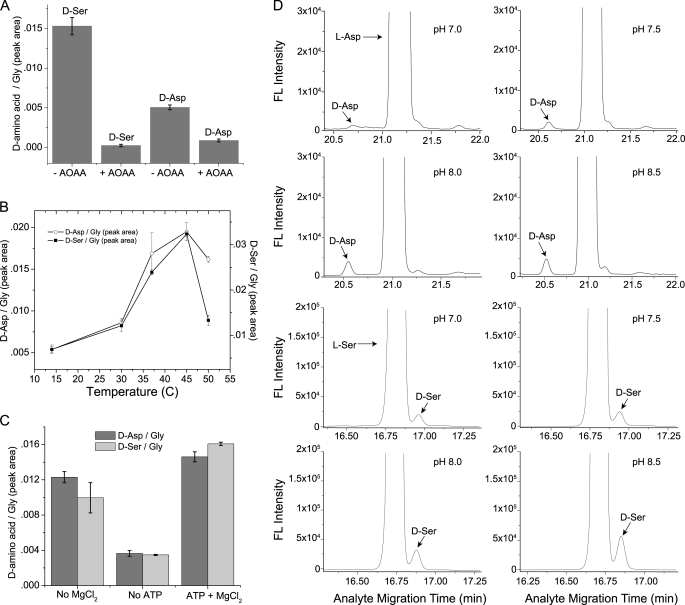
DAR1, like other PLP-dependent racemases, requires the cofactor PLP for its activity. Purified enzyme appeared greenish yellow and absorbed light at 420 nm, a characteristic of PLP-bound enzyme (33). The absorption peak disappeared after the enzyme was incubated for 10 min at room temperature with 2 μm aminooxyacetic acid (AOAA), a PLP-inactivating reagent (data not shown). The enzyme did not lose color after a 48 h dialysis, indicating a high affinity between the enzyme and PLP. AOAA inhibited the conversion by DAR1 of d-Ser and d-Asp from their corresponding enantiomer (Fig. 4A). The thermal stability profile of DAR1 is shown in Fig. 4B. The enzyme activity increased between 14–45 °C and started to decrease at 50 °C for both the Ser and Asp conversions. We examined the modulating effects of ATP and MgCl2 on DAR1 racemase activity (Fig. 4C). MgCl2 (2 mm) had only a small effect on enzyme racemase activity but ATP (4 mm) significantly increased enzyme activity. When both effectors were present, the enzyme showed the largest activity for the d-Asp and d-Ser conversions. The ATP and MgCl2 concentrations were not optimized for DAR1; we chose the concentrations based on published research on Ser and Asp racemases (34, 35). Finally, Fig. 4D depicts the pH-dependent DAR1 activity. The enzyme showed little racemase activity below pH 6.5 (data not shown) and an increasing activity between pH 7.0–8.5 in 50 mm Tris-HCl buffer.
FIGURE 4.
DAR1 racemase activity characterization. A, AOAA inhibition assay: l-Asp (50 mm) and l-Ser (20 mm) were each incubated with 1 mm AOAA, 4 μg of DAR1 and cofactors in a 150-μl reaction containing 25 mm Tris-HCl, pH 8.5, at 30 °C for 2 h. The substrate starting concentration of 100 μm l-Asp or 40 μm l-Ser was injected into the CE capillary. B, thermal stability of DAR1 racemase activity: l-Asp (80 mm) and l-Ser (50 mm) were each incubated with 1 μg of DAR1 and cofactors in a 100-μl reaction in 50 mm Tris-HCl, pH 8.5, at 14, 30, 37, 45, and 50 °C for 3 h (Asp) or 1 h (Ser). The substrate starting concentration (100 μm for l-Ser or 160 μm for l-Asp) was injected into the capillary. C, effects of ATP and MgCl2 on DAR1 racemase activity: l-Ser (20 mm) and l-Asp (200 mm) were each incubated with 2 μg each of DAR1, PLP, and DTT with either 4 mm ATP or 2 mm MgCl2 or both effectors, in a 100-μl reaction in 50 mm Tris-HCl, pH 8.5, at 30 °C for 1 h. The substrate starting concentration (40 μm l-Ser or 100 μm l-Asp) was injected into the capillary. Controls with heat-inactivated enzymes were subtracted from the data shown. For panels A–C, the data from three batches of enzymes were averaged and the standard deviations represented by error bars. D, pH dependence of DAR1 racemase activity: CE-LIF electropherograms of a DAR1 racemase reaction with l-Asp (top 4 panels) or l-Ser (lower 4 panels) substrate. l-Ser (13.3 mm) and l-Asp (26.7 mm) were each incubated with 1 μg of DAR1 and cofactors in a 150-μl reaction in 50 mm Tris-HCl, at pH 7, 7.5, 8.0, and 8.5 at 30 °C for 3 h. The substrate starting concentration of 1.77 μm l-Ser or 17.7 μm l-Asp was injected into the capillary.
Enzyme Kinetics
The DAR1 Asp and Ser racemase kinetics linear curves are shown in Lineweaver-Burk Plots (supplemental Fig. S3). Analyzing the linear fits, the Km (mm) was determined to be 8 ± 2 (S.D.) for l-Asp, 94 ± 30 for d-Asp, and 15 ± 4 and 16 ± 2 for l-Ser and d-Ser, respectively; the Vmax (μmol h−1 mg−1) for the same substrate order was determined to be 3.3 ± 0.8, 7.9 ± 1, 11 ± 1.6, and 15 ± 3, respectively.
Co-localization of DAR1 and d-Amino Acids in the A. californica CNS
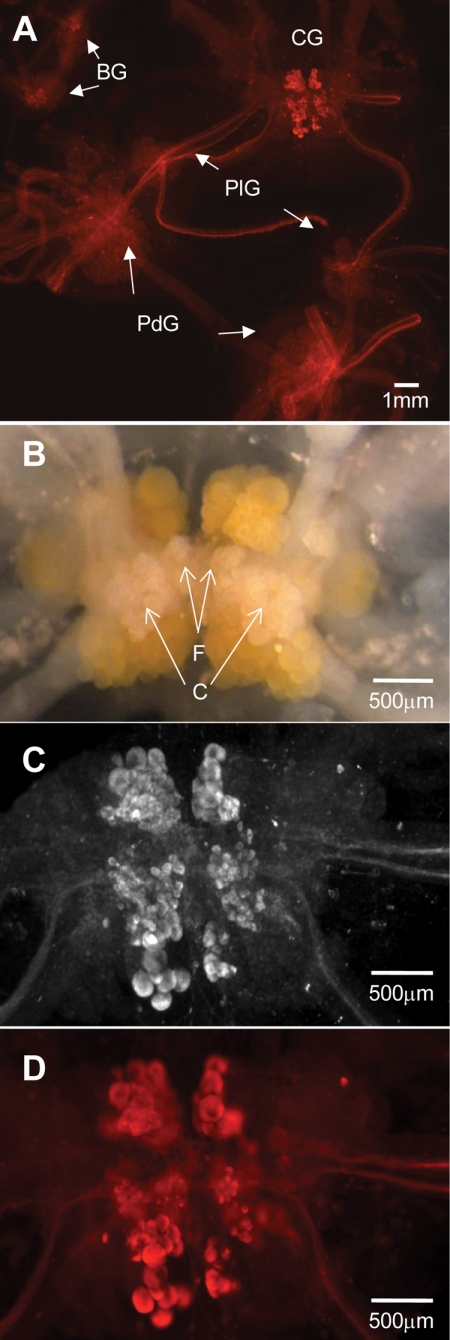
We raised a rabbit polyclonal antibody against DAR1 and used it to perform whole-mount (versus section) immunohistochemistry with A. californica CNS material. We stained the isolated CNS, including the buccal, pleural, pedal, abdominal, and cerebral ganglia, and their connective and peripherally extending nerves. The immunohistochemistry experiments revealed a highly localized distribution of DAR1 protein in the CNS (Fig. 5). The antibody exclusively stained F-, C-, G-, and B-cluster cells located in the middle region of the fused cerebral ganglia. The staining was specific; controls without primary or secondary antibody produced only background signal (data not shown). The other ganglia showed only background staining, except for a group of 3–4 unidentifiable neurons in the vicinity of B1 and B2 landmark neurons in the buccal hemiganglia and a lone neuron in the abdominal hemiganglia.
FIGURE 5.
DAR1-like immunoreactivity localization in the A. californica CNS. A, whole-mount staining of the CNS (except abdominal ganglia) with rabbit anti-DAR1 serum shown as a rhodamine fluorescence micrograph; BG: buccal ganglia; CG: cerebral ganglia; PdG: pedal ganglia; PlG: pleural ganglia. B, light micrograph of desheathed but not stained CG control; enlarged CG staining image from (A) showing (C) a rhodamine fluorescence micrograph and (D) a bright-field fluorescence micrograph.
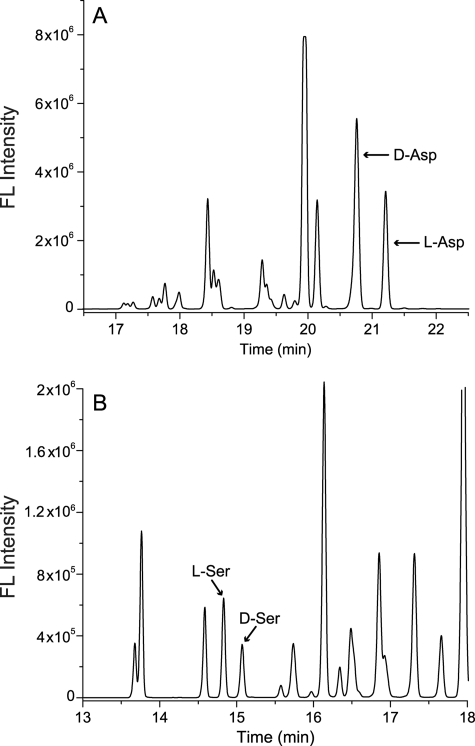
Whereas our prior research characterized d-Asp, we had not performed d-Ser separations; the optimum CE conditions are distinct for the two amino acid pairs. Thus, we have re-measured both d/l-Asp and d/l-Ser within several CNS structures. As demonstrated in the CE-LIF electropherograms in Fig. 6, not only was a high level of d-Asp (more than l-Asp) once again detected, a significant amount of d-Ser (less than l-Ser) was also found for the first time in the F/C-clusters.
FIGURE 6.
CE-LIF electropherograms of d-Asp and d-Ser in CNS neuronal F/C clusters. A. californica cerebral ganglion F/C cluster neurons were dissected and subjected to amino acid separation with CE-LIF using optimal buffer for (A) Asp separation and (B) Ser separation.
DISCUSSION
We have characterized a novel racemase enzyme from A. californica. Homology data suggest that the known mammalian and our new molluskan amino acid racemase share a conserved structural similarity. Intriguingly, although a similar enzyme has not been found in mammals, a distinct aspartate racemase (DR) was recently identified (13). DR has less than 20% identity to the other five racemase proteins, and it lacks the similar PLP binding and stabilizing sequences shared by other racemase groups. Based on our phylogenetic analysis, Ser racemases are closer to serine dehydratase (EC 4.3.1.17), while DR is closer to glutamate-oxaloacetate transaminase enzyme (EC 2.6.1.1). It appears that DAR1 and DR are distinct Asp racemases from animal brains. Intriguingly, DAR1 and mammalian SerR are closely related and yet the mammalian DR is distinct. Are there other animal amino acid racemases that are still unknown? Given recent reports of high levels of d-Glu (36) and d-Ala (37) in rat brain, this appears a certainty. It will be interesting to determine their structures and how they fit within the growing Metazoan CNS amino acid racemase family.
After expressing and purifying DAR1, we tested its activity and ability for racemizing the amino acids that have previously been shown to be present at high levels in animal brains: Asp, Ser, Glu, and Ala (38, 39). The enzyme was active toward Ser and Asp enantiomers but showed no activity toward Glu and Ala. To our best knowledge, DAR1 is the first characterized eukaryotic racemase that can catalyze the racemization of two substrates. The enzyme is PLP-dependent and appears to require dimerization for activity. Homodimers have been observed with mouse SerR and SbAspR (32, 34, 40). The recombinant DAR1 appears to be heat-tolerant under the assay conditions used. As with most protein enzymes, its activity increases with increasing temperatures. The enzyme shows a wide temperature range of activity, between 14 and 45 °C.
We examined ATP and MgCl2 effects on DAR1 racemase activity and found that both reagents, especially ATP, are important for promoting enzyme racemase activity, and they act more effectively together than they do alone, as shown in Fig. 4C. It has been proposed that ATP and MgCl2 exert allosteric regulatory effects on mouse SerR by lowering enzyme Km (34). Although not examined, the same regulatory mechanisms might be at work for DAR1. Studies by other groups have also shown that other nucleotides and bivalent cations could regulate animal Asp and Ser racemase activities. As examples, besides Mg2+, both Ca2+ and Mn2+ have been shown to promote mouse SerR activity (34) and AMP promoted SbAspR activity, but ATP reduced it (35). It is interesting that while ATP has a positive effect on DAR1 activity, the same nucleotide showed an inhibitory effect on SbAspR activity. The opposite effects of ATP on the two closely related mollusk aspartate racemases indicate that the enzymes may have different physiological functions. In addition, SbAspR might have a role in energy metabolism under anoxic conditions in bivalve muscle tissues (35).
The enzyme is more active under alkaline conditions. Both Ser and Asp activities were low below pH 6.5, while increasing between pH 7.0 and pH 8.5 in 50 mm Tris-HCl. The pH profile is similar to that of a mouse SerR (3) and distinct from that reported for SbAspR (23), where the enzyme activity started to decrease above pH 8.5. We found that the optimal buffers were 25 mm Tris-HCl, pH 8.5, for l-Asp conversion and 50 mm Tris-HCl, pH 9.0, for l-Ser conversion (data not shown), suggesting that ion strength and pH can affect enzyme efficiency for a particular substrate. We chose 50 mm Tris-HCl, pH 8.5, for most of our assays to simplify the experimental procedures.
We determined the Km and Vmax for the four racemase reactions catalyzed by DAR1 (supplemental Fig. S3). The kinetic data revealed the following. First, the enzyme favored l-Asp over d-Asp, the smaller the Km, the higher affinity a substrate has for an enzyme, and the Km for l-Asp substrate was about 10× smaller than that for d-Asp substrate. Although the enzyme had twice as large a Vmax for l-Asp production, l-Asp is 10× more competitive than d-Asp for binding to the enzyme. Therefore, the enzyme was more efficient in producing d-Asp from l-Asp than vice versa. Differing from the Asp kinetics, Ser kinetics are comparable with both substrates. But the Km values for both forms of Ser were still 4× larger than that for l-Asp. Based on the Km values, the enzyme had the highest affinity toward l-Asp, followed by l-Ser/d-Ser, and then d-Asp. However, more studies are required to understand the physiological meaning of these kinetics parameters.
The selected presence and relatively sparse distribution of DAR1 within the A. californica CNS, specifically several cerebral ganglion clusters and isolated single neurons in the buccal and abdominal ganglia, suggests that the enzyme serves a specific function within these areas (and does not have a more general “cellular housekeeping” role). At this point, we have determined that DAR1 is capable of synthesizing d-Asp and it is localized to specific structures. Using CE-LIF we measured the presence of d-Ser and d-Asp in the cells containing the enzyme to determine if they co-localize. It is particularly telling that the enzyme was present in the F-, C-, and G-clusters where high levels of d-Asp content were previously reported (21, 24); we now confirm the co-localization of d-Ser and d-Asp in these structures. The correlation of DAR1 and its products supports our determination that the enzyme is involved in the actual biosynthesis of d-Asp and d-Ser. Several years ago, we hypothesized that d-Asp was a neurotransmitter in the A. californica CNS (24). Since then, we have accumulated evidence of d-Asp release from and uptake by the cerebral ganglia. The finding of a d-Asp biosynthesizing enzyme in the cerebral ganglia substantiates this hypothesis.
CONCLUSIONS
We have isolated a novel racemase from A. californica neuronal tissues, characterized its biochemical features, and validated its ability to form d-Asp and d-Ser. DAR1 can catalyze racemization reactions between Asp and Ser enantiomers with differing kinetics. The enzyme Km and Vmax values suggest a substrate preference order of l-Asp > l-Ser ≥ d-Ser > d-Asp. DAR1 is the first Asp and Ser racemase discovered in an invertebrate brain and the first eukaryotic dual Asp/Ser racemase described. Using whole-mount CNS tissue staining with DAR1 antibody, we localized DAR1 to the central region of the cerebral ganglia where the F- and C-clusters are situated. These clusters contained a high level of d-Asp and a significant amount of d-Ser. The co-localization of the enzyme with the four amino acids demonstrates that DAR1 plays a role in the biosynthesis of d-Asp and/or d-Ser in the F/C-clusters. Future work will examine the roles of d-Asp and d-Ser in cell-to-cell communication and study the interactions between these two putative signaling molecules.
Acknowledgments
We thank Dr. Gary Olsen (UIUC) for bioinformatics consultation, Peter Yau (UIUC) for protein purification advice, and Dr. Leonid Moroz (University of Florida) for discussions concerning the nascent A. californica genome, the University of Illinois Roy J. Carver Biotechnology Center for oligo synthesis, DNA sequencing, and antibody production, and the University of Illinois Beckman Institute Imaging Technology Group for fluorescence imaging assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS, National Institutes of Health or NSF.
This work was supported, in whole or in part, by Award No. NS031609 from the NINDS, National Institutes of Health, and by Award Nos. CHE-04-00768 and CHE-05-26692 from the National Science Foundation (NSF).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- PLP
- pyridoxal-5′-phosphate
- CE
- capillary electrophoresis
- CE-LIF
- capillary electrophoresis laser-induced fluorescence detection
- DAO
- d-amino acid oxidase
- DAR1
- A. californica d-amino acid racemase 1
- DR
- mammalian aspartate racemase
- GSP
- gene-specific primer
- ID
- inner diameter
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- KCN
- potassium cyanide
- NiNTA
- nickel-nitrilotriacetic acid
- nt
- nucleotide
- OD
- outer diameter
- PMT
- photomultiplier tube
- SerR
- serine racemase
- SDC
- sodium deoxycholate.
REFERENCES
- 1. Schell M. J., Molliver M. E., Snyder S. H. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3948–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolosker H., Dumin E., Balan L., Foltyn V. N. (2008) FEBS J. 275, 3514–3526 [DOI] [PubMed] [Google Scholar]
- 3. Wolosker H., Blackshaw S., Snyder S. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13409–13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hashimoto A., Nishikawa T., Konno R., Niwa A., Yasumura Y., Oka T., Takahashi K. (1993) Neurosci. Lett. 152, 33–36 [DOI] [PubMed] [Google Scholar]
- 5. Mothet J. P., Parent A. T., Wolosker H., Brady R. O., Jr., Linden D. J., Ferris C. D., Rogawski M. A., Snyder S. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4926–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Panatier A., Theodosis D. T., Mothet J. P., Touquet B., Pollegioni L., Poulain D. A., Oliet S. H. (2006) Cell 125, 775–784 [DOI] [PubMed] [Google Scholar]
- 7. Habl G., Zink M., Petroianu G., Bauer M., Schneider-Axmann T., von Wilmsdorff M., Falkai P., Henn F. A., Schmitt A. (2009) J. Neural. Transm. 116, 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Labrie V., Fukumura R., Rastogi A., Fick L. J., Wang W., Boutros P. C., Kennedy J. L., Semeralul M. O., Lee F. H., Baker G. B., Belsham D. D., Barger S. W., Gondo Y., Wong A. H., Roder J. C. (2009) Hum. Mol. Genet. 18, 3227–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verrall L., Walker M., Rawlings N., Benzel I., Kew J. N., Harrison P. J., Burnet P. W. (2007) Eur. J. Neurosci. 26, 1657–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Aniello A. (2007) Brain Res. Rev. 53, 215–234 [DOI] [PubMed] [Google Scholar]
- 11. Neidle A., Dunlop D. S. (1990) Life Sci. 46, 1517–1522 [DOI] [PubMed] [Google Scholar]
- 12. Errico F., Napolitano F., Nisticò R., Centonze D., Usiello A. (2009) Rev. Neurosci. 20, 429–440 [DOI] [PubMed] [Google Scholar]
- 13. Kim P. M., Duan X., Huang A. S., Liu C. Y., Ming G. L., Song H., Snyder S. H. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3175–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schell M. J., Cooper O. B., Snyder S. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaar K., Köst H. P., Schad A., Völkl A., Baumgart E., Fahimi H. D. (2002) J. Comp. Neurol. 450, 272–282 [DOI] [PubMed] [Google Scholar]
- 16. D'Aniello S., Spinelli P., Ferrandino G., Peterson K., Tsesarskia M., Fisher G., D'Aniello A. (2005) Biochem. J. 386, 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hashimoto A., Oka T. (1997) Prog. Neurobiol. 52, 325–353 [DOI] [PubMed] [Google Scholar]
- 18. Sakai K., Homma H., Lee J. A., Fukushima T., Santa T., Tashiro K., Iwatsubo T., Imai K. (1998) Brain Res. Rev. 808, 65–71 [DOI] [PubMed] [Google Scholar]
- 19. Topo E., Soricelli A., Di Maio A., D'Aniello E., Di Fiore M. M., D'Aniello A. (2010) Amino Acids 38, 1561–1569 [DOI] [PubMed] [Google Scholar]
- 20. Miao H., Rubakhin S. S., Sweedler J. V. (2006) J. Chromatogr. A 1106, 56–60 [DOI] [PubMed] [Google Scholar]
- 21. Scanlan C., Shi T., Hatcher N. G., Rubakhin S. S., Sweedler J. V. (2010) J. Neurochem. 115, 1234–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spinelli P., Brown E. R., Ferrandino G., Branno M., Montarolo P. G., D'Aniello E., Rastogi R. K., D'Aniello B., Baccari G. C., Fisher G., D'Aniello A. (2006) J. Cell Physiol. 206, 672–681 [DOI] [PubMed] [Google Scholar]
- 23. Shibata K., Watanabe T., Yoshikawa H., Abe K., Takahashi S., Kera Y., Yamada R. H. (2003) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 134, 307–314 [DOI] [PubMed] [Google Scholar]
- 24. Miao H., Rubakhin S. S., Scanlan C. R., Wang L., Sweedler J. V. (2006) J. Neurochem. 97, 595–606 [DOI] [PubMed] [Google Scholar]
- 25. Lapainis T., Sweedler J. V. (2008) J. Chromatogr. A 1184, 144–158 [DOI] [PubMed] [Google Scholar]
- 26. Páez X., Hernández L. (2001) Biopharm. Drug Dispos. 22, 273–289 [DOI] [PubMed] [Google Scholar]
- 27. Ueda T., Mitchella R., Kitamura F., Metcalfa T., Kuwanab T., Nakamoto A. (1992) J. Chromatogr. A 593, 265–274 [Google Scholar]
- 28. Zhao S., Song Y., Liu Y. M. (2005) Talanta 67, 212–216 [DOI] [PubMed] [Google Scholar]
- 29. Kirschner D. L., Jaramillo M., Green T. K. (2007) Anal. Chem. 79, 736–743 [DOI] [PubMed] [Google Scholar]
- 30. Llewellyn-Smith I. J., Costa M., Furness J. B. (1985) J. Histochem. Cytochem. 33, 857–866 [DOI] [PubMed] [Google Scholar]
- 31. Fujisawa Y., Furukawa Y., Ohta S., Ellis T. A., Dembrow N. C., Li L., Floyd P. D., Sweedler J. V., Minakata H., Nakamaru K., Morishita F., Matsushima O., Weiss K. R., Vilim F. S. (1999) J. Neurosci. 19, 9618–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abe K., Takahashi S., Muroki Y., Kera Y., Yamada R. H. (2006) J. Biochem. 139, 235–244 [DOI] [PubMed] [Google Scholar]
- 33. Yoshimura T., Goto M. (2008) FEBS J. 275, 3527–3537 [DOI] [PubMed] [Google Scholar]
- 34. Neidle A., Dunlop D. S. (2002) Neurochem. Res. 27, 1719–1724 [DOI] [PubMed] [Google Scholar]
- 35. Shibata K., Watanabe T., Yoshikawa H., Abe K., Takahashi S., Kera Y., Yamada R. H. (2003) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 134, 713–719 [DOI] [PubMed] [Google Scholar]
- 36. Mangas A., Coveñas R., Bodet D., Geffard M., Aguilar L. A., Yajeya J. (2007) Neuroscience 144, 654–664 [DOI] [PubMed] [Google Scholar]
- 37. Errico F., Rossi S., Napolitano F., Catuogno V., Topo E., Fisone G., D'Aniello A., Centonze D., Usiello A. (2008) J. Neurosci. 28, 10404–10414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morikawa A., Hamase K., Zaitsu K. (2003) Anal. Biochem. 312, 66–72 [DOI] [PubMed] [Google Scholar]
- 39. Quan Z., Liu Y. M. (2003) Electrophoresis 24, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 40. Yamashita T., Ashiuchi M., Ohnishi K., Kato S., Nagata S., Misono H. (2004) Eur. J. Neurosci. 271, 4798–4803 [DOI] [PubMed] [Google Scholar]