Abstract
Translesion synthesis (TLS) is a DNA damage tolerance mechanism in which DNA lesions are bypassed by specific polymerases. To investigate the role of TLS activities in ultraviolet light-induced somatic mutations, we analyzed Arabidopsis (Arabidopsis thaliana) disruptants of AtREV3, AtREV1, and/or AtPOLH genes that encode TLS-type polymerases. The mutation frequency in rev3-1 or rev1-1 mutants decreased compared with that in the wild type, suggesting that AtPolζ and AtRev1 perform mutagenic bypass events, whereas the mutation frequency in the polh-1 mutant increased, suggesting that AtPolη performs nonmutagenic bypass events with respect to ultraviolet light-induced lesions. The rev3-1 rev1-1 double mutant showed almost the same mutation frequency as the rev1-1 single mutant. The increased mutation frequency found in polh-1 was completely suppressed in the rev3-1 polh-1 double mutant, indicating that AtPolζ is responsible for the increased mutations found in polh-1. In summary, these results suggest that AtPolζ and AtRev1 are involved in the same (error-prone) TLS pathway that is independent from the other (error-free) TLS pathway mediated by AtPolη.
Plants are continuously exposed to various environmental stresses including UV light and other DNA-damaging agents. UV light induces DNA lesions such as the formation of cyclobutane pyrimidine dimers and pyrimidine (6-4) pyrimidone photoproducts that block DNA replication and transcription (Umlas et al., 1985). To remove the DNA lesions, plants possess various DNA repair mechanisms such as photorepair, excision repair, and recombination repair. Some lesions, however, are not removed by the repair processes (Hidema et al., 1999) and therefore can endanger faithful DNA replication.
Translesion synthesis (TLS) is an important damage tolerance mechanism in which damaged DNA is bypassed by the action of specific polymerases, thus avoiding potential replication arrest (Friedberg et al., 2005). Many organisms ranging from bacteria to humans possess TLS activities (Woodgate, 1999; Baynton and Fuchs, 2000), and loss of TLS often results in survival rate reduction and increased sensitivity to DNA-damaging agents (Lawrence, 2004; Waters et al., 2009). DNA synthesis during TLS often causes mutations due to the low fidelity of replication that occurs when the DNA lesions are bypassed (Friedberg et al., 2005).
The specific polymerases involved in TLS are conserved among species (Waters et al., 2009). For example, DNA Polymerase ζ (Polζ), Rev1, and Polη from yeast and mammals have been well characterized (Waters et al., 2009) and show high conservation. Genes for REV3 and REV7, the subunits of Polζ, were originally isolated from yeast using a screening process based on the reduction of UV light-induced reversions (Lemontt, 1971; Lawrence et al., 1985). Polζ belongs to the B-family of DNA polymerases, possessing mismatch-extension activity (Nelson et al., 1996; Johnson et al., 2000a). Rev1 was also isolated from a yeast reversionless mutant (Lawrence and Christensen, 1976) and possesses deoxycitidyl transferase activity (Nelson et al., 1996). Based on several lines of evidence, Polζ and Rev1 are thought to promote damage-induced mutations (Kunz et al., 2000). On the other hand, Polη was originally identified in yeast from its homology with the Escherichia coli DinB protein (McDonald et al., 1997). Lack of Polη in humans causes xeroderma pigmentosum variant (XPV), a disease characterized by high susceptibility to sunlight-induced cancer (Kraemer, 2003). Therefore, Polη is thought to possess a function that involves the prevention of UV light-induced mutations in humans (Pagès and Fuchs, 2002).
In Arabidopsis (Arabidopsis thaliana), we and other groups have isolated several genes encoding TLS-type polymerase homologs: AtREV3, AtREV7, AtREV1, AtPOLH, and AtPOLK (Sakamoto et al., 2003; García-Ortiz et al., 2004; Takahashi et al., 2005; Santiago et al., 2006). AtREV3 and AtREV7 encode a catalytic subunit and a regulatory subunit of AtPolζ, respectively. AtRev1, encoded by the AtREV1 gene, possesses deoxynucleotidyl transferase activity with low fidelity and inserts a nucleotide opposite apurinic/apyrimidinic sites in vitro (Takahashi et al., 2007). AtPolη, encoded by AtPOLH, bypasses cyclobutane pyrimidine dimers with comparable activity to human or yeast Polη (Anderson et al., 2008; Hoffman et al., 2008). AtREV3-, AtREV7-, AtREV1-, or AtPOLH-disrupted plants are more sensitive to UV light exposure compared with wild-type plants, although the levels of sensitivity differ (Sakamoto et al., 2003; Takahashi et al., 2005; Curtis and Hays, 2007; Anderson et al., 2008), suggesting that these genes play a role in conferring tolerance to UV light-induced damage.
In this study, we analyzed UV light-induced somatic mutation frequencies in disruptants of the AtREV3, AtREV1, and AtPOLH genes. Point-mutated, and thus inactivated, uidA genes were employed to score UV light-induced mutations. Nucleotide substitutions that may occur by error during TLS restore the uidA genes, whose activity can be detected by examination of the blue sectors following GUS staining. We demonstrate that the mutation frequency in the disruptants was significantly altered compared with that of the wild type. This result suggests that AtPolζ, AtRev1, and AtPolη, encoded by AtREV3, AtREV1, and AtPOLH, respectively, are involved in TLS processes related to UV light mutagenesis.
RESULTS
Detection of Somatic Mutations in Arabidopsis
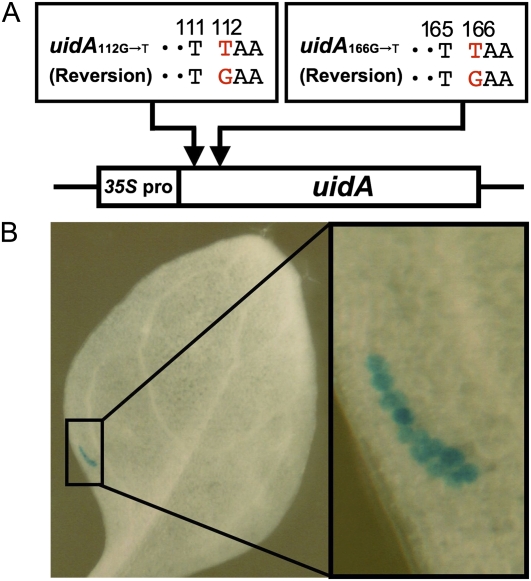
To examine whether TLS pathways are employed in UV light resistance in higher plants, we analyzed the mutation frequency in Arabidopsis somatic tissues following UV-C irradiation. The reporter genes used for this analysis comprised uidA genes containing a nonsense mutation, uidA112G-T or uidA166G-T, generated by replacing the 112th or 166th guanine with thymine, respectively (Fig. 1A; Kovalchuk et al., 2000). The substituted thymine and the flanking 111th or 165th thymine generate a potential thymine-thymine (TT) dimer target. The reporter gene will become active when a T-to-G reversion occurs at the 112th or 166th thymine position. Transgenic plants carrying the reporter gene were treated with or without UV-C light and then grown for an additional 10 d, which allows cells with an active uidA gene to proliferate and produce a detectable blue sector on somatic tissues (Fig. 1B). Therefore, the number of blue sectors present is indicative of the number of reversions that have actually taken place (Kovalchuk et al., 2000).
Figure 1.
Detection of reversion using nonsense codon-introduced uidA genes. A, Design of reporter genes used for the detection of base substitutions. Two reporter genes, uidA112G-T and uidA166G-T, were generated by substituting thymine (T) for guanine (G) at positions 112 or 166 of the uidA gene (Kovalchuk et al., 2000). B, Visualization of a GUS reversion sector in Arabidopsis following histochemical staining with 5-bromo-4-chloro-3-indolyl-β-d-GlcUA cyclohexylammonium salt. The box indicates GUS-positive cells with reversion of the uidA gene. Left, whole leaf; right, higher magnification image of stained cells.
To investigate the role of TLS-type polymerases in UV light-induced mutation, the mutation frequency in disruptants of the AtREV3, AtREV1, and AtPOLH genes, rev3-1, rev1-1, and polh-1, respectively, was compared with that of the wild type.
The reversion frequencies were initially measured without UV light exposure to determine the level of spontaneous mutations. Twenty-four-day-old nonirradiated plants were examined by counting the active GUS sectors. In the wild type, the number of reversion events per 100 plants was 1.48 ± 0.46. This result is comparable to that of a previous report (Kovalchuk et al., 2000). We also examined the mutation frequencies in rev3-1, rev1-1, and polh-1. The frequencies in the rev3-1 and rev1-1 mutants were slightly lower compared with that of the wild type (P < 0.05; Table I). By contrast, the mutation frequency in polh-1 plants did not change significantly compared with that of the wild type. This result suggests that AtPolζ and AtRev1 play some role in the generation of spontaneous mutations in Arabidopsis somatic cells.
Table I. Spontaneous mutation frequency in AtREV3-, AtREV1-, and AtPOLH-disrupted plants.
Each experiment was repeated at least three times.
| Reporter | Background | Reversion Event per 100 Plants | Fresh Wt per Plant |
| mg | |||
| uidA166G-T | Wild type | 1.48 ± 0.46 | 14.76 ± 3.44 |
| rev3-1 | 0.46 ± 0.05 | 11.32 ± 3.44 | |
| rev1-1 | 0.64 ± 0.54 | 12.93 ± 2.05 | |
| polh-1 | 1.97 ± 1.58 | 11.39 ± 3.51 |
UV-Induced Mutation Frequency in AtREV3, AtREV1, or AtPOLH Disruptants
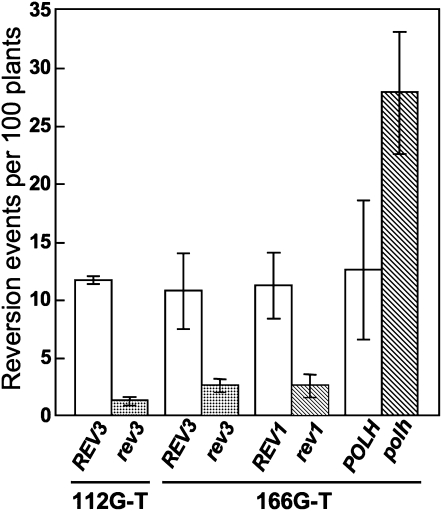
The mutation frequencies in UV-irradiated rev3-1, rev1-1, polh-1, and wild-type plants were then examined. With the uidA166G-T reporter gene, the average reversion in wild-type plants was approximately 11 per 100 plants (Fig. 2; Supplemental Table S1). By contrast, the average reversion in rev3-1 plants was approximately one-quarter that of wild-type siblings (P < 0.05; Fig. 2; Supplemental Table S1). A similar result was obtained with the uidA112G-T gene, where the reversion frequency in rev3-1 was significantly lower than that of the wild type (P < 0.05; Fig. 2). These results indicate that AtPolζ plays a role in promoting mutagenesis following UV light exposure. Since two independent reporter genes showed similar results, we concluded that these two inactivated genes were similarly useful in the detection of reversion events, and hereafter only uidA166G-T was used for further analysis. The UV light-induced mutation frequency in rev1-1 plants was less than one-quarter that of wild-type siblings (P < 0.05; Fig. 2). This result suggests that AtRev1 also promotes UV light-induced mutagenesis.
Figure 2.
UV light-induced mutation frequencies in AtREV3-, AtREV1-, and AtPOLH-disrupted plants. Bars represent average frequencies per 100 plants derived from multiple experiments. Error bars indicate sd.
By contrast, disruption of the AtPOLH gene significantly increased the UV light-induced mutation frequency, where the average reversion in polh-1 plants was approximately 2.5 times that of wild-type siblings (P < 0.05; Fig. 2). This result indicates that AtPolη plays a role in suppressing the induction of mutations following UV light exposure.
The mutation frequency in the wild type was 7.7 times higher with UV light exposure than without UV light exposure. Similarly, the frequency in rev3-1, rev1-1, or polh-1 was 5.6, 4.0, or 14 times higher with UV light exposure. This result indicates that mutations are greatly induced by UV-C light treatment. Under these assay conditions, the average fresh weights of mutant plants and wild-type siblings grown side by side under the same conditions were not significantly different (Supplemental Table S1). Therefore, the decreased or increased reversion frequencies in the mutants are not likely due to the reduced or enhanced growth of these plants.
Mutation Frequency in rev3 polh Double Mutants
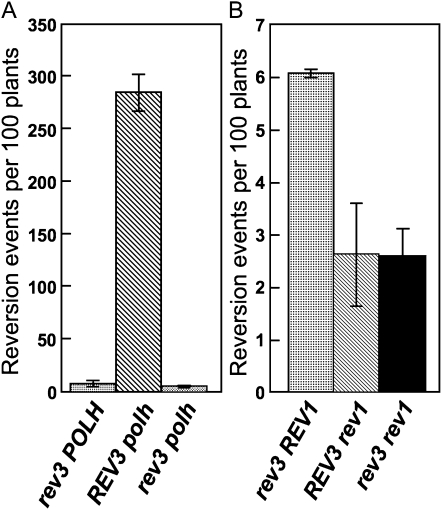
We and other groups previously reported that the AtREV3 and AtPOLH double disruptant was more sensitive to UV irradiation than either of the single mutants (Curtis and Hays, 2007; Anderson et al., 2008). This observation suggests that AtPolζ and AtPolη are involved in independent pathways pertaining to UV light tolerance. On the other hand, analysis of the UV light-induced mutation frequency in rev3-1 and polh-1 mutants showed that AtPolζ and AtPolη seem to possess opposite functions with respect to UV light-induced mutagenesis. To explore the relationship between AtPolζ and AtPolη with respect to UV light-induced mutagenesis, we prepared the rev3-1 polh-1 double mutant and compared the UV light-induced mutation frequency in the double mutant with that of the single mutant siblings rev3-1 and polh-1. The high reversion frequency in the polh-1 mutant was significantly reduced in the rev3-1 polh-1 double mutant, and the frequency in rev3-1 polh-1 was almost similar to that of the rev3-1 mutant (Fig. 3A; Supplemental Table S2). This result indicates that the mutations observed in polh-1 are mainly induced in an AtPolζ-dependent manner.
Figure 3.
UV light-induced mutation frequencies in AtREV3, AtREV1, and AtPOLH double disruptions. A, Mutation frequencies in rev3, polh, and rev3 polh mutants. B, Mutation frequencies in rev3, rev1, and rev3 rev1 mutants. Bars represent average frequencies per 100 plants derived from multiple experiments. Error bars indicate sd.
Mutation Frequency in rev1 rev3 Double Mutants
In a previous report, we showed the possibility that AtPolζ and AtRev1 might cooperate to bypass certain types of DNA damage (Takahashi et al., 2007). When the rev3-1 and rev1-1 mutants were grown under long-term UV-B light exposure, their fresh weights were similarly inhibited (Takahashi et al., 2005). Furthermore, the reversion frequencies in both rev3-1 and rev1-1 mutants were lower compared with that of the wild type (Fig. 2). Based on these data, we further hypothesized that AtPolζ and AtRev1 may operate in the same pathway that confers tolerance to UV damage. In an effort to examine this possibility, we prepared the rev3-1 rev1-1 double mutant and compared its UV light-induced mutation frequency with that of the single mutants rev3-1 and rev1-1. It was found that the reversion frequency in the rev3-1 rev1-1 mutant was slightly lower than that in the rev3-1 mutant (P < 0.05) and similar to that in the rev1-1 mutant (Fig. 3B; Supplemental Table S2). This result is consistent with the notion that AtPolζ and AtRev1 are involved in the same pathway and work cooperatively in mutagenesis. The almost identical mutation frequencies of the rev1-1 and rev3-1 rev1-1 mutants suggest that the pathway requires the presence of AtRev1. By contrast, the slight difference in mutation frequency between the rev3-1 and rev3-1 rev1-1 mutants may suggest that AtRev1 could function, at least partly, in an AtPolζ-independent manner.
DISCUSSION
AtPolζ, AtRev1, and AtPolη Play Important Roles in UV Light-Induced Mutagenesis
TLS is an important damage tolerance mechanism that is conserved in many organisms. It is thought that two TLS pathways exist for the bypass of UV light-induced DNA damage: an error-prone TLS pathway and an error-free TLS pathway. In yeast, where disruption of REV3 or REV1 suppresses the formation of UV light-induced reversions (Lemontt, 1971; Lawrence and Christensen, 1976) and both REV3 and REV1 encode proteins containing DNA polymerase domains (Larimer et al., 1989; Morrison et al., 1989), UV damage is believed to be bypassed in a mutagenic manner (error-prone TLS). In this study, disruption of AtREV3 or AtREV1 greatly reduced the generation of UV-induced mutations (Fig. 2). Thus, the results suggest that AtREV3 and AtREV1 play a similar role in UV-induced mutagenesis as with yeast REV3 and REV1. Additionally, we have shown that the mutation frequency of the rev3-1 rev1-1 double mutant was almost identical to that of the rev1-1 single mutant (Fig. 3B). This result supports the notion that AtPolζ and AtRev1 work cooperatively to bypass UV damage, which is required for UV light tolerance and UV light-induced mutagenesis.
By contrast, Polη was originally identified from analysis of the yeast rad30 mutant and its homology with the bacterial UmuC or DinB protein (McDonald et al., 1997; Roush et al., 1998). Polη deficiency in humans is associated with the inherited disorder XPV, a disease characterized by hypersensitivity to UV light and an increased incidence of skin cancer (Johnson et al., 1999a; Masutani et al., 1999). Based on these facts and in vitro bypass activities (Johnson et al., 1999b), Polη is thought to be involved in the error-free TLS pathway. Polη-deficient yeast strains showed elevated UV light-induced mutation frequencies (Yu et al., 2001; Kozmin et al., 2003), and the high UV light-induced mutation frequency in XPV cells was suppressed by the presence of an intact POLH gene (Stary et al., 2003). In Arabidopsis, disruption of AtPOLH resulted in an increase in the mutation frequency (Fig. 2). This result suggests that AtPolη, encoded by AtPOLH, is also involved in the bypass of UV light-induced damage in an error-free manner.
In a previous study, we found that disruption of both AtREV3 and AtPOLH resulted in an additive inhibitory effect on root growth (Anderson et al., 2008), suggesting that AtPolζ and AtPolη act via two independent pathways. Furthermore, disruption of AtREV3 decreased the mutation frequency while disruption of AtPOLH increased the mutation frequency in this study (Fig. 2). These results suggest that two pathways, an error-prone pathway involving AtPolζ and an error-free pathway involving AtPolη, compete in part for the bypass of UV light-induced DNA damage. The high mutation frequency of the polh-1 mutant was completely suppressed by the rev3-1 mutation (Fig. 3A). This result could be accounted for if the error-prone pathway involving AtPolζ predominates in the bypass of UV damage when AtPolη is unavailable. By contrast, the mutation frequency of the rev3-1 mutant was not lower than that of the rev3-1 polh-1 mutant (Fig. 3A). This might indicate that the error-free pathway involving AtPolη does not complement the error-prone pathway even when AtPolζ is unavailable.
Template Preference, Replication Fidelity, and Mutagenesis
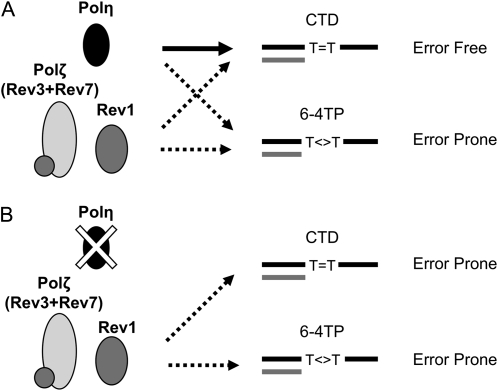
Results of mutational analyses suggested the mutagenic roles of AtPolζ and AtRev1 and the antimutagenic role of AtPolη. However, a series of biochemical analyses of Arabidopsis and other organisms do not support such roles in mutagenesis. For example, yeast and human Polη have lower replication fidelity than yeast Polζ when replicating undamaged DNA (McCulloch and Kunkel, 2008). This contradiction is mostly due to differences in template preference and assay conditions. Since our reporter genes are designed to detect a base substitution at 3′T of TT, we focused here on the bypass activity and fidelity for cyclobutane TT dimers (CTDs) and (6-4) TT photoproducts (6-4TPs). Analysis of yeast and mammals revealed that Polη bypasses CTDs efficiently, although the fidelity is quite low (Johnson et al., 2000b; Washington et al., 2001). Any misincorporation by Polη, however, is corrected by other polymerases (Bebenek et al., 2001; Washington et al., 2001) to effect bypass in an error-free manner. In Arabidopsis, AtPolη bypassed CTDs efficiently (Anderson et al., 2008) and disruption of AtPOLH increased the mutation frequency (Fig. 2). Therefore, we hypothesize that the bypass of CTDs by AtPolη is still more efficient and accurate compared with other polymerases and that AtPolη “prevents” mutagenesis by other polymerases (Fig. 4A). By contrast, in vitro replication analysis using recombinant polymerases revealed that human, yeast, and Arabidopsis Polη poorly bypass 6-4TPs (Hoffman et al., 2008).
Figure 4.
Schematic representation of error-free and error-prone TLS pathways in Arabidopsis. Black and gray lines indicate template and newly synthesized DNA strands, respectively. The solid arrow indicates accurate, efficient bypass activity, whereas the broken arrows indicate inaccurate, inefficient bypass activity. A, TLS activities in wild-type plants. B, TLS activities in AtPOLH-disrupted plants.
On the other hand, although several reports have detailed the biochemical activities of Polζ and Rev1, most of these suggest that neither Polζ nor Rev1 can efficiently bypass CTDs and 6-4TPs (Johnson et al., 2000a; Guo et al., 2001; Zhang et al., 2002). Recombinant AtRev1 showed no insertion activity against templates with this type of DNA damage (Takahashi et al., 2007). However, in vivo replication assays in other organisms revealed that although Polζ and Rev1 are required for the bypass of 6-4TPs (Otsuka et al., 2005), they are dispensable for the bypass of CTDs (Nelson et al., 2000; Gibbs et al., 2005). If this were also true in Arabidopsis, most of the mutations observed in the wild-type background would be caused by the mutagenic bypass of 6-4TPs by AtPolζ and AtRev1 (Fig. 4A). It is conceivable that AtRev1 plays a role in recruiting other TLS-type polymerase(s) such as AtPolζ, AtPolκ, and AtPolη, thereby allowing these enzymes to perform the bypass of damaged DNA, as suggested in our previous report (Takahashi et al., 2007).
Several lines of evidence indicate that the mutagenic bypass of CTDs occur in a Polζ-dependent manner if Polη is unavailable (Gibbs et al., 2005; Shachar et al., 2009). We suppose that the increased mutation frequency in polh-1 was caused by AtPolζ (and AtRev1), which complements AtPolη for the bypass of CTDs (Fig. 4B).
Spontaneous and Other Mutation Sources
Employment of the GUS assay without UV-C light exposure revealed that Arabidopsis plants carried somatic mutations even under normal growth conditions (Table I). Reduction of the spontaneous mutation frequency in rev3-1 and rev1-1 indicates that AtPolζ and AtRev1 are involved in spontaneous mutagenesis, as reported for yeast (Kalinowski et al., 1995; Harfe and Jinks-Robertson, 2000). In our previous report, we suggested that AtPolζ and AtRev1 are involved in the bypass of apurinic/apyrimidinic sites that are generated under normal physiological conditions (Takahashi et al., 2007). Additionally, our preliminary results suggested the involvement of AtPolζ and AtRev1 in γ-ray-induced mutations (M. Nakagawa, S. Takahashi, A. Tanaka, I. Narumi, and A.N. Sakamoto, unpublished data). These lines of evidence indicate that TLS-type polymerases are required to overcome various types of DNA damage under a variety of plant life conditions.
Plant Development and Mutations
In experiments employing the GUS assay, 2-week-old plants possessing two to four true leaves were irradiated with UV-C light and then subjected to GUS staining 10 d after irradiation, by which time plants possessed six to nine true leaves. Interestingly, active GUS sectors were mainly detected on relatively mature leaves or leaf stalks but never on meristems or first to third leaves from inside (data not shown). Furthermore, almost all blue sectors were relatively small and consisted of several cells resulting from a few cell divisions (Fig. 1B). If the mutation had occurred at the initial stage of leaf development, the blue sector should have covered the large area of the leaf. However, such large GUS sectors were never observed. This observation suggests that reversion events do not arise in undifferentiated cells at the time of irradiation. That is, mutagenic TLS activities might be avoided in undifferentiated tissues that could lead to germline cells in plants. In rice, several excision repair genes are expressed predominantly in shoot and root meristems but not in mature leaves (Kimura et al., 2004). It is known that homologous recombinations, an error-free repair pathway, take place more frequently at younger stages during plant development in Arabidopsis (Boyko et al., 2006). It is possible that higher plants employ error-free pathways to maintain an intact genome in undifferentiated tissue, whereas mutagenic bypass is utilized in differentiated tissues notwithstanding the risk of alterations to the genetic information. Further analyses investigating the possible temporal- and tissue-specific activity of TLS are required in an effort to delineate plant strategies that operate to maintain genome integrity.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia accession was used as the wild type in this study. The rev3-1 (Sakamoto et al., 2003), rev1-1 (SALK_011334; Takahashi et al., 2005), polh-1 (SALK_129731; Curtis and Hays, 2007; Anderson et al., 2008), and GUS reporter transgenic lines uidA112G-T and uidA166G-T (Kovalchuk et al., 2000) were in the Columbia background. The rev1-1 and polh-1 lines were provided by the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu). The uidA transgenic lines were crossed with rev3-1, rev1-1, polh-1, rev3-1 rev1-1, and rev3-1 polh-1, and F2 plants homozygous for the mutant rev3-1, rev1-1, polh-1, rev3-1 rev1-1, and rev3-1 polh-1 were selected by PCR. F2 lines homozygous for the reporter gene were selected by BASTA resistance. From the same sibling of the cross, homozygous REV3, REV1, POLH, REV3 rev1-1, rev3-1 REV1, REV3 polh-1, and rev3-1 POLH plants harboring the uidA gene homozygously were obtained and used as controls.
Plants were grown on Rockwool (Nichias) with 0.1% (v/v) commercial nutrient under a 16-h-light/8-h-dark photoperiod at 120 μmol m–2 s–1 and 21°C in a growth chamber. To minimize experimental errors caused by uneven growth or physiological conditions, the wild type and mutant derived from a single F1 plant were grown in the same tray and examined side by side.
Reversion Assay
Two-week-old seedlings carrying the reporter gene were irradiated with 1 kJ m–2 UV-C light supplied by a UV lamp (CSL-30C; COSMO BIO) and then incubated in the dark for 1 d. Ten days following UV-C irradiation, plants were vacuum infiltrated twice for 10 min with 100 mm sodium phosphate buffer (pH 7.2) containing 0.1% (v/v) Triton X-100, 0.05% (w/v) sodium azide, 0.5 mm each of potassium hexacyanoferrate (III) and potassium hexacyanoferrate (II) trihydrate, and 0.95 m 5-bromo-4-chloro-3-indolyl-β-d-GlcUA cyclohexylammonium salt. Following vacuum infiltration, plants were incubated at 37°C for 2 d in the dark and then bleached with 70% (v/v) ethanol.
The number of blue sectors on each plant was determined visually using a stereomicroscope. For each experiment, 300 to 500 plants were analyzed. All experiments were repeated multiple times to obtain average numbers with sd. The significance level was calculated by the t test.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. UV-induced mutation frequency in AtREV3-, AtREV1-, or AtPOLH-disrupted plants.
Supplemental Table S2. UV-induced mutation frequency in double disruptant in comparison with its siblings.
Supplementary Material
Acknowledgments
We are grateful to I. Kovalchuk for his kind gift of the uidA transgenic lines, technical advice, and critical reading of the manuscript. We also thank C. Suzuki for her skillful technical assistance and Y. Oono and Y. Hase for their critical reading of the manuscript.
References
- Anderson HJ, Vonarx EJ, Pastushok L, Nakagawa M, Katafuchi A, Gruz P, Di Rubbo A, Grice DM, Osmond MJ, Sakamoto AN, et al. (2008) Arabidopsis thaliana Y-family DNA polymerase η catalyses translesion synthesis and interacts functionally with PCNA2. Plant J 55: 895–908 [DOI] [PubMed] [Google Scholar]
- Baynton K, Fuchs RP. (2000) Lesions in DNA: hurdles for polymerases. Trends Biochem Sci 25: 74–79 [DOI] [PubMed] [Google Scholar]
- Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel TA. (2001) Proofreading of DNA polymerase η-dependent replication errors. J Biol Chem 276: 2317–2320 [DOI] [PubMed] [Google Scholar]
- Boyko A, Zemp F, Filkowski J, Kovalchuk I. (2006) Double-strand break repair in plants is developmentally regulated. Plant Physiol 141: 488–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Hays JB. (2007) Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: requirements for DNA translesion polymerases η and ζ. DNA Repair (Amst) 6: 1341–1358 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. (2005) DNA Repair and Mutagenesis, Ed 2 ASM Press, Washington, DC [Google Scholar]
- García-Ortiz MV, Ariza RR, Hoffman PD, Hays JB, Roldán-Arjona T. (2004) Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J 39: 84–97 [DOI] [PubMed] [Google Scholar]
- Gibbs PE, McDonald J, Woodgate R, Lawrence CW. (2005) The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Wu X, Rajpal DK, Taylor JS, Wang Z. (2001) Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res 29: 2875–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S. (2000) DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol Cell 6: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Hidema J, Kanga HS, Kumagai T. (1999) Changes in cyclobutyl pyrimidine dimer levels in rice (Oryza sativa L.) growing indoors and outdoors with or without supplemental UV-B radiation. J Photochem Photobiol B 52: 7–13 [Google Scholar]
- Hoffman PD, Curtis MJ, Iwai S, Hays JB. (2008) Biochemical evolution of DNA polymerase η: properties of plant, human, and yeast proteins. Biochemistry 47: 4583–4596 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. (1999a) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. (1999b) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 283: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. (2000a) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. (2000b) Fidelity of human DNA polymerase η. J Biol Chem 275: 7447–7450 [DOI] [PubMed] [Google Scholar]
- Kalinowski DP, Larimer FW, Plewa MJ. (1995) Analysis of spontaneous frameshift mutations in REV1 and rev1-1 strains of Saccharomyces cerevisiae. Mutat Res 331: 149–159 [DOI] [PubMed] [Google Scholar]
- Kimura S, Tahira Y, Ishibashi T, Mori Y, Mori T, Hashimoto J, Sakaguchi K. (2004) DNA repair in higher plants: photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res 32: 2760–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Hohn B. (2000) Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J 19: 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin SG, Pavlov YI, Kunkel TA, Sage E. (2003) Roles of Saccharomyces cerevisiae DNA polymerases Poleta and Polzeta in response to irradiation by simulated sunlight. Nucleic Acids Res 31: 4541–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH. (2003) Xeroderma pigmentosum. Pagon RA, Bird TC, Dolan CR, Stephens K, , Gene Reviews. University of Washington, Seattle, http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=xp [PubMed] [Google Scholar]
- Kunz BA, Straffon AF, Vonarx EJ. (2000) DNA damage-induced mutation: tolerance via translesion synthesis. Mutat Res 451: 169–185 [DOI] [PubMed] [Google Scholar]
- Larimer FW, Perry JR, Hardigree AA. (1989) The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J Bacteriol 171: 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW. (2004) Cellular functions of DNA polymerase ζ and Rev1 protein. Adv Protein Chem 69: 167–203 [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Christensen R. (1976) UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82: 207–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Das G, Christensen RB. (1985) REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet 200: 80–85 [DOI] [PubMed] [Google Scholar]
- Lemontt JF. (1971) Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. (2008) The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 18: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. (1989) REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol 171: 5659–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JR, Gibbs PEM, Nowicka AM, Hinkle DC, Lawrence CW. (2000) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol 37: 549–554 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Otsuka C, Kunitomi N, Iwai S, Loakes D, Negishi K. (2005) Roles of the polymerase and BRCT domains of Rev1 protein in translesion DNA synthesis in yeast in vivo. Mutat Res 578: 79–87 [DOI] [PubMed] [Google Scholar]
- Pagès V, Fuchs RP. (2002) How DNA lesions are turned into mutations within cells? Oncogene 21: 8957–8966 [DOI] [PubMed] [Google Scholar]
- Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. (1998) Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol Gen Genet 257: 686–692 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Lan VT, Hase Y, Shikazono N, Matsunaga T, Tanaka A. (2003) Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15: 2042–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago MJ, Alejandre-Durán E, Ruiz-Rubio M. (2006) Analysis of UV-induced mutation spectra in Escherichia coli by DNA polymerase η from Arabidopsis thaliana. Mutat Res 601: 51–60 [DOI] [PubMed] [Google Scholar]
- Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, et al. (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J 28: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary A, Kannouche P, Lehmann AR, Sarasin A. (2003) Role of DNA polymerase η in the UV mutation spectrum in human cells. J Biol Chem 278: 18767–18775 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Sakamoto A, Sato S, Kato T, Tabata S, Tanaka A. (2005) Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol 138: 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Sakamoto AN, Tanaka A, Shimizu K. (2007) AtREV1, a Y-family DNA polymerase in Arabidopsis, has deoxynucleotidyl transferase activity in vitro. Plant Physiol 145: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlas ME, Franklin WA, Chan GL, Haseltine WA. (1985) Ultraviolet light irradiation of defined-sequence DNA under conditions of chemical photosensitization. Photochem Photobiol 42: 265–273 [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S. (2001) Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc Natl Acad Sci USA 98: 8355–8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev 73: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev 13: 2191–2195 [DOI] [PubMed] [Google Scholar]
- Yu SL, Johnson RE, Prakash S, Prakash L. (2001) Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol 21: 185–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z. (2002) Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res 30: 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.